Summary
The final degradation product of the complement protein C3, C3d, has been used as a molecular adjuvant to various antigens. Chimera proteins of the antigen and multiple copies of C3d were developed to test the adjuvant effect of this molecule. The main mechanism by which C3d enhances the immune response is interaction with CR2. In-vitro studies showed that the avidity of C3d for CR2 is affected by residues located at the interacting surface (e.g. 170N) as well as by residues located in other areas. The role of the latter residues has been proposed to depend on the electrostatic nature of the C3d-CR2 interaction, where the charges of the whole molecules are responsible for their binding. C3d is primarily a negatively charged molecule, while CR2 is a positive one. Previous experiments demonstrated that elimination of a positive charge (K162A) in C3d enhanced its avidity for CR2, while elimination of negative charges or addition positives ones (D163A N170R, respectively), impaired the avidity for CR2. Despite the extensive in-vitro research, the role of these residues in the adjuvant effect of C3d is unclear. To study the role of residues at the interacting and non-interacting surface of C3d on the adjuvanticity, single as well as a double residue substitutions were engineered in the murine C3d (R162A, D163A, N170R and D163A-N170R) gene. Two copies of these mutant molecules were fused to HIV-1 Envgp120 and the proteins were tested for their avidity to bind CR2 (sCR2). Later, these DNA constructs were tested in mice to determine their adjuvant capability. Mutation at residue 162 (R162A) neither enhanced nor impaired the avidity of Envgp120-C3d2 for sCR2 in-vitro. Mutations at residues D163A and N170R, on the other hand, reduced the binding affinity of Envgp120- C3d2 for sCR2. Furthermore, these mutations synergized and abolished the interaction of C3d for CR2. The data correlated with the adjuvant capability of these molecules in the mouse model. In summary, residues that alter the electronegative status of C3d (D163A and N170R) impair the binding of chimera proteins to CR2, reducing the adjuvant activity of this molecule.
Keywords: Molecular adjuvant, C3d, CR2, DNA vaccine
Introduction
The complement system is composed of more than 20 soluble and membrane proteins that are activated in cascade. The three pathways that can activate the complement system (classic, alternative and mannose-binding lectin) converge in the formation of C3-convertases, which cleave the third component of complement, C3. This event leads to the effector functions of the complement system, including formation of anaphylotoxins (e.g. C3a and C5a), formation of the membrane attack complex (C5b, C6, C7, C8 and multiple C9) and opsonization of microorganisms (e.g. C3b). The cleavage of C3 initially leads to the formation of C3a and C3b. C3d attaches to foreign pathogens (opsonization) by covalent attachment through a cysteine (C) residue that gets exposed only after proteolytic cleavage or hydrolysis. This C is present in all the subsequent cleavage fragments including iC3b, C3dg and C3d.
Once C3d is formed, it can simultaneously bind the foreign antigen and CR2 (CD21)[1,2]. CR2 is part of the B-cell receptor complex that also involves CD19 and Tapa-1 (CD81) [3]. Binding of the antigen to CR2 facilitates its uptake by the B-cell receptor (membrane-bound IgM plus Igα and Igβ). Additionally, cross-linking the B-cell receptor and CR2 amplifies B-cell activation by simultaneously triggering the signaling pathways of the molecules associated with these receptors (Igα-Igβ and CD19, respectively) [4–8]. The C3d-CR2 complex links the innate with the adaptive immune responses resulting in C3d as a natural adjuvant that amplifies the signal required for B-cell activation and therefore enhances antigen processing, presentation and antibody production [9].
In order to exploit the natural adjuvant properties of C3d, our laboratory and others have engineered chimera proteins composed of an antigen fused to multiple copies of C3d (Ag-C3d) [10–19]. These constructs have been shown to enhance antibody titers to a variety of viral antigens in DNA as well as protein immunizations [20–24]. The primary mechanism by which C3d enhances the immune response is CR2-dependent; however, CR2-independent mechanisms have also been described [25].
The interaction between C3d and CR2 has been an area of great debate. The publication of the crystal structure of C3d and the description of a negatively charged channel led to the hypothesis that this region on C3d was the surface contact with CR2 [26]. To support this hypothesis, elimination of negatively charged residues (amino acids) in this channel altered the binding of C3d to full length CR2 expressed on Raji cells (rabbit B-cells). Two clusters of amino acid residues important for the C3d-CR2 interaction were identified (cluster I: 36 Aspartate (D), 37 Glutamate (E) and 39E; cluster II: 160E, 162 Lysine (K), 163D, 164 Isoleucine (I), 166E and 167E) [27]. However, publication of the crystal structure of C3d coupled to CR2 (short consensus repeats 1 and 2) demonstrated that the surface contact area of C3d did not involve the negatively charged channel. Furthermore, mutations in residue 170 Asparagine (N), located in the newly described surface contact of C3d, altered the interaction with CR2 in a competition binding assay [28]. Therefore, there was not a clear explanation of why mutations in the negatively charged channel altered the interaction with CR2. A feasible explanation came from a publication by Morikis and Lambris [29], which suggested that the interaction between C3d and CR2 was of an electrostatic nature. C3d is primarily a negatively charged molecule, while CR2 is positively charged. It was hypothesized that the whole charge of the each molecule was responsible for the interaction. In order to prove this hypothesis, theoretical mutants using the previously described residues were generated and the effect of these mutations on the charges of the whole molecule (C3d) was evaluated. These results demonstrated that elimination of negative charges in C3d (mutations in the negatively charged groove) altered the whole electric potential of the molecule and explained the role of these residues. Furthermore, elimination of a positive charge in C3d (162K) enhanced the avidity of C3d for CR2. Finally, a theoretical mutation at residue 170 increased the positive charge in C3d explaining the importance of this residue [29].
The data generated to date has relied on in-vitro experiments. The in-vivo relevance, especially for the adjuvant properties of C3d, has not been determined. In order to explore the importance of the residues in the negatively charged channel and in the contact surface of C3d for the adjuvant effect, mutations in C3d at residues 162, 163 and 170 were generated. Each mutation in the murine C3d protein was fused in frame with a model antigen, HIV-1 Envgp120, and used for DNA vaccination of mice to determine the in-vivo effect of each residue on the adjuvant properties.
Materials and Methods
Site directed mutagenesis of C3d expression plasmids
A plasmid encoding a single copy of wild-type murine C3d with a histidine (His) (6×) tag was generated by PCR amplification of the wild-type gene [13] and cloned behind the tPA leader sequence in the expression vector TR600 [16]. This plasmid was used as template to generate C3d mutant genes.
