Abstract
The consequences of injuries to the CNS are profound and persistent, resulting in substantial burden to both the individual patient and society. Existing treatments for CNS injuries such as stroke, traumatic brain injury and spinal cord injury have proved inadequate, partly owing to an incomplete understanding of post-injury cellular and molecular changes. MicroRNAs (miRNAs) are RNA molecules composed of 20–24 nucleotides that function to inhibit mRNA translation and have key roles in normal CNS development and function, as well as in disease. However, a role for miRNAs as effectors of CNS injury has recently emerged. Use of bioinformatics to assess the mRNA targets of miRNAs enables high-order analysis of interconnected networks, and can reveal affected pathways that may not be identifiable with the use of traditional techniques such as gene knock-in or knockout approaches, or mRNA microarrays. In this Review, we discuss the findings of miRNA microarray studies of spinal cord injury, traumatic brain injury and stroke, as well as the use of gene ontological algorithms to discern global patterns of molecular and cellular changes following such injuries. Furthermore, we examine the current state of miRNA-based therapies and their potential to improve functional outcomes in patients with CNS injuries.
Introduction
CNS injuries such as spinal cord injury (SCI), traumatic brain injury (TBI) and stroke are common pathologies, collectively affecting over 13.5 million people in the USA alone.1–3 These injuries constitute a major cause of morbidity and mortality, as beneficial treatments for these complex pathologies are lacking, and obstacles remain with regard to development of effective therapies. Successful treatments for CNS injury must minimize both short-term and long-term cellular damage. Furthermore, identification of factors that lead to irreversible damage requires analyses at both the cellular and molecular level, from disruptions in tissue to alterations in signalling pathways. Microarray technology has been used to uncover some of these factors, and has revealed transcriptome changes in tissue from patients with—as well as in animal models of—CNS injuries.4 Through microarray studies, a role for protein-coding mRNAs in CNS injury has been revealed; however, the family of noncoding RNAs (ncRNAs) has been increasingly implicated in many diseases of the CNS.5,6
microRNAs (miRNAs)—short ncRNAs that modulate protein expression levels by antagonizing mRNA translation—are emerging as powerful regulators of cellular function.7 As a single miRNA can target and block translation of numerous mRNAs, these molecules are capable of altering cellular function at a network level and are, therefore, a particularly potent means of governing cell biology and behaviour. In light of the central roles of miRNAs in CNS development and disease, microarray studies of these molecules in CNS injuries have recently been performed.8,9 These studies provide the highest-order view to date of the molecular and cellular changes that occur after CNS injury. From this view, global patterns of change in the affected tissue as well as the circulating blood and cerebrospinal fluid (CSF) can be determined, providing insight into pathways that can be therapeutically targeted.
In this Review, we provide an overview of miRNA biogenesis and its role in CNS development, as well as summarizing data from recent miRNA microarray studies of SCI, TBI and stroke. Furthermore, common themes of miRNA biology and function that have arisen from these studies are discussed, and conserved pathways in the CNS injury response are identified. Finally, the potential of miRNA-based therapies as a novel approach to ameliorate the morbidity and mortality associated with CNS injury is highlighted.
