Abstract
Somatic mutation of the tumor suppressor gene LKB1 occurs frequently in lung cancer where it causes tumor progression and metastasis, but the underlying mechanisms remain mainly unknown. Here, we show that the oncogene NEDD9 is an important downstream mediator of lung cancer progression evoked by LKB1 loss. In de novo mouse models, RNAi-mediated silencing of Nedd9 inhibited lung tumor progression, whereas ectopic NEDD9 expression accelerated this process. Mechanistically, LKB1 negatively regulated NEDD9 transcription by promoting cytosolic translocation of CRTC1 from the nucleus. Notably, ectopic expression of either NEDD9 or CRTC1 partially reversed the inhibitory function of LKB1 on metastasis of lung cancer cells. In clinical specimens, elevated expression of NEDD9 was associated with malignant progression and metastasis. Collectively, our results decipher the mechanism through which LKB1 deficiency promotes lung cancer progression and metastasis, and provide a mechanistic rationale for therapeutic attack of these processes.
Introduction
Lung cancer is one of the most deadly diseases worldwide, mainly attributing to the high frequency of metastasis (1). Somatic inactivating mutation of LKB1 is frequently observed in approximately 30% of non–small cell lung cancer (NSCLC; refs. 2–8) as well as several other epithelial carcinomas including prostate cancer, cervix cancer, and pancreas cancer (9). Our previous work in mouse models has uncovered an essential role of LKB1 loss in promoting lung cancer progression and metastasis (4). However, the molecules mediating lung cancer progression and metastasis triggered by LKB1 loss remain elusive. Identifying such mediators and deciphering the underlying mechanism are important for better understanding the cancer progression and metastasis process, as well as finding potential therapeutic targets for effective lung cancer treatment in clinic.
As a master serine/threonine kinase and tumor suppressor, LKB1 phosphorylates more than a dozen of downstream kinases including the well-known energy gauge kinase adenosine monophosphate kinase involved in mTOR signaling pathway (10). We have recently shown that the activation of mTOR signaling pathway by LKB1 loss turns on lysyl oxidase (LOX) gene expression, which promotes lung cancer progression and metastasis through extracellular matrix remodeling (11). However, only partial inhibition of tumor progression was achieved by either LOX enzymatic inhibitor treatment or the combinational therapy using phosphatidylinositol 3-kinase-mTOR and mitogen-activated protein–extracellular signal-regulated kinase inhibitors (11, 12). Considering the limited efficacy of targeting either LOX or mTOR signaling pathway, we reason there may exist other pathways or molecules playing essential roles in lung cancer progression and metastasis induced by LKB1 loss.
Through microarray data analysis, we have previously identified several potential candidates associated with metastasis in Lkb1-deficient murine lung cancer, including Vegf and Nedd9 (neural precursor cell expressed, developmentally downregulated 9, also known as HEF1, Cas-L; ref. 4). While we have previously shown in mice that blocking Vegfr signaling is not effective in inhibition of Lkb1-deficient lung tumor metastasis (13), additional data support a potential role for Nedd9. NEDD9 is a noncatalytic scaffold protein (14–16), and has been implicated in the metastatic behavior of several types of solid tumors (17, 18). In melanoma, NEDD9 interacts with focal adhesion kinase (FAK) to promote cell invasion and metastasis (19). Similar observation has been reported in glioblastoma studies (20). These data support an important role of NEDD9 in cancer progression and metastasis of a number of malignancies. However, the interactive regulation between LKB1 and NEDD9, and the potential functional contribution of such a network to lung cancer progression and metastasis has not been explored yet.
Here, we provide strong evidences for NEDD9 as an important downstream mediator in LKB1-deficient lung cancer progression, and uncover a novel mechanism of LKB1 in contribution to lung cancer progression through CRTC1-NEDD9 axis. Our data provide novel functional evidences and mechanistic insights into lung cancer progression and metastasis evoked by LKB1 loss and define a potential biomarker for lung cancer prognosis in clinic.
Materials and Methods
Mouse colony, mouse treatment, and mouse tumor analyses
KrasG12D and Lkb1L/L mice were generously provided by Drs. T. Jacks and R. Depinho, respectively (4). Nude mice (6 weeks old, male) were purchased from Shanghai SLAC Laboratory Animal Co. Ltd. All mice were housed in a specific pathogen-free environment at Shanghai Institute of Biochemistry and Cell Biology and treated in strict accordance with protocols approved by the Institutional Animal Use Committee of the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences.
The KrasG12D or KrasG12D, Lkb1L/L mice at 6 to 8 weeks old were virally infected with either 2 × 106 PFU Adeno-Cre or lentivirus (Lenti-Cre, NEDD9-Cre, Ctrl-Cre, or shNedd9-Cre) via nasal inhalation as previously described (21, 22). The lentiviruses were prepared as previously described (4). All mice were sacrificed for gross inspection and pathologic examination. Lung tumors were dissected for molecular analyses. The number of total tumor, well- or poorly differentiated tumors as well as the total tumor area were analyzed for each group from sectioned H&E slides as described before (23, 24) and the data were presented as mean ± SEM. As for lung seeding assay, nude mice were injected with 1 × 106 cells via tail veins. After 10 weeks of inoculation, all mice were sacrificed and mouse lungs were weighted and lung metastasis was detected by H&E staining from series section.
Human lung cancer specimen analyses
All 175 human lung cancer samples were collected with patient consents in Fudan University Shanghai Cancer Center (Shanghai, China) from October 2007 to February 2008 with approval from the Institute Research Ethics Committee.
Cell culture, plasmids, and antibodies
Plasmids construction and the detailed information for antibodies are described in Supplementary Materials and Methods. Human cell lines including A549, CRL-5907, CRL-5866, HEK-293, and HEK-293T were purchased from American Type Culture Collection (ATCC) and authenticated by ATCC by short tandem repeat (STR) profiling and used for functional studies within 6 months after thawn from liquid nitrogen tank. A549, CRL-5907, CRL-5866 and HEK-293, and HEK-293T cells were, respectively, cultured in RPMI-1640 medium (Hyclone) or Dulbecco’s Modified Eagle’s Media (Hyclone) supplemented with 10% FBS (Biochrom AG) and antibiotics (100 U/mL streptomycin and 100 μg/mL penicillin; Invitrogen).