Single amino acid substitutions were engineered in the murine C3d gene using a QuickChange XL- Site Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA), using unique oligonucleotide primers following the directions of the manufacturer. The amino acid substitutions introduced in the C3d gene were selected based on previous reports [27,28]. Four different C3d mutants were generated (C3d R162A, C3d D163A, C3d N170R and C3d D163A-N170R) using the next set of oligo primers: FRT880S 5’-atcgcactgcaggaagccgcggacatctgtgagggg-3’, FRT881AS 5’cccctcacagatgtccgcggcttcctgcagtgcgat-3’(to generate C3d R162A); FRT882S 5’-ctgcaggaagccagggccatctgtgaggggcagatc-3’, FRT883AS 5’-gatctgcccctcacagatggccctggcttcctgcag-3’ (to generate C3d D163A); FRT884S 5’-tgtgaggggcaggtcagaagccttcctgggagc-3’, FRT885AS 5’-gctcccaggaaggcttctgacctgcccctcaca-3’ (to generate C3d R170R) (Table 1). The double mutant (C3d D163A-N170R) was generated by introducing the N170R substitution on the initially generated C3d D163A. All the C3d mutant plasmids generated were tested for expression as described before.
Table 1.
| Mutation | Sense | Anti-sense |
|---|---|---|
| R162A | 5′-atcgcactgcaggaagccgcggacatctgtgagggg-3′ | 5′-cccctcacagatgtccgcggcttcctgcagtgcgat-3′ |
| D163A | 5′-ctgcaggaagccagggccatctgtgaggggcagatc-3′ | 5′-gatctgcccctcacagatggccctggcttoctgcag-3′ |
| N170R | 5′-tgtgaggggcaggtcagaagccttcctgggagc-3′ | 5′-gctcccaggaaggcttctgacctgcccctcaca-3′ |
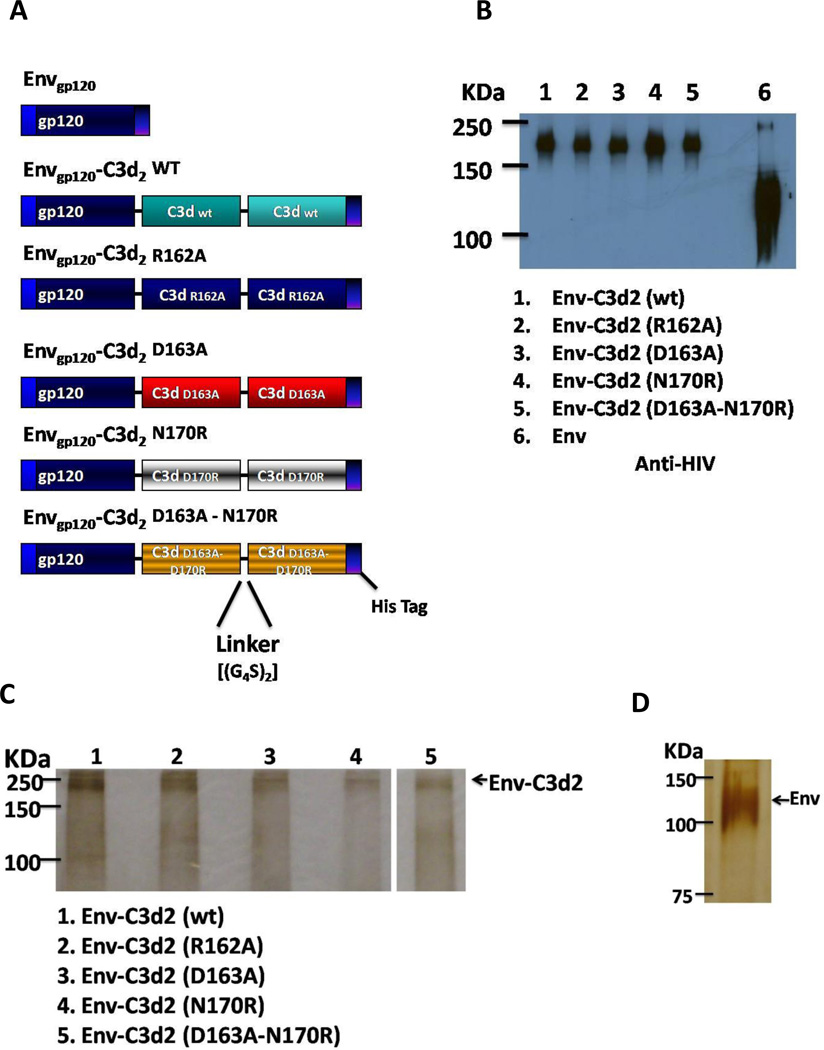
Once single C3d mutants were generated and expression tested, constructs of Envgp120 (89.6 isolate) fused to two copies of C3d (wild-type or mutants) were generated (Envgp120- C3d2). Linkers composed of two repeats of four glycines and a serine [(G4S)2] were fused at the junctures of each C3d repeat. Additionally, a His (6×) Tag was introduced at the 3’ end of each fusion protein (Fig. 2A)
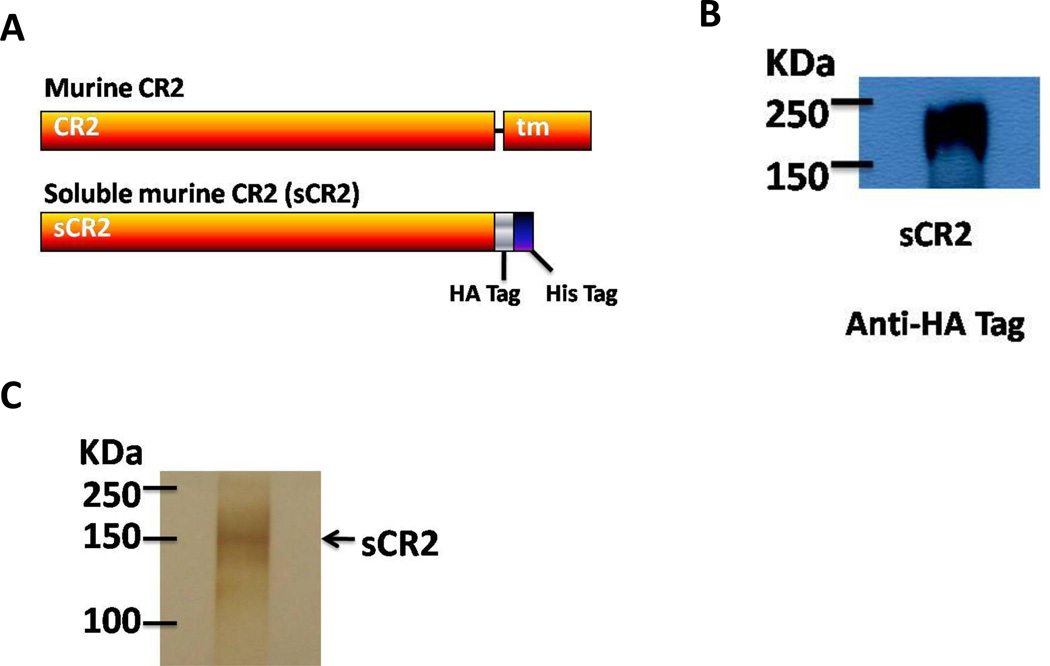
Figure 2. Construction and expression of soluble CR2 gene.