miRNA structure and function
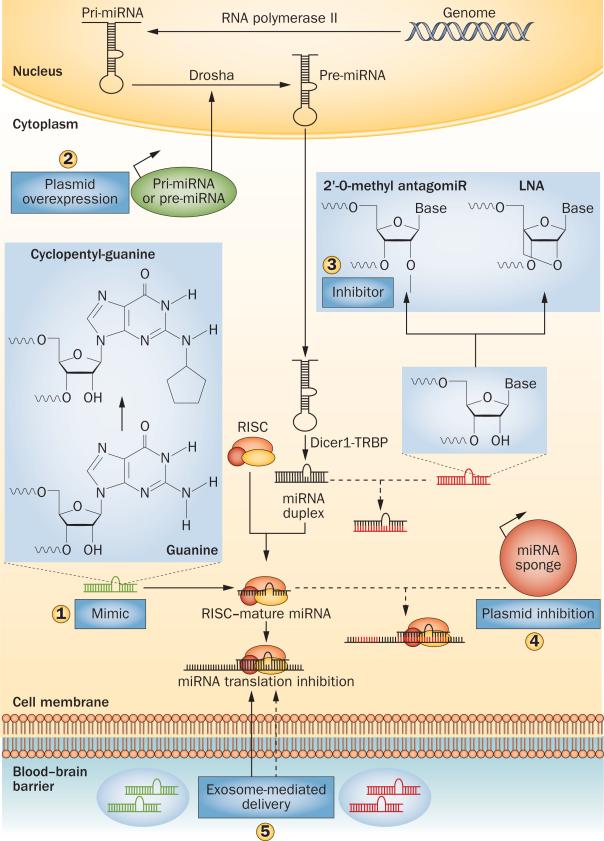
miRNAs are single-stranded ncRNA molecules composed of 20–24 nucleotides that regulate protein expression at the epigenetic level.10 They are initially transcribed from genomic DNA, largely by RNA polymerase II, in the form of primary miRNA transcripts (pri-miRNA; Figure 1).11 Pri-miRNAs, which can be thousands of base pairs in length, form functional secondary structures that contain stem-loops consisting of inverted repeats.11,12 These loops are recognized and cleaved by the endonuclease Drosha (also known as ribonuclease III), which leads to liberation of a precursor miRNA (pre-miRNA) from the pri-miRNA.13,14 Through the function of exportin-5 (a protein that is involved in nuclear export of RNA), pre-miRNA is transported from the nucleus into the cytoplasm, where Dicer and TAR RNA-binding protein process it to form a duplex of the mature miRNA strand and a complementary strand of similar length (designated miRNA*).15–17 The mature miRNA strand is generally biologically active, whereas the miRNA* strand is not. Consequently, the latter strand is degraded, although exceptions to this rule have been noted.18,19
Figure 1.
Therapeutic modulation of miRNA activity. miRNA mimics are molecules that can be used to overexpress miRNAs (1). They contain modified cyclopentylguanine bases, which enable association with the RISC within the cytoplasm to inhibit mRNA translation. Pri-miRNAs and pre-miRNAs can be expressed within cells by a plasmid, and the transcript is then processed by endogenous enzymes into mature miRNA (2). 2′-O-methyl antagomiRs and LNAs inhibit miRNA function by binding to mature miRNAs, thereby preventing their interaction with target mRNAs (3). Plasmids encoding an miRNA sponge transcript can be used for stable miRNA inhibition (4). The sponge transcript contains repeats of complementary sequences to the target miRNA (red regions of transcript), thereby sequestering miRNAs away from endogenous targets. Exosomes are BBB-crossing lipid vesicles that can deliver either miRNA mimics or inhibitors to modulate miRNA activity (5). Solid arrows indicate pathways that lead to increased miRNA activity; dashed arrows indicate pathways that lead to inhibition of miRNA activity. Abbreviations: BBB, blood–brain barrier; LNA, locked nucleic acid; miRNA, microRNA; pre-miRNA, precursor miRNA; pri-miRNA, primary miRNA; RISC, RNA-induced silencing complex; TRBP, TAR RNA-binding protein.