Immunohistochemical analysis
Immunohistochemistry was conducted as previously described (24). The proliferation rate was assessed by counting Ki-67 positive nuclear staining at high-power field for more than 1,000 cells. The apoptosis was assessed by analysis of cleaved caspase 3 immunostaining. The NEDD9 immunostaining was reviewed and scored blindly into 2 criteria as low and high expression as previously described (25). The correlation between clinical features and NEDD9 expression in human lung cancer was analyzed by SPSS 13.0 statistical software, and P values were calculated by Pearson χ2 test. A value of P < 0.05 was considered as significant (2 tailed).
RT-PCR, real-time RT-PCR, and Western blot analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions and retrotranscribed into cDNA (Ferments). [Primers for reverse transcriptase (RT-PCR) and real-time RT-PCR are listed in Supplementary Table S1]. Western blot analysis was done as previously described (26). The samples of cytoplasm and nuclear protein were obtained using proteoextract subcellular extraction regents (Calbiochem) according to the manufacturer’s instruction.
Reporter gene assay
Luciferase activities were measured 48 hours after transfection using the Dual-Luciferase Assay kit (Promega) on a GloMax 20/20 luminometer (Promega). pRL-SV40 was cotransfected as internal control. Experiments were conducted in triplicates and repeated at least 3 times.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was conducted as previously described (27). Briefly, 5 μg antibody of rabbit anti-cAMP-responsive element binding protein (CREB) or rabbit anti-CRTC1 was added into each lytic sample at 4°C overnight. Rabbit IgG was used as negative control. Fifty microliter Protein A (Invitrogen) was added into each sample for immunoprecipitation, and then samples were rotated at 4°C for 2 hours. An aliquot of sonicated cleared extract (input) and the immunoprecipitated material were decross-linked in TE plus 1% SDS for at least 8 hours at 65°C. Primers for ChIP assay were listed in Supplementary Table S1. NR4A2 and GAPDH served as positive and negative controls, respectively (28). Representative data were shown from 3 independent experiments.
Wound healing assay
Wound healing assay was conducted as described before (29). Cells at 100% confluence were starved in serum-free medium for more than 12 hours dependent on different cell lines. Cells were then lightly and quickly scratched with a pipette tip. The cell migration kinetics were closely monitored, photographed, and calculated by IncuCyte 2008B.
Three-dimensional cell culture
Cells were seeded in medium contain 2% Matrigel (BD Biosciences) on the top of another layer of solidified Matrigel. Cells were then cultured at 37°C incubators and monitored for the appearing morphologic changes in 1 to 2 weeks. Photos were taken using a light microscope (Leica).
Statistical analysis
Data were analyzed by Student t test. P < 0.05 was considered to be significant (2 tailed). Error bars represent SEM.
Results
Transcriptional regulation of NEDD9 by LKB1 in mouse and human lung tumors
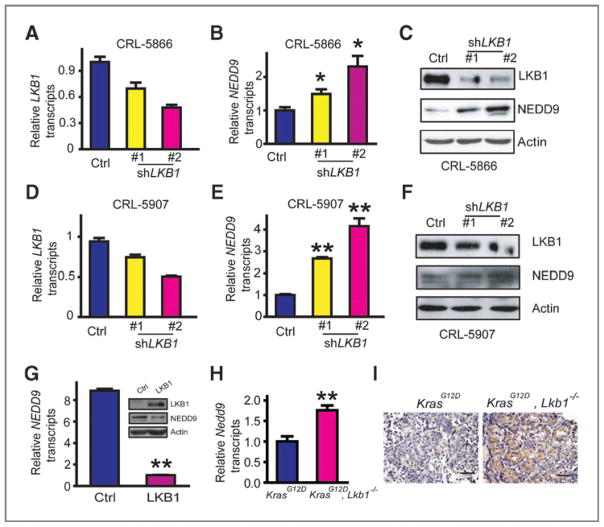
Our previous gene expression profiling analyses of mouse KrasG12D lung tumors with and without Lkb1 deficiency identified Nedd9 as a potential target gene downstream of Lkb1 (4). To extend this finding, we investigated the functional correlation of NEDD9 and LKB1 in human lung cancer. We found that knockdown of LKB1 in human NSCLC cell lines CRL-5866 and CRL-5907, which express wild-type LKB1, resulted in increased NEDD9 mRNA and protein levels (Fig. 1A–F). Reciprocally, ectopic LKB1 expression in the LKB1-mutant A549 human NSCLC cells inhibited NEDD9 expression (Fig. 1G). The regulation was associated with marked induction of Nedd9 expression in Lkb1 deficient mouse tumors in vivo (Fig. 1H and I). These data show that LKB1 is a negative regulator of NEDD9 transcription in mouse and human lung tumors.
Figure 1.
NEDD9 is transcriptionally regulated by LKB1 in human and mouse lung cancer. A, real-time RT-PCR analysis of LKB1 knockdown efficiency in CRL-5866 cells. B and C, knockdown of LKB1 upregulated NEDD9 expression at mRNA level (B) and protein level (C) in CRL-5866 cells assessed by real-time RT-PCR and Western blot analysis, respectively. Data are shown as mean ± SEM.*, P < 0.05, compared with the control (Ctrl) group (B). D, real-time RT-PCR analysis of LKB1 knockdown efficiency in CRL-5907 cells. E and F, knockdown of LKB1 upregulated NEDD9 expression at mRNA level (E) and protein level (F) in CRL-5907 cells assessed by real-time RT-PCR and Western blot analysis, respectively. Data are shown as mean ± SEM. **, P < 0.01, compared with the control (Ctrl) group (E). G, NEDD9 mRNA and protein levels were downregulated in A549 cells by ectopic LKB1 expression assessed by real-time RT-PCR and Western blot analysis, respectively. Data are shown as mean ± SEM. **, P < 0.01. H and I, real-time RT-PCR (H) and immunostaining analyses (I) of Nedd9 expression level in KrasG12D mouse tumors with or without Lkb1 deficiency. Data are shown as mean ± SEM. **, P < 0.01 (3 tumors in each group). Scale bar, 50 μm (I).