In order to generate a soluble form of CR2 for the binding assay, the transmembrane region of this gene was truncated (nt 3009) (A). The product of the gene was detected in the supernatants of transfected HEK 293T cells (B) and the 2µg of purified protein were tested for purity by Silver Staining (C).
A soluble form of Envgp120 (89.6 isolate) with a His (6×) tag alone was also engineered as previously described. Finally, a soluble form of murine CR2 (sCR2) was generated by truncating the gene at position 3009 (residue 965 in the protein), this removes the trans-membrane portion of the gene. The truncated gene was then cloned into the expression vector TR600. His (6×) and HA tags were introduced at the 3’ end of the gene (Fig. 3A).
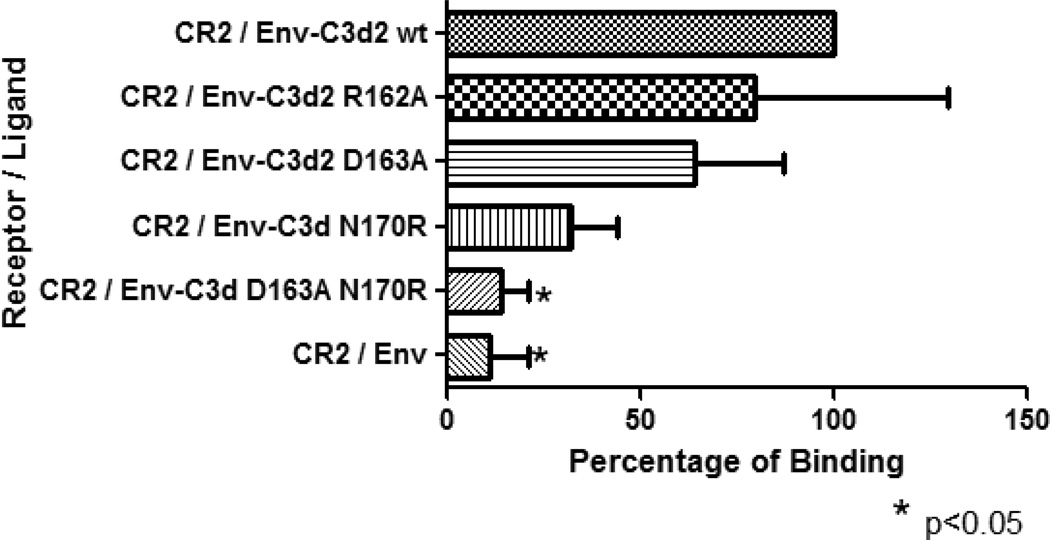
Figure 3. ELISA binding assay.
Envgp120 alone and fused to two copies of wild-type or mutant C3d were assayed for their binding capacity to sCR2. Envgp120- C3d2 wild-type was used as the standard control. Mutation at residue 162 did not alter the binding capacity of Envgp120- C3d2 (R162A) to CR2. Mutations at residues D163A and N170R reduced but not statistically significant the binding of C3d to CR2. Introduction of the double mutation (D163A-N170R) in C3d, synergized and eliminated the binding avidity of C3d for CR2 (p<0.05). As expected, Envgp120 alone showed only background levels of binding to CR2. The experiment was repeated twice in triplicate.
All DNA plasmids were amplified in Escherichia coli DH5-α; purified using endotoxin-free, anion-exchange resin columns (Qiagen, Valencia, CA, USA), resuspended in distilled water and stored at −20 °C.
Plasmid Expression
Each plasmid was transfected into HEK 293T cells (~500,000 cells) performed in six-well plates using Lipofectamine 2000 reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) and Opti-MEM I reduced serum media (Gibco, Grand Island, NY, USA). Seventy-two hours post-transfection, supernatants (1%) (Fig. 2B and 3B) and cell lysates (2%) were collected and diluted in SDS sample buffer and loaded onto SDS-polyacrylamide gels (5% stacking; 10% resolving) and run for 2h (100 Volts). The resolved proteins were transferred onto 0.2mA PVDF membranes (Millipore, Bedford, MA, USA) and incubated with the appropriate anti-sera (1:1000–5000). Bound antibodies were detected using goat appropriate anti-secondary antibodies conjugated to HRP (1:7000) (Southern Biotechnology, Birmingham, AL, USA), followed by enhanced chemiluminescence (Pierce Biotechnology, Rockford, IL, USA).
Protein Purification
For protein purifications, ~8 × 106 HEK 293T cells were transfected with plasmids in T-75 flasks. Briefly, cells were grown in complete Dubelcco’s Modified Media (cDMEM) [DMEM supplemented to contain 10% heat-inactivated fetal bovine serum (FBS) (Atlanta Biologicals, Atlanta, GA, USA), 4 mM L-glutamine (Invitrogen Life Technologies, Carlsbad, CA, USA), and 0.4 mg/L gentamicine (Gibco, Grand Island, NY, USA)] until 90% confluency. DNA plasmids (15 µg) were incubated with Lipofectamine 2000 (40 µl) and Opti-MEM I reduced serum media (1 ml) for 25 min (Gibco, Grand Island, NY, USA). cDMEM was removed from HEK 293T cells, replaced with 5 ml of Opti-MEM I and the DNA - Lipofectamine 2000 mixture added. Cells were incubated for 6h at 37 °C/ 5% CO2 and 5 ml extra of fresh, warmed Opti-MEM I reduced serum media were added. Flasks were incubated for 48h (37°C/ 5% CO2) and 5 ml extra of supernatants added. The cells were incubated for an additional 48h and the supernatants harvested. A protease inhibitor cocktail for purification of His-tagged proteins was added (2.5 µl/ml) (Sigma, St. Louis, MO, USA). Supernatants were stored at −80 °C until protein purification.