The RNA-induced silencing complex (RISC), which contains the Argonaute family of proteins that have a key role in mediation of RNA interference, facilitates targeting and binding of the miRNA to mRNA.20–22 The resultant effect of the miRNA–mRNA complex is decreased protein production, as translation and stabilization of the mRNA is inhibited.23 Quantitative proteomics studies have found that under physiological conditions, a single miRNA can downregulate expression of hundreds of proteins, but the magnitude of change is small (less than fourfold).24 Moreover, reduction of protein levels is predominately due to mRNA destabilization rather than inhibition of translation.25,26 These findings suggest that although a given miRNA may have broad effects, multiple miRNAs targeting genes of similar pathways are needed to cause functionally relevant changes in the cell.
miRNAs target mRNAs containing regions that are complementary to the miRNA ‘seed sequence’, a region encompassing nucleotides 2–7 at the 5′ end of the miRNA that has a critical role in miRNA–mRNA binding.27 The miRNA binding sites on mRNA are typically conserved between species, and are found in the 3′ untranslated region (UTR), although other binding locations have been described.10 A given miRNA can have multiple binding sites in the same mRNA, thereby enhancing its overall effect. Not all mRNAs are targeted by miRNA, however: it is estimated that only one-third of mRNAs contain miRNA binding sites.28 Some mRNA transcripts that lack miRNA binding sites were found to contain a short 3′ UTR and, therefore, lacked an miRNA binding domain. Such transcripts escape miRNA-mediated repression, and many of these ‘short’ mRNAs were found to regulate key cell functions such as DNA repair, protein synthesis and proliferation.29,30
With bioinformatics algorithms such as PicTar, miRanda and TargetScan, the mRNA targets of miRNAs can be predicted. As described above, one miRNA can bind to numerous different mRNAs, and a given mRNA can be bound by different miRNAs. Consequently, a given miRNA can modulate many members of a family of genes that are functionally related, thereby enhancing the overall effect of miRNA-mediated inhibition. Conversely, a single mRNA can be strongly repressed through concerted upregulation of the multiple miRNAs that target that mRNA. The various modes through which miRNAs can regulate gene networks highlight the potency of these molecules.
miRNAs in CNS development
Numerous sequencing studies in organisms ranging from Caenorhabditis elegans to humans have revealed that expression of miRNAs is ubiquitous.15,31–34 In mice, global depletion of miRNAs, which is achieved through deletion of the gene that encodes Dicer, leads to arrested development at embryonic day 7.5,35 highlighting the vital role of these molecules in general development. miRNAs are also indispensable for development of the CNS, as targeted ablation of the Dicer gene in nervous system tissue during development yields a smaller cortex,36,37 and deletion of Argonaute proteins (and, therefore, miRNA function) prevents neural tube closure.38
Numerous miRNAs are found in the CNS, and many are specific to a given lineage or cell type.32,39–43 For example, miRNA-9 (miR-9) is expressed in neurogenic regions of the brain where it regulates neural stem cell numbers and functions by binding the 3′ UTR of TLX, which encodes a nuclear receptor that maintains proliferation of neural stem cells while inhibiting their differentiation.44,45 Neuronal miRNAs include miR-124, expression of which increases on neuronal differentiation and reaches maximal levels in mature neurons.33,46 Targets of miR-124 include protein jagged-1 (Jag-1, also known as Notch-ligand receptor), SOX-9 (an adult neurogenesis transcription factor), and DLX2 (a transcription factor involved in neuronal subtype specification).46,47 Oligodendrocyte-specific miRNAs include miR-219, miR-338 and miR-17.48 miR-219 and miR-338 promote differentiation of oligodendrocyte precursor cells (OPCs) by repressing expression of proteins that promote OPC proliferation, such as platelet-derived growth factor receptor α, and the transcription factors SOX-6 and HES-5.49,50 By contrast, miR-17 promotes OPC proliferation by targeting inhibitors of the AKT pathway.51 Astrocytic miRNAs have also been identified and include miR-23, miR-26 and miR-29.52,53 Deletion of Dicer from the astrocytic lineage results in aberrant astrogliogenesis and astrocytic function;43,54,55 however, the specific mRNA targets of these miRNAs in astrocytes are less well-delineated than in other neural cell types.
miRNA modulation following CNS injury
CNS injury causes profound cellular changes that occur as a result of dysregulation of signalling pathways and structural proteins. As miRNAs have emerged as important regulators of molecular networks, insight into their modulation following tissue damage could provide a global perspective