NEDD9 is an important downstream mediator of lung cancer progression and metastasis evoked by Lkb1 deficiency
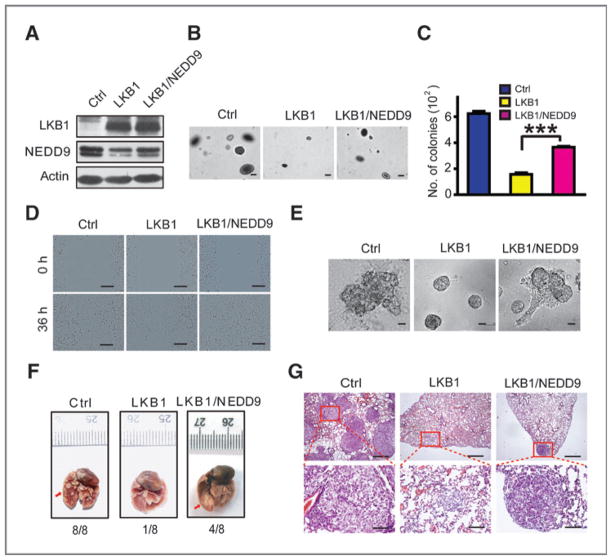
We next investigated the functional interaction between NEDD9 and LKB1 in lung cancer progression and metastasis. We found that LKB1 overexpression in A549 cells blocked colony formation in soft agar, migration in wound healing assays, invasion in Matrigel as well as in boyden chamber assay whereas these effects were reversed by ectopic NEDD9 expression (Fig. 2A–E and Supplementary Fig. S1). Similarly, the inhibition of tumor formation by LKB1 overexpression in A549 lung seeding assay was partially overcome by ectopic NEDD9 expression; large nodules were detectable in all mice from the control group, in 1/8 mice from the LKB1 group, and in 4/8 mice from the LKB1/NEDD9 group (Fig. 2F and G). These data showed that NEDD9 can partially rescue the inhibitory function of LKB1 upon lung cancer invasion and metastasis.
Figure 2.
Ectopic NEDD9 expression partially reverses the inhibitory function of LKB1 upon colony formation, migration, and invasion of human lung cancer cells. A, Western blot detection of LKB1 and NEDD9 expression in A549 cells with ectopic LKB1 and/or NEDD9 expression. B and C, colony formation abilities of A549 cells with ectopic LKB1 and/or NEDD9 expression were assessed in soft agar. Representative photos (B) and the number of colonies (C) are shown. Scale bar, 100 μm (B). Data are shown as mean ± SEM. ***, P < 0.001. D and E, representative photos for migration in wounding assay (D) and invasiveness in Matrigel (E) of A549 cells with ectopic LKB1 and/or NEDD9 expression. Scale bar, 200 μm (D) and 40 μm (E). F and G, ectopic NEDD9 expression partially reverses the inhibitory function of LKB1 upon metastasis of A549 cells. Representative photos show the gross metastatic nodules on lung surface (F, indicated by the red arrows) as well as the histology of lung metastasis (G) from nude mice received with A549 cells with ectopic LKB1 and/or NEDD9 expression via tail vein injection. Metastatic incidence is shown on the bottom. Scale bars, 500 μm (top) and 100 μm (bottom).
Nedd9 knockdown inhibits de novo lung cancer progression in KrasG12D, Lkb1L/L mice model
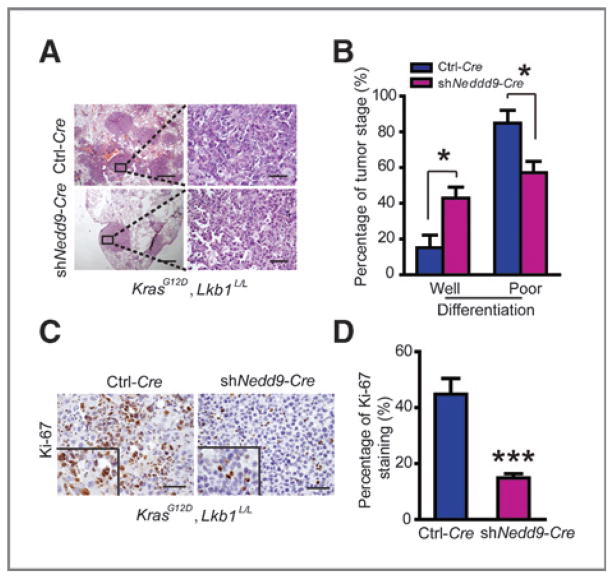
We next asked if Nedd9 knockdown affects de novo lung cancer progression (4). We infected KrasG12D, Lkb1L/L mice with lentivirus carrying Cre expression (Ctrl-Cre) or Cre expression together with Nedd9 knockdown (shNedd9-Cre) as previously described (21). Mice were sacrificed for gross inspection and pathologic analyses at 21 weeks post viral administration. Knockdown of Nedd9, confirmed by real-time RT-PCR and immunostaining analyses (Supplementary Fig. S2A and S2B), resulted in a slight decrease of the number of tumors on lung surface but not total tumor number and area (Supplementary Fig. S2C–S2E). More importantly, we found that the percentage of poorly differentiated tumors significantly decreased in Nedd9-knockdown group in comparison with control group (Fig. 3A and B). Consistently, Nedd9 knockdown also resulted in a decrease of cell proliferation rate (Fig. 3C and D) while had no effect on cell apoptosis (Supplementary Fig. S2F). These data further support the essential role of Nedd9 in regulating de novo lung cancer progression in context with Lkb1 deficiency.