Following a single freeze thaw supernatants were pooled and Envgp120 or Envgp120-C3d2 wild-type or mutants proteins were purified at 4°C using a 5 ml HiTrap chelating nickel column (Amersham Biosciences, Piscataway, NJ, USA). Briefly, the column was loaded with 0.1 M NiSO4 (5 ml) (Fisher Scientific, Fair Lawn, NJ, USA) and subsequently washed with distilled water (15 ml). The HiTrap column was equilibrated with 30 ml of Binding Buffer [20mM phosphate, 0.5 M NaCl and 10 mM imidazole (Sigma, St. Louis, MO, USA)]. Supernatants containing His-tagged proteins were loaded into the equilibrated HiTrap column at a rate of 2 ml/min. The column was then washed with Binding Buffer (30 ml). Subsequently, the proteins were eluted using 15 ml of Elution Buffer (20 mM phosphate, 0.5 M NaCl and 500 mM imidazole). The eluted fraction, containing the purified protein, was concentrated immediately using 100,000 molecular weight cut-off columns (Vivascience, Hannover, Germany) (3,000 g, 14°C, 30–45 min). The protein was washed twice with 1× PBS (15 ml) (buffer exchange) and resuspended in a total volume of 0.5 ml. The purified, concentrated and buffer exchanged protein was stored at −80°C. Protein concentration was determined using a Micro BCA Protein Assay Kit (Pierce, Rockford, IL, USA).
Silver Staining
The purified proteins were verified by silver staining using a ProteoSilver Stain Kit (Sigma, St. Louis, MO, USA), following the manufacturer’s protocol (Fig. 1C, D and 2C). Briefly, purified protein samples stored at −80 °C were gently thawed and prepared for electrophoresis. 500 ng of Envgp120-C3d2 and 2 µg of sCR2 or Envgp120 alone were resuspended in a total volume of 10ul of ddH2O. Protein samples were diluted (1:2) in sodium dodecyl sulfate (SDS) Laemmil sample buffer (Bio-Rad, Hercules, CA, USA) and the mixture was boiled for 5 min. The samples were loaded onto a 5–10% SDS-polyacrylamide gel (Stacking Gel: 30% acrylamide:bis, 10% SDS, 10% ammonium persulfate, 1% TEMED, 0.5 M Tris-HCl pH 6.8 or Resolving Gel: 1.5 M Tris-HCl, pH 8.8). After electrophoresis of the proteins by SDS-PAGE, the gel was placed in a clean tray with 100 ml of Fixing Solution (50% ethanol, 10% acetic acid in ultrapure water) for 1h. The Fixing Solution was removed and the gel was washed with 100 ml of Ethanol Solution (30% ethanol in ultrapure water) for 10 min. The Ethanol Solution was decanted and the gel was washed twice (10 min) with 200 ml of ultrapure water. The gel was then incubated for 10 min with 100 ml of Sensitization Solution (1% ProteoSilver Sensitizer in ultrapure water). Following removal of the sensitizing solution, the gel was washed twice (10 min) with 200 ml of ultrapure water. The water was decanted and 100 ml of Silver Equilibration Solution (1% ProteoSilver Silver solution in ultrapure water) was added to the gel for 10 min. After the Silver Equilibration Solution was removed, the gel was washed for 1 min with 200 ml of ultrapure water. The water was decanted and 100 ml of Developer Solution (5% of Proteo Silver Developer 1, 0.1% of ProteoSilver Developer 2 in ultrapure water) were added to the gel. The gel was carefully rocked back and forth for 3–7 min until the desired staining intensity was observed. 5 ml of the ProteoSilver Stop Solution was added to the Developer Solution to stop the reaction (5 min). Finally, the Developer / ProteoSilver Stop solution was decanted and the gel was washed with 200 ml of ultrapure water for 15 minutes.
Figure 1. Construction and expression of Envgp120-C3d.
The gene encoding for HIV-1 Envgp120 was fused to two copies of wild type or mutant C3d (R162A, D163A, N170R or D163A-N170R) and cloned into the expression vector TR600 (A). Linkers composed of two repeats of four glycines and a serine [(G4S)2] were fused at the junctures of each C3d copy. These constructs were tested for expression in HEK 293T cells. The amino acid substitutions introduced in C3d did not alter the expression of the fusion proteins (B). Similar to previous reports, Envgp120 alone produced more protein than the chimera constructs. (C and D) Purity of the purified proteins was assessed by silver staining (500 ng of Envgp120-C3d2 and 2 µg of Envgp120 alone were evaluated).
C3d-CR2 binding assay
Plates (96-well) were coated with sCR2 recombinant protein (5 µg/ml – 100µl/well) (1h, 37 °C). The plate was then blocked for 1h (5% non-fat dry milk in 1× PBS-0.05 tween − 1× PBS-T-) at RT. Envgp120 alone and Envgp120- C3d2 wild-type and mutants (1 µg/ml – 100 µl/well) were added to the corresponding wells and allowed to bind for 1h at RT. Following 4 washed with 200µl of 1× PBS-T, 100µl of a 1:5,000 dilution (5% non-fat milk 1× PBS-T) of HIV-Ig was added to each well (1h, RT). The plate was washed (4×) and 100ul of a 1:10,000 dilution of goat anti-human IgG-HRP was incubated for 1h at RT. TMB substrate (100ul) were added to each well and the plate was incubated in the dark for 20 min. The reaction was stopped by adding 50ul of a 2N sulfuric acid solution. The plate was read immediately and the O.D. was determined at 450 nm. Each experiment was run in triplicate. The O.D. data was normalized to percentage and the data analyzed by one-way ANOVA to compare all groups. Then each group was compared to the sCR2/Envgp120- C3d2 wt using a Dunnett’s multiple comparison tests.
Mice immunizations
Five female BALB/c mice (12–14 weeks-old) (Harlan-Sprague, Indianapolis, IN, USA) per group were gene gun immunized on shaved abdominal skin by using the hand-held Bio-Rad gene delivery system. Mice were immunized with two gene gun doses containing 2 µg of DNA per 0.5 mg of approximately 1-µm gold beads (Bio-Rad, Hercules, CA) at a helium pressure setting of 400 lb/in2. Immunizations were performed three weeks apart (weeks 0 and 3) and sera samples were collected by retro-orbital bleeding, two weeks after each immunization (weeks 2 and 5). For blood sample collection animals were anesthetized with a mixture of ketamine / xylazine.