Figure 3.
shNedd9 alleviates de novo lung cancer progression evoked by Lkb1 deficiency. A, pathologic photos of lung tumors from KrasG12D, Lkb1L/L mice at 21 weeks post viral infection of Ctrl-Cre or shNedd9-Cre (7 mice per group). Scale bars, 500 μm (top) and 50 μm (bottom). B, statistic analyses of well- or poorly differentiated lung tumors from KrasG12D, Lkb1L/L mice receiving Ctrl-Cre or shNedd9-Cre infection. Data are shown as mean ± SEM. *, P < 0.05 (n = 7). C, Ki-67 immunostaining of lung sections from KrasG12D, Lkb1L/L mice virally infected with Ctrl-Cre or shNedd9-Cre. Scale bars, 50 μm. D, bar diagrams illustrate the percentage of Ki-67–positive staining in lung tumors from KrasG12D, Lkb1L/L mice virally infected with either Ctrl-Cre or shNedd9-Cre. The percentage of Ki-67 staining was quantified by counting more than 1,000 nuclei staining tumor cells. Data are shown as mean ± SEM. ***, P < 0.001.
Ectopic NEDD9 expression promotes de novo lung cancer progression in KrasG12D mouse model
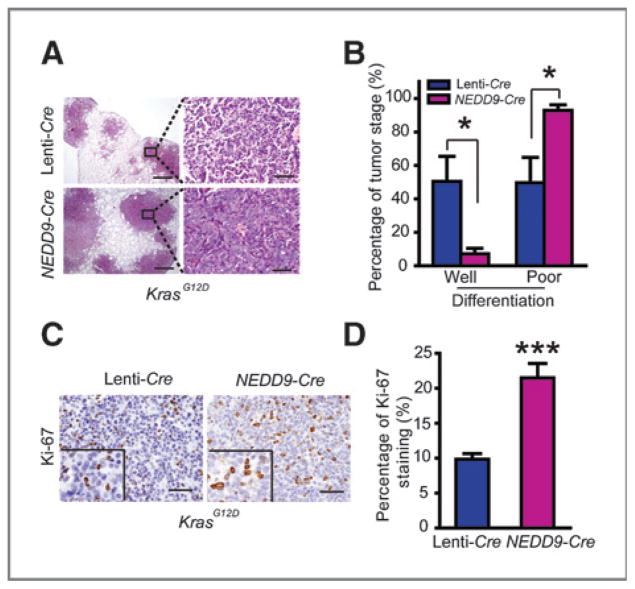
We further investigated whether ectopic Nedd9 expression is sufficient to promote lung cancer progression in de novo KrasG12D mouse model. We treated KrasG12D mice with lentivirus carrying either Cre (Lenti-Cre) or Cre with NEDD9 expression (NEDD9-Cre) via nasal inhalation and all mice were sacrificed for analyses at 24 weeks post viral administration. We found that ectopic NEDD9 expression, confirmed by real-time RT-PCR and immunostaining analyses (Supplementary Fig. S3A and S3B), did not affect the total number and area of lung tumors, whereas more nodules were observed on lung surface from NEDD9-Cre group than control (Supplementary Fig. S3C–S3E). Detailed pathologic analyses further revealed that the percentage of poorly differentiated tumors was significantly higher in NEDD9-Cre group (Fig. 4A and B). Consistently, a higher proliferation rate was observed in lung tumors with ectopic NEDD9 expression (Fig. 4C and D). However, no significant change of apoptosis was found in NEDD9 over-expressed lung tumors (Supplementary Fig. S3F). Taken together, these data show that ectopic NEDD9 expression significantly promotes lung cancer progression in de novo mouse model.
Figure 4.
NEDD9 promotes de novo lung cancer progression of KrasG12D mice. A, pathologic photos of lung tumors from KrasG12D mice at 24 weeks post Lenti-Cre or NEDD9-Cre infection (5 mice per group). Scale bars, 500 μm (top) and 50 μm (bottom). B, statistic analyses of well-or poorly differentiated lung tumors from KrasG12D mice receiving Lenti-Cre or NEDD9-Cre. Data are shown as mean ± SEM. *, P < 0.05 (n = 5). C, Ki-67 immunostaining of lung sections from KrasG12D mice virally infected with either Lenti-Cre or NEDD9-Cre. Scale bars, 50 μm. D, bar diagrams illustrate the percentage of Ki-67–positive staining in lung tumors from KrasG12D mice treated with Lenti-Cre or NEDD9-Cre infection. The percentage of Ki-67 staining was quantified by counting more than 1,000 nuclei staining tumor cells. Data are shown as mean ± SEM. ***, P < 0.001.
CRTC1 regulates NEDD9 transcription and contributes to LKB1-deficient lung tumor progression
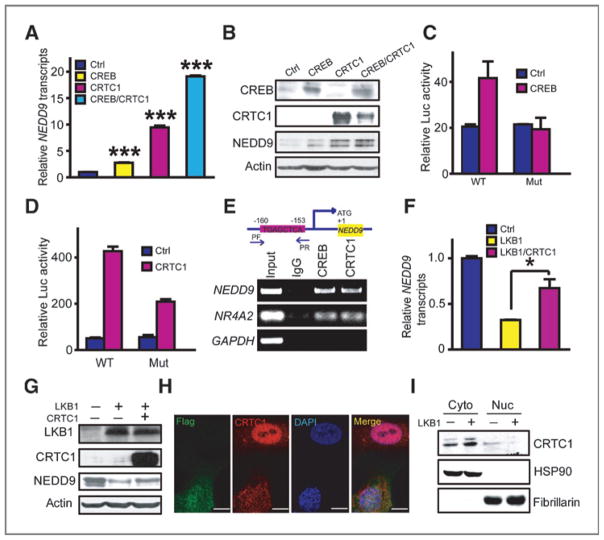
To determine how LKB1 negatively controls NEDD9 expression, we initially screened seven reporters including CRE, serum response element (SRE), NF-κB, glucocorticoid response element (GRE), heat-shock element (HSE), TATA-like promoter (TAL), and AP1 for their response to LKB1 knockdown in CRL-5866 cells. Interestingly, CRE reporter was the only one regulated by LKB1 (Supplementary Fig. S4), consistent with the inhibitory role of LKB1 upon the transcription activity of CREB and its coactivator CRTC1 (30, 31). We found that ectopic expression of CREB and/or CRTC1 significantly upregulated NEDD9 mRNA and protein levels (Fig. 5A and B). Conversely, knockdown of CRTC1 dramatically decreased the NEDD9 mRNA level (Supplementary Fig. S5). To further explore the regulation of NEDD9 by CRTC1/CREB, we cloned the 3kb genomic DNA fragment containing NEDD9 promoter for reporter gene assay analyses. Through a series of deletion/mutation analyses (Supplementary Fig. S6), we had narrowed down the CRTC1/CREB -responsive element to a 20bp fragment (−146 to −166) with a nonclassical CRE site “TGAGCTCA” (32), which could not be predicted by TFSCAN (http://www-bimas.cit.nih.gov/molbio/proscan/; ref. 33). Mutation of this non-classical CRE site abolished the response of NEDD9 promoter to either CREB or CRTC1 expression (Fig. 5C and D). Results from ChIP assay confirmed the de novo binding of either CREB or CRTC1 to NEDD9 promoter region (Fig. 5E). These data convincingly show that CRTC1/CREB participates in regulating NEDD9 gene transcription.
Figure 5.
LKB1 regulates NEDD9 expression by modulating CRTC1 nuclear translocation. A and B, both NEDD9 mRNA (A) and protein level (B) were upregulated by expression of CRTC1 and/or CREB in CRL-5907 cells assessed by real-time RT-PCR and Western blot analysis. Data are shown as mean ± SEM. ***, P < 0.001. C and D, ectopic expression of either CREB (C) or CRTC1 (D) significantly increased the transcriptional activity of NEDD9 promoter with wild-type but not mutated nonclassical CRE site in CRL-5866 cells. E, ChIP assay confirmed the binding of CREB or CRTC1 to NEDD9 promoter in A549 cells. PF, forward primer; PR, reverse primer. NR4A2 and GAPDH served as positive and negative controls respectively. F and G, ectopic expression of CRTC1 reversed the downregulation of NEDD9 mRNA (F) and protein levels (G) by LKB1 in A549 cells. Data are shown as mean ± SEM. *, P < 0.05. H, immunofluorescence assay showed that ectopic Flag-LKB1 expression promoted CRTC1 translocation into cytoplasm. Flag-LKB1 (green), CRTC1 (red), and DAPI (blue). Scale bar, 10 μm. I, Western blot analyses showed that ectopic LKB1 expression decreased the nuclear CRTC1 but increased the amount of CRTC1 in cytoplasm. HSP90 and fibrillarin served as loading controls for cytoplasm and nuclear fractions, respectively.
Moreover, we found that LKB1 knockdown or ectopic LKB1 expression regulated the transcriptional activity of NEDD9 promoter with wild-type nonclassical CRE site but not with mutated site (Supplementary Fig. S7). Ectopic CRTC1 expression partially reversed the decrease of NEDD9 mRNA and protein levels caused by ectopic LKB1 expression in A549 cells (Fig. 5F and G). Immunofluorescence analysis showed that ectopic LKB1 expression resulted in CRTC1 subcellular translocation from nucleus to cytoplasm (Fig. 5H). This was further confirmed by Western blot analysis using cytoplasmic and nuclear fractions (Fig. 5I). Previous studies have shown that salt inducible kinase (SIK), a downstream substrate of LKB1, is important in linking LKB1 and CRTC1/CREB transcription activity (31). Our data showed that ectopic LKB1 expression induced nuclear translocation of SIK2, which results in CRTC1 nuclear exportation in human lung cancer cells (Supplementary Fig. S8). This regulation was further confirmed by LKB1 knockdown experiment (Supplementary Fig. S9). Moreover, SIK2 knockdown resulted in CRTC1 nuclear retention which eventually upregulates NEDD9 transcript (Supplementary Fig. S10). These data convincingly showed that LKB1 regulates NEDD9 gene expression through CRTC1 cellular translocation via SIK2 in lung cancer.
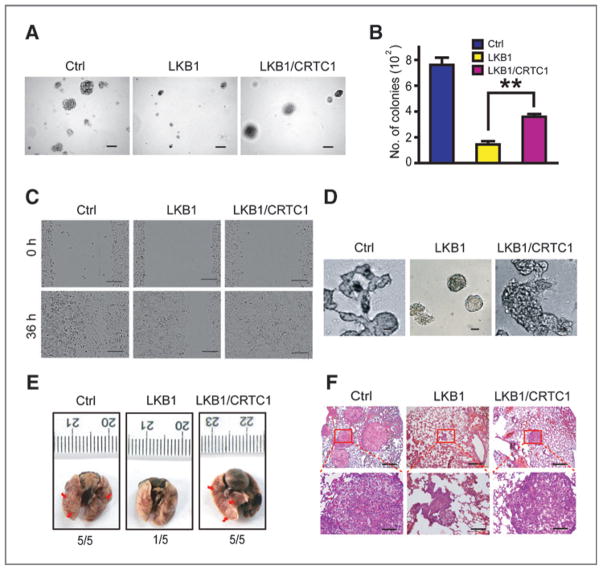
We further found that ectopic CRTC1 expression partially rescued the inhibition of colony formation, cell migration, and invasion caused by LKB1 expression in A549 cells (Fig. 6A–D and Supplementary Fig. S11), as well as the inhibitory effect of LKB1 upon lung cancer metastasis in lung seeding assay (Fig. 6E and F). Taken together, these data show that CRTC1 regulates NEDD9 gene expression and contributes to LKB1-deficient lung tumor progression.