Evaluation of the immune response by ELISA
Sera samples were stored at −80 °C until analysis. Antigen-specific antibody titers were assessed by ELISA. To determine anti-Env antibodies, microtiter plates (96-well) were coated with Envgp120 (YU2) containing supernatants from transiently transfected HEK 293T cells (≈ 100 ng of Envgp120/well). Plates were incubated overnight at 4°C and then blocked with 5% nonfat dry milk in 1× PBS/0.05% Tween (PBS-T) (2 h). After extensive washing with PBS-T, antiserum collected from vaccinated mice was serially diluted (initial dilution 1:50) in 5% nonfat dry milk in PBS-T. Serum was allowed to bind to antigen coated plates (2 h), followed by thorough washing with PBS-T. The plates were then incubated (25° C for 1 h) with 100 µl of goat anti-mouse IgG conjugated to horseradish peroxidase (HRP) (1:5,000) (Southern Biotechnology Associates, Inc., Birmingham, AL) diluted in PBS-T containing 5% non-fat dry milk. The unbound antibody was removed, and the wells were washed (3×) with PBS-T. 100 µl of TMB substrate (1 TMB tablet per 10 ml of phosphate-citrate pH 5.0 buffer; 2 µl 30% H202) (Sigma, St Louis, MO, USA) were added to each well (25°C for 30 min). Following 30 min incubation, the reaction was stopped with 50 µl / well of 2N Sulfuric Acid. The colorimetric change was measured as the O.D. at 450 nm using a spectrophotometer (Dynex Technologies, Chantilly, VA, USA).
Results
Generation of C3d mutants
Two clusters of residues were identified as important for the human C3d-CR2 interaction by crystallography (cluster I: 36D, 37E and 39E; cluster II: 160E, 162K, 163D, 164I, 166E and 167E) [27]. These residues were located in a negatively charged channel. However, when C3d was coupled to CR2, the surface contact area of C3d did not involve the negatively charged channel. Furthermore, within the newly described surface contact, residue 170N was identified as critical for the interaction of these two molecules [28]. This was surprising since N is a neutral residue and a major role for this residue was not expected. Among these 10 residues, amino acid substitutions at positions 162, 163 and 170 in C3d significantly changed the binding of C3d to CR2 in two different studies, therefore these residues were selected to perform in vivo studies. Human and murine C3d have an overall sequence similarity of 90% and while residues 163 (D) and 170 (N) are the identical, there is a K at residue 162 in mice and an R at this position in humans. K and R are similar amino acids (branched, hydrophilic, basic); therefore, a similar role for these residues can be speculated.
In order to determine the role of these residues in adjuvant function of C3d, single amino acid substitutions were introduced in the murine C3d molecule. These substitutions included an R for A substitution at residue 162 (R162A), D for A at position 163 (D163A) and N for R at residue 170 (N170R). Additionally, double substitutions at positions 162 and 170 (D163A-N170R) were engineered in a single molecule. None of these mutations altered the expression of the C3d gene (data not shown). Two copies of each C3d protein (wild-type and mutants) were cloned in frame to the HIV-1 Envgp120 (Fig. 1A). Each chimera protein efficiently secreted and expressed into the supernatant of transiently transfected HEK 293T cells (Fig 1B).
Role of amino acids 162, 163 and 170 in the binding to sCR2
Envgp120-C3d2 with wild-type and individual mutant C3d proteins (R162A, D163A, N170R) were tested for the binding affinity to immobilized murine sCR2. Mutation K162A in the human C3d molecule was reported to enhance the binding avidity of C3d for CR2 [27]. In the murine C3d, however, the R162A mutation neither impaired nor enhanced the binding affinity for sCR2 compared to wild-type C3d (Fig. 3). Mutations D163A and N170R have both been reported to reduce the binding affinity for CR2 in the human molecules [27,28]. Consistent with this report, Envgp120 fused to C3d D163A and N170R had a reduced, albeit not significantly, binding affinity for sCR2 (30% and 60% reduction, respectively). There was a significant reduction on the binding affinity of the double mutant D163A-N170R for sCR2 (90% reduction) (p<0.05) (Fig. 3). Envgp120 alone had only background binding to sCR2.
Role of amino acids 162, 163 and 170 in the enhancing the immune response to Env
C3d is an effective molecular adjuvant [11,13–18,21,22,30–35]. To determine if C3d residues 162, 163 and, 170 affect the adjuvant properties of C3d, mice were vaccinated with DNA plasmids expressing Envgp120 fused to either the wild-type C3d or the mutant C3d proteins. Consistent with our previous results, Envgp120 fused to two copies of wild-type C3d enhanced the development of anti-Env antibody titers, compared to Envgp120 alone (Fig. 4A and B). Consistent with the results of the binding assay, Envgp120-C3d2 R162A did not enhance or reduced anti-Env antibody titers (Fig. 4A and B). Both sets of mice had similar endpoint dilution titers (1:12,500). In contrast, both Envgp120-C3d2 D163A and Envgp120-C3d2 N170R reduced the anti-Env antibodies titers compared to wild-type C3d. Mice vaccinated with either of these DNA vaccines elicited ~1 log lower anti-Env antibody titer than Envgp120 with wild-type C3d. Interestingly, similar to the binding assay results, the combined mutations at residues 162 and 170 (Envgp120-C3d2 D163A-N170R) reduced the anti-Env titers by 87% (1:200) (Fig. 4A and B).