Figure 6.
Ectopic expression of CRTC1 partially reverses the inhibitory function of LKB1 upon colony formation, migration, invasion, and metastasis of lung cancer cells. A and B, representative photos (A) and statistic analyses (B) of colonies formed in soft agar of three different groups of A549 cells as indicated. Scale bar, 100 μm (A). Data are shown as mean ± SEM. **, P < 0.01. C and D, ectopic expression of CRTC1 partially reversed the inhibitory effect of LKB1 upon A549 cell migration ability (C) and cell invasiveness in Matrigel (D). Scale bar, 200 μm (C) and 40 μm (D). E and F, ectopic expression of CRTC1 partially reverses the inhibitory function of LKB1 upon metastasis of A549 cells. Representative photos show the gross metastatic nodules on lung surface (E, indicated by the red arrows) as well as the histology of lung metastasis (F) from nude mice received with A549 cells with ectopic LKB1 and/or CRTC1 expression via tail vein injection (5 mice per group). Metastatic incidence is also shown on the bottom. Scale bars, 500 μm (top) and 100 μm (bottom).
High NEDD9 expression correlates with human lung cancer malignancy progression and metastasis
Finally, we examined the clinical relevance of NEDD9 expression in human lung cancer. A cohort of 175 human NSCLC samples was used for NEDD9 immunostaining (Supplementary Fig. S12). Remarkably, high NEDD9 expression was strongly correlated with lymph node metastasis and advanced clinical stage, as well as with smoking and low differentiation (Table 1). No significant correlations were observed between NEDD9 expression and other clinical features, including NSCLC subtypes and tumor size (Table 1). Therefore, high NEDD9 level appears to be an indicator of poor prognosis for human lung cancer.
Table 1.
The clinical correlation of NEDD9 expression in human lung cancer
| Characteristics | n | NEDD9
|
P | |
|---|---|---|---|---|
| Low | High | |||
| Smoking | ||||
| No | 77 | 32 (41.6%) | 45 (58.4%) | 0.025a |
| Yes | 98 | 25 (25.5%) | 73 (74.5%) | |
| Histology | ||||
| ADC | 121 | 39 (32.2%) | 82 (67.8%) | 0.886 |
| SCC | 54 | 18 (33.3%) | 36 (66.7%) | |
| Differentiation degree | ||||
| High | 19 | 12 (63.2%) | 7 (36.8%) | 0.001b |
| Moderate | 104 | 36 (34.6%) | 68 (65.4%) | |
| Low | 52 | 9 (17.3%) | 43 (87.2%) | |
| T classification | ||||
| T1–2 | 146 | 49 (33.6%) | 97 (66.4%) | 0.531 |
| T3–4 | 29 | 8 (27.6%) | 21 (72.4%) | |
| LN metastasis | ||||
| Negative | 90 | 38 (42.2%) | 52 (57.8%) | 0.005b |
| Positive | 85 | 19 (22.4%) | 66 (77.6%) | |
| Clinical stage | ||||
| I/II | 102 | 42 (41.2%) | 60 (58.8%) | 0.004b |
| III/IV | 73 | 15 (20.5%) | 58 (79.5%) | |
Abbreviations: ADC, adenocarcinoma; SCC, squamous cell carcinoma; LN metastasis, lymph node metastasis; n, number.
P < 0.05.
P < 0.01.
Discussion
Metastasis is a major factor contributing to the high mortality of lung cancer. Here, we discover an important novel pathway involved in lung cancer progression and metastasis triggered by LKB1 loss: LKB1 loss triggers the nuclear translocation of CRTC1, which in turn upregulates NEDD9 and promotes lung cancer progression and metastasis. Our study highlights NEDD9 as an important downstream mediator in lung cancer progression evoked by LKB1 loss. High NEDD9 expression strongly associates with poor differentiation, advanced clinical stage, as well as lymph node metastasis. Collectively, these data provide a novel mechanism by which LKB1 loss promotes cancer progression and metastasis as well as the potential biomarker for human NSCLC prognosis.
Our previous work showed that NEDD9 was upregulated by LKB1 loss in lung cancer metastasis but did not resolve how NEDD9 potentially links LKB1 loss to the malignant phenotype (4). Recent studies have shown that activation of hypoxia-inducible factor-1α (HIF-1α), downstream of mTOR pathway, regulates NEDD9 expression in colon, and renal cancers (34, 35). We have previously shown that the mTOR-HIF-1α axis actually upregulates LOX, which in turn cross-links collagen to remodel extracellular matrix and triggers β1-integrin signaling through FAK activation, and thus promotes lung cancer metastasis (11). However, no evidence of the involvement of mTOR activation was found in NEDD9 regulation (4). Interestingly, CRTC1/CREB transcription activation is critical for mediating NEDD9 expression and potentially promotes lung tumor malignancy. Nuclear translocation of CRTC1 regulated by LKB1 loss promotes NEDD9 gene expression in lung cancer, which may indicate a cell type–specific pattern of NEDD9 gene regulation. As a coactivator of CREB, CRTC1 forms complex with CREB in nucleus and turns on the CREB transcriptional activity (30, 31, 36), which plays important roles in glucose and lipid metabolism (31, 37–39). However, apart from its role in metabolic regulation, less is known about its potential contribution in lung cancerigenesis. Previous studies have identified SIK as an important mediator of CRTC1/CREB transcription activity downstream of LKB1 (31). In LKB1-deficient cells, SIK is incapable of phosphorylating CRTC1, resulting in CRTC1 nucleus retention where it interacts with CREB and promotes certain gene expression (30, 31, 36, 40, 41). Previous study has shown that NR4A2 is transcriptionally upregulated by CRTC1/CREB pathway in LKB1 null lung cancer cells and promotes cell growth (42). Our data here identify NEDD9 as another important downstream mediator of CRTC1/CREB pathway in lung cancer progression and metastasis caused by LKB1 loss. Further dissection of this circuitry may provide important hints for targeted therapy in LKB1-deficient lung cancer.