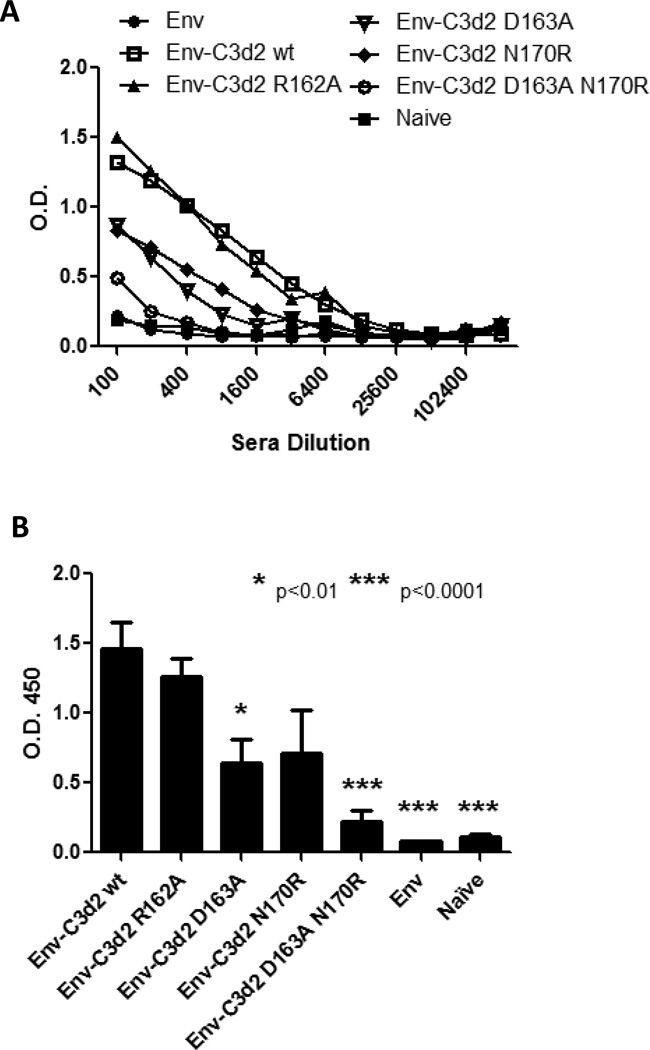
Figure 4. Anti-Env antibody titers.
Mice (5 mice per group) were DNA immunized (weeks 0 and 3) with Envgp120 alone or fused to two copies of C3d wild-type or mutants. Sera collected two weeks after the last immunization (week 5) was tested for anti-Env antibody titers by ELISA. Different sera dilutions were assayed and the O.D. reported (A). Envgp120- C3d2 wild-type (white square) enhanced the anti-Env antibody titers compared to Envgp120 alone (black circle). Envgp120- C3d2 (R162A) did not enhance or reduce the adjuvant effect of C3d. Envgp120- C3d2 (D163A) (inverted white triangle) and Envgp120-C3d (N170R) (black rhomboid) reduced the adjuvant effect of C3d. However, these mutations did not completely eliminate it, since higher Ab titers than in the Envgp120 alone group were detected. Introduction of the double mutation (D163A-N170R) in C3d (white circle) resulted on almost complete elimination of the adjuvant effect. (B) The results of a 1:200 dilution are shown. Columns indicate the mean and bars ± SEM. The different groups were compared to Envgp120- C3d2 wild-type for statistical analysis (* p<.01, ** P,0.0001).
Discussion
C3d is an efficient adjuvant for several antigens derived from microorganisms [11,13–17,22,30–34,36,37]. Interaction with its natural ligand, CR2, is the primary mechanism for its adjuvant effect and despite the intense in-vitro research of the C3d-CR2 interaction, the translation of these findings for the adjuvant effect in-vivo has mainly remained unexplored. Identification of critical residues for the C3d-CR2 interaction might provide clues for improving the adjuvant properties of C3d and improve vaccine design. In the current study, previously reported mutations that alter the binding avidity of C3d (R162A, D163A, N170R) for CR2 were engineered. These mutations were selected due to the significantly higher effect on the interaction with CR2 that has been reported and include 1) a mutation that enhanced the binding affinity for CR2 (K162R), 2) a single mutation that significantly impairs the avidity for CR2 and located in the non-interacting surface (>95% impairment) and 3) a mutation that reduced the affinity for CR2 and located on the interacting surface [27,28]. These C3d mutant molecules were then fused to HIV-1 Envgp120, a well characterized model antigen, and these chimera proteins (Envgp120-C3d2) used in in-vitro binding assays to CR2 as well as in-vivo vaccine studies.
Previous reports showed that mutations of residue 162K in the human C3d enhanced the avidity for CR2 [27]. The mutation that enhanced the binding avidity of C3d (K162A) replaced a basic (positively charged) for a neutral residue. In the mouse C3d molecule, position 162 is occupied by an R instead of K. These residues, however, share similar characteristics (basic, branched), therefore a similar function was expected. Despite this, the mutation at this residue (R162A) did not improve or impair the affinity of C3d for CR2. It is possible that these amino acids play different roles in the human and mouse molecules. However, it is not clear how these might differ; especially considering that mathematical models suggest that the electrostatic potential of the whole molecule is responsible for the interaction with CR2. Another possibility is that the generation of the chimera (Envgp120-C3d2 R162A) molecule “obstructed” this residue, reducing its natural role. However, this is very unlikely, since mutations at the neighboring residue (D163A) did have an effect of the binding affinity of Envgp120-C3d2 for CR2.
In contrast, mutations at residues D163A and N170R reduced the binding affinity of Envgp120-C3d2 for CR2, by 30% and 70% respectively. Interestingly, the substitution that involved the interacting surface (170N) had a more significant effect than the mutation located in the negative channel. These differences, however, were not statistically significant. Mutations in these residues have previously been tested by two different teams using different binding assays. The D163A mutation was tested for its binding avidity to the full length CR2 molecule on Raji cells [27]. Meanwhile, mutation R170N was tested on for its binding avidity to full-length soluble CR2 on a competition ELISA [28]. Therefore, a direct comparison was lacking and the results of the Envgp120-C3d2 suggested that residues on the interacting surface play a slightly more important role than residues in the adjacent areas. A synergistic impairment (>90% in binding impairment) in the C3d-CR2 interaction was shown by the double C3d mutant (D163A-N170R) (Fig. 4). This reinforces the hypothesis that residues at the interacting and non-interacting surfaces of C3d are important for binding to CR2 and also confirms the electrostatic nature of the interaction between these proteins, since the mutations removed a negative charge (D163A) and added a positive one (N170R), respectively, therefore increasing the overall positive charge in C3d.
To evaluate the effect of these mutations on the adjuvant effect by C3d (in-vivo), mice were immunized with DNA expressing the wild-type Envgp120-C3d2 or one of three Envgp120-C3d2 mutant chimera proteins. Consistent with previous reports [13,16], wild-type Envgp120-C3d2 enhanced the immune response compared to Envgp120 alone (Fig. 4A and B). Consistent with the lack of C3d binding to CR2 in-vitro, Envgp120-C3d2 R162A did not enhance the anti- Envgp120 antibody titer. Both Envgp120-C3d2 D163A and Envgp120-C3d2 N170R were impaired in their ability to elicit anti-Envgp120 antibodies, reducing the anti- Envgp120 antibodies by ~50% compared to wild-type Envgp120-C3d2 (Fig. 4). However, combining these two mutations (D163A and N170R) almost completely eliminated the elicitation of anti- Envgp120 antibodies, which correlated with the ablation of C3d binding to CR2.
This study has demonstrated a strong correlation between the C3d-CR2 interaction and the adjuvant effect of C3d. One question that remains unanswered however, is the role of these mutations in the CR2-independent mechanism. Various hypotheses for the CR2-independent mechanism of enhancement of the immune response have been proposed 1) enhanced half-life of the fused antigen, 2) carrier protein effect and 3) additional C3d receptor(s). A previous publication suggested that C3d works as a carrier protein, since the adjuvant effect was similar to CGG (known CR2-independent adjuvant) in CR2 KO mice [25]. In the current study, mutations that reduced (D163A or N170R) or eliminated (D163A-N170R) the interaction between C3d and CR2 resulted in reduced or complete elimination of the C3d-induced adjuvant effect. Therefore, the data presented here does not favor a C3d carrier effect for the CR2-independent mechanism and may favor a secondary C3d receptor. It is possible that in the CR2 KO mice C3d bound to the secondary and this resulted in the enhancement of the immune response. Furthermore, since the mutations introduced eliminate the overall negative charge (D163A and/or N170R) of C3d, the data also suggest that the interaction with the possible secondary receptor also depends on the electrostatic nature of the molecules.
In summary, the present study demonstrates that the interaction of C3d with CR2 remains the primary mechanism for enhancement of antibodies to linked proteins. Mutations on the interacting surface (N170R) or adjacent areas (D163A) that alter the negatively charged nature of C3d impair the binding of C3d with CR2 and alter the adjuvant potential of C3d. Finally, the data suggest the presence of a secondary receptor, which identity remains unknown. A complete understanding of the mechanism of interaction between C3d and CR2 might contribute to better vaccine design for poorly antigenic molecules.
Acknowledgments
The authors would like to thank to Hermancia Eugene and Brendan Giles for technical assistance. We are also thankful to Dr. Douglas Fearon, who provided the initial C3d construct. This research was supported by an award from NIH R01 GM083602-01 to TMR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weis JJ, Tedder TF, Fearon DT. Identification of a 145,000 Mr membrane protein as the C3d receptor (CR2) of human B lymphocytes. Proc Natl Acad Sci U S A. 1984;81:881–885. doi: 10.1073/pnas.81.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iida K, Nadler L, Nussenzweig V. Identification of the membrane receptor for the complement fragment C3d by means of a monoclonal antibody. J Exp Med. 1983;158:1021–1033. doi: 10.1084/jem.158.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsumoto AK, Kopicky-Burd J, Carter RH, Tuveson DA, Tedder TF, Fearon DT. Intersection of the complement and immune systems: a signal transduction complex of the B lymphocyte-containing complement receptor type 2 and CD19. J Exp Med. 1991;173:55–64. doi: 10.1084/jem.173.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prechl J, Baiu DC, Horvath A, Erdei A. Modeling the presentation of C3d-coated antigen by B lymphocytes: enhancement by CR1/2-BCR co-ligation is selective for the co-ligating antigen. Int Immunol. 2002;14:241–247. doi: 10.1093/intimm/14.3.241. [DOI] [PubMed] [Google Scholar]
- 5.Lyubchenko T, dal Porto J, Cambier JC, Holers VM. Coligation of the B cell receptor with complement receptor type 2 (CR2/CD21) using its natural ligand C3dg: activation without engagement of an inhibitory signaling pathway. J Immunol. 2005;174:3264–3272. doi: 10.4049/jimmunol.174.6.3264. [DOI] [PubMed] [Google Scholar]
- 6.Koch M, Frazier J, Sodroski J, Wyatt R. Characterization of antibody responses to purified HIV-1 gp120 glycoproteins fused with the molecular adjuvant C3d. Virology. 2005;340:277–284. doi: 10.1016/j.virol.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 7.Mongini PK, Vilensky MA, Highet PF, Inman JK. The affinity threshold for human B cell activation via the antigen receptor complex is reduced upon co-ligation of the antigen receptor with CD21 (CR2) J Immunol. 1997;159:3782–3791. [PubMed] [Google Scholar]
- 8.Rickert RC. Regulation of B lymphocyte activation by complement C3 and the B cell coreceptor complex. Curr Opin Immunol. 2005;17:237–243. doi: 10.1016/j.coi.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Carroll MC. The role of complement in B cell activation and tolerance. Adv Immunol. 2000;74:61–88. doi: 10.1016/s0065-2776(08)60908-6. [DOI] [PubMed] [Google Scholar]
- 10.Liu D, Niu ZX. Construction, expression and immunoassay detection of recombinant plasmid encoding fusion protein of Roman chicken complement C3d and Newcastle disease virus F gene. Scandinavian journal of immunology. 2008;68:598–606. doi: 10.1111/j.1365-3083.2008.02177.x. [DOI] [PubMed] [Google Scholar]
- 11.Bower JF, Green TD, Ross TM. DNA vaccines expressing soluble CD4-envelope proteins fused to C3d elicit cross-reactive neutralizing antibodies to HIV-1. Virology. 2004;328:292–300. doi: 10.1016/j.virol.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 12.Gor DO, Ding X, Li Q, Greenspan NS. Genetic fusion of three tandem copies of murine C3d sequences to diphtheria toxin fragment B elicits a decreased fragment B-specific antibody response. Immunol Lett. 2006;102:38–49. doi: 10.1016/j.imlet.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Green TD, Montefiori DC, Ross TM. Enhancement of antibodies to the human immunodeficiency virus type 1 envelope by using the molecular adjuvant C3d. Journal of virology. 2003;77:2046–2055. doi: 10.1128/JVI.77.3.2046-2055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green TD, Newton BR, Rota PA, Xu Y, Robinson HL, Ross TM. C3d enhancement of neutralizing antibodies to measles hemagglutinin. Vaccine. 2001;20:242–248. doi: 10.1016/s0264-410x(01)00266-3. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell JA, Green TD, Bright RA, Ross TM. Induction of heterosubtypic immunity to influenza A virus using a DNA vaccine expressing hemagglutinin-C3d fusion proteins. Vaccine. 2003;21:902–914. doi: 10.1016/s0264-410x(02)00539-x. [DOI] [PubMed] [Google Scholar]