With integrative studies using de novo animal model, lung seeding assay as well as in vitro migration and invasion systems, we have convincingly shown that NEDD9 plays a positive role in lung cancer progression and metastasis. Previous studies have supported that NEDD9 promotes solid tumor metastasis via FAK and Src activation (19, 20, 43) through direct interaction (18). Interestingly, 2 recent studies have implicated the potential involvement of NEDD9 in epithelial–mesenchymal transition (EMT) process in breast cancer (44, 45). EMT is thought to be an important event involved in early stages of cancer metastasis (46). It is attempting to ask if Nedd9 mediates the EMT process observed in Lkb1-deficienct lung cancer progression and metastasis (12).
We further investigated the clinical correlation of NEDD9 expression level in 175 lung cancer specimens. Interestingly, we have found that high NEDD9 expression is positively correlated with lymph node metastasis and malignant progression, suggesting that NEDD9 is an indicator for poor prognosis of lung cancer patients. Future studies with an even larger sample number will be needed to establish a correlation between NEDD9 expression and survival.
In summary, our work has provided mechanistic insights into how LKB1 loss-of-function mutations promote lung cancer progression. Identification of CRTC1/CREB as important regulators of NEDD9 gene expression may support mechanistic insights into lung cancer progression and metastasis evoked by LKB1 loss. NEDD9 also serves as a potential biomarker for lung cancer prognosis.
Supplementary Material
Acknowledgments
The authors thank Drs. Ronald A. DePinho, Lynda Chin, Tyler Jacks, and Hiroshi Takemori for material contribution and Drs. Diego H. Castrillon, Jun-Lin Guan, Boyi Gan, Gaoxiang Ge, and Dangsheng Li for helpful discussion and technical supports.
Grant Support
This work was supported by the National Basic Research Program of China (2010CB912102; 2012CB910800), the National Natural Science Foundation of China (81101583 and 30971461). T. Xiao was supported by the postdoctoral foundation (2010KIP504) and Shanghai postdoctoral foundation (Y15CS11371 and Y149S11371). F. Li was supported by the postdoctoral foundation (2011KIP505).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
K.-K. Wong has a commercial research grant from AstraZenca and Millenium, and is a consultant/advisory board member of Molecular MD. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: Y. Feng, Z.-q. Xiong, H. Ji
Development of methodology: Y. Feng, H. Ji
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): Y. Feng, Y. Wang, Z. Wang, F. Li, Y. Gao, Y. Zhou, Q. Zhai, Y. Sun, N. Bardeesy, H. Chen, H. Ji
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): Y. Feng, Z. Fang, N. Bardeesy, K.-K. Wong, H. Ji
Writing, review, and/or revision of the manuscript: Y. Feng, Y. Gao, T. Xiao, F. Li, N. Bardeesy, K.-K. Wong, H. Ji
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): Z. Wang, Z. Fang, Y. Gao, H. Liu, F. Li, X. Liu, H. Chen, H. Ji
Study supervision: H. Ji
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Carretero J, Medina PP, Pio R, Montuenga LM, Sanchez-Cespedes M. Novel and natural knockout lung cancer cell lines for the LKB1/STK11 tumor suppressor gene. Oncogene. 2004;23:4037–40. doi: 10.1038/sj.onc.1207502. [DOI] [PubMed] [Google Scholar]
- 3.Gao B, Sun Y, Zhang J, Ren Y, Fang R, Han X, et al. Spectrum of LKB1, EGFR, and KRAS mutations in chinese lung adenocarcinomas. J Thorac Oncol. 2010;5:1130–5. doi: 10.1097/JTO.0b013e3181e05016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 5.Kim MJ, Jin G, Jheon HS, Lee SY, Cha SI, Kim CH, et al. LKB1 mutations are extremely rare in Korean non-small cell lung cancers. Cancer Genet Cytogenet. 2010;196:204–6. doi: 10.1016/j.cancergencyto.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 6.Matsumoto S, Iwakawa R, Takahashi K, Kohno T, Nakanishi Y, Matsuno Y, et al. Prevalence and specificity of LKB1 genetic alterations in lung cancers. Oncogene. 2007;26:5911–8. doi: 10.1038/sj.onc.1210418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onozato R, Kosaka T, Achiwa H, Kuwano H, Takahashi T, Yatabe Y, et al. LKB1 gene mutations in Japanese lung cancer patients. Cancer Sci. 2007;98:1747–51. doi: 10.1111/j.1349-7006.2007.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Cespedes M, Parrella P, Esteller M, Nomoto S, Trink B, Engles JM, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res. 2002;62:3659–62. [PubMed] [Google Scholar]
- 9.Sanchez-Cespedes M. A role for LKB1 gene in human cancer beyond the Peutz-Jeghers syndrome. Oncogene. 2007;26:7825–32. doi: 10.1038/sj.onc.1210594. [DOI] [PubMed] [Google Scholar]
- 10.Katajisto P, Vallenius T, Vaahtomeri K, Ekman N, Udd L, Tiainen M, et al. The LKB1 tumor suppressor kinase in human disease. Biochim Biophys Acta. 2007;1775:63–75. doi: 10.1016/j.bbcan.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Xiao Q, Ma H, Li L, Liu J, Feng Y, et al. LKB1 inhibits lung cancer progression through lysyl oxidase and extracellular matrix remodeling. Proc Natl Acad Sci U S A. 2010;107:18892–7. doi: 10.1073/pnas.1004952107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17:547–59. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stahmann N, Woods A, Spengler K, Heslegrave A, Bauer R, Krause S, et al. Activation of AMP-activated protein kinase by vascular endothelial growth factor mediates endothelial angiogenesis independently of nitric-oxide synthase. J Biol Chem. 2010;285:10638–52. doi: 10.1074/jbc.M110.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar S, Tomooka Y, Noda M. Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem Biophys Res Commun. 1992;185:1155–61. doi: 10.1016/0006-291x(92)91747-e. [DOI] [PubMed] [Google Scholar]