- 16.Ross TM, Xu Y, Green TD, Montefiori DC, Robinson HL. Enhanced avidity maturation of antibody to human immunodeficiency virus envelope: DNA vaccination with gp120-C3d fusion proteins. AIDS Res Hum Retroviruses. 2001;17:829–835. doi: 10.1089/088922201750252025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toapanta FR, Ross TM. Mouse strain-dependent differences in enhancement of immune responses by C3d. Vaccine. 2004;22:1773–1781. doi: 10.1016/j.vaccine.2003.10.050. [DOI] [PubMed] [Google Scholar]
- 18.Dempsey PW, Allison ME, Akkaraju S, Goodnow CC, Fearon DT. C3d of complement as a molecular adjuvant: bridging innate and acquired immunity. Science. 1996;271:348–350. doi: 10.1126/science.271.5247.348. [DOI] [PubMed] [Google Scholar]
- 19.Movsesyan N, Mkrtichyan M, Petrushina I, Ross TM, Cribbs DH, Agadjanyan MG, Ghochikyan A. DNA epitope vaccine containing complement component C3d enhances anti-amyloid-beta antibody production and polarizes the immune response towards a Th2 phenotype. Journal of neuroimmunology. 2008;205:57–63. doi: 10.1016/j.jneuroim.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Oriol Sunyer J, Bello LJ. Immunogenicity of a bovine viral diarrhea virus E2-C3d fusion protein containing a bovine homolog of C3d. Dev Comp Immunol. 2005;29:907–915. doi: 10.1016/j.dci.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Sunyer JO, Bello LJ. Fusion to C3d enhances the immunogenicity of the E2 glycoprotein of type 2 bovine viral diarrhea virus. J Virol. 2004;78:1616–1622. doi: 10.1128/JVI.78.4.1616-1622.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Test ST, Mitsuyoshi J, Connolly CC, Lucas AH. Increased immunogenicity and induction of class switching by conjugation of complement C3d to pneumococcal serotype 14 capsular polysaccharide. Infect Immun. 2001;69:3031–3040. doi: 10.1128/IAI.69.5.3031-3040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrault DV, Steward M, Cox VF, Smith RA, Knight AM. Efficient production of complement (C3d)3 fusion proteins using the baculovirus expression vector system. J Immunol Methods. 2005;304:158–173. doi: 10.1016/j.jim.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 24.Henson SE, Smith D, Boackle SA, Holers VM, Karp DR. Generation of recombinant human C3dg tetramers for the analysis of CD21 binding and function. J Immunol Methods. 2001;258:97–109. doi: 10.1016/s0022-1759(01)00471-9. [DOI] [PubMed] [Google Scholar]
- 25.Haas KM, Toapanta FR, Oliver JA, Poe JC, Weis JH, Karp DR, Bower JF, Ross TM, Tedder TF. Cutting edge: C3d functions as a molecular adjuvant in the absence of CD21/35 expression. J Immunol. 2004;172:5833–5837. doi: 10.4049/jimmunol.172.10.5833. [DOI] [PubMed] [Google Scholar]
- 26.Nagar B, Jones RG, Diefenbach RJ, Isenman DE, Rini JM. X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2. Science. 1998;280:1277–1281. doi: 10.1126/science.280.5367.1277. [DOI] [PubMed] [Google Scholar]
- 27.Clemenza L, Isenman DE. Structure-guided identification of C3d residues essential for its binding to complement receptor 2 (CD21) J Immunol. 2000;165:3839–3848. doi: 10.4049/jimmunol.165.7.3839. [DOI] [PubMed] [Google Scholar]
- 28.Szakonyi G, Guthridge JM, Li D, Young K, Holers VM, Chen XS. Structure of complement receptor 2 in complex with its C3d ligand. Science. 2001;292:1725–1728. doi: 10.1126/science.1059118. [DOI] [PubMed] [Google Scholar]
- 29.Morikis D, Lambris JD. The electrostatic nature of C3d-complement receptor 2 association. J Immunol. 2004;172:7537–7547. doi: 10.4049/jimmunol.172.12.7537. [DOI] [PubMed] [Google Scholar]
- 30.Bower JF, Sanders KL, Ross TM. C3d enhances immune responses using low doses of DNA expressing the HIV-1 envelope from codon-optimized gene sequences. Current HIV research. 2005;3:191–198. doi: 10.2174/1570162053506937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bower JF, Yang X, Sodroski J, Ross TM. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J Virol. 2004;78:4710–4719. doi: 10.1128/JVI.78.9.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ross TM, Xu Y, Bright RA, Robinson HL. C3d enhancement of antibodies to hemagglutinin accelerates protection against influenza virus challenge. Nat Immunol. 2000;1:127–131. doi: 10.1038/77802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terrazzini N, Hannesdottir S, Delves PJ, Lund T. DNA immunization with plasmids expressing hCGbeta-chimeras. Vaccine. 2004;22:2146–2153. doi: 10.1016/j.vaccine.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe I, Ross TM, Tamura S, Ichinohe T, Ito S, Takahashi H, Sawa H, Chiba J, Kurata T, Sata T, Hasegawa H. Protection against influenza virus infection by intranasal administration of C3dfused hemagglutinin. Vaccine. 2003;21:4532–4538. doi: 10.1016/s0264-410x(03)00510-3. [DOI] [PubMed] [Google Scholar]
- 35.Mitsuyoshi JK, Hu Y, Test ST. Role of complement receptor type 2 and endogenous complement in the humoral immune response to conjugates of complement C3d and pneumococcal serotype 14 capsular polysaccharide. Infect Immun. 2005;73:7311–7316. doi: 10.1128/IAI.73.11.7311-7316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu F, Mboudjeka I, Shen S, Chou TH, Wang S, Ross TM, Lu S. Independent but not synergistic enhancement to the immunogenicity of DNA vaccine expressing HIV-1 gp120 glycoprotein by codon optimization and C3d fusion in a mouse model. Vaccine. 2004;22:1764–1772. doi: 10.1016/j.vaccine.2003.09.054. [DOI] [PubMed] [Google Scholar]
- 37.Wang LX, Xu W, Guan QD, Chu YW, Wang Y, Xiong SD. Contribution of C3d-P28 repeats to enhancement of immune responses against HBV-preS2/S induced by gene immunization. World J Gastroenterol. 2004;10:2072–2077. doi: 10.3748/wjg.v10.i14.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]