- 15.Law SF, Estojak J, Wang B, Mysliwiec T, Kruh G, Golemis EA. Human enhancer of filamentation 1, a novel p130cas-like docking protein, associates with focal adhesion kinase and induces pseudohyphal growth in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3327–37. doi: 10.1128/mcb.16.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minegishi M, Tachibana K, Sato T, Iwata S, Nojima Y, Morimoto C. Structure and function of Cas-L, a 105-kD Crk-associated substrate-related protein that is involved in beta 1 integrin-mediated signaling in lymphocytes. J Exp Med. 1996;184:1365–75. doi: 10.1084/jem.184.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh M, Cowell L, Seo S, O’Neill G, Golemis E. Molecular basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator of invasion, apoptosis and cell cycle. Cell Biochem Biophys. 2007;48:54–72. doi: 10.1007/s12013-007-0036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Neill GM, Seo S, Serebriiskii IG, Lessin SR, Golemis EA. A new central scaffold for metastasis: parsing HEF1/Cas-L/NEDD9. Cancer Res. 2007;67:8975–9. doi: 10.1158/0008-5472.CAN-07-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–81. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 20.Natarajan M, Stewart JE, Golemis EA, Pugacheva EN, Alexandropoulos K, Cox BD, et al. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene. 2006;25:1721–32. doi: 10.1038/sj.onc.1209199. [DOI] [PubMed] [Google Scholar]
- 21.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–72. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, et al. Suppression of lung adenocarcinoma progression by Nkx2–1. Nature. 2011;473:101–4. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji H, Li D, Chen L, Shimamura T, Kobayashi S, McNamara K, et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–95. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 25.Allred DC, Clark GM, Elledge R, Fuqua SA, Brown RW, Chamness GC, et al. Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J Natl Cancer Inst. 1993;85:200–6. doi: 10.1093/jnci/85.3.200. [DOI] [PubMed] [Google Scholar]
- 26.Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene. 2004;23:379–85. doi: 10.1038/sj.onc.1207074. [DOI] [PubMed] [Google Scholar]
- 27.Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu LJ, et al. The polycomb group protein Bmi-1 represses the tumor suppressor PTEN and induces epithelial-mesenchymal transition in human nasopharyngeal epithelial cells. J Clin Invest. 2009;119:3626–36. doi: 10.1172/JCI39374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amelio AL, Miraglia LJ, Conkright JJ, Mercer BA, Batalov S, Cavett V, et al. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc Natl Acad Sci U S A. 2007;104:20314–9. doi: 10.1073/pnas.0707999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez LG, Wu X, Guan JL. Wound-healing assay. Methods Mol Biol. 2005;294:23–9. doi: 10.1385/1-59259-860-9:023. [DOI] [PubMed] [Google Scholar]
- 30.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, et al. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–23. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Katoh Y, Takemori H, Lin XZ, Tamura M, Muraoka M, Satoh T, et al. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. FEBS J. 2006;273:2730–48. doi: 10.1111/j.1742-4658.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- 32.Kumar KU, Reddy DL, Pater MM, Pater A. Human JC virus cAMP response elements functional for enhanced glial cell expression in differentiating embryonal carcinoma cells. Virology. 1996;215:178–85. doi: 10.1006/viro.1996.0020. [DOI] [PubMed] [Google Scholar]
- 33.Prestridge DS. Predicting Pol II promoter sequences using transcription factor binding sites. J Mol Biol. 1995;249:923–32. doi: 10.1006/jmbi.1995.0349. [DOI] [PubMed] [Google Scholar]
- 34.Kim SH, Xia D, Kim SW, Holla V, Menter DG, Dubois RN. Human enhancer of filamentation 1 Is a mediator of hypoxia-inducible factor-1alpha-mediated migration in colorectal carcinoma cells. Cancer Res. 2010;70:4054–63. doi: 10.1158/0008-5472.CAN-09-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Li H, Wang B, Xu Y, Yang J, Zhang X, et al. VHL inactivation induces HEF1 and Aurora kinase A. J Am Soc Nephrol. 2010;21:2041–6. doi: 10.1681/ASN.2010040345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iourgenko V, Zhang W, Mickanin C, Daly I, Jiang C, Hexham JM, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci U S A. 2003;100:12147–52. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12:141–51. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–11. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Vera L, Fischer WH, Montminy M. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature. 2009;460:534–7. doi: 10.1038/nature08111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 41.Katoh Y, Takemori H, Min L, Muraoka M, Doi J, Horike N, et al. Salt-inducible kinase-1 represses cAMP response element-binding protein activity both in the nucleus and in the cytoplasm. Eur J Biochem. 2004;271:4307–19. doi: 10.1111/j.1432-1033.2004.04372.x. [DOI] [PubMed] [Google Scholar]
- 42.Komiya T, Coxon A, Park Y, Chen WD, Zajac-Kaye M, Meltzer P, et al. Enhanced activity of the CREB co-activator Crtc1 in LKB1 null lung cancer. Oncogene. 2010;29:1672–80. doi: 10.1038/onc.2009.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Izumchenko E, Singh MK, Plotnikova OV, Tikhmyanova N, Little JL, Serebriiskii IG, et al. NEDD9 promotes oncogenic signaling in mammary tumor development. Cancer Res. 2009;69:7198–206. doi: 10.1158/0008-5472.CAN-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kong C, Wang C, Wang L, Ma M, Niu C, Sun X, et al. NEDD9 is a positive regulator of epithelial-mesenchymal transition and promotes invasion in aggressive breast cancer. PLoS One. 2011;6:e22666. doi: 10.1371/journal.pone.0022666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tikhmyanova N, Golemis EA. NEDD9 and BCAR1 negatively regulate E-cadherin membrane localization, and promote E-cadherin degradation. PLoS One. 2011;6:e22102. doi: 10.1371/journal.pone.0022102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–29. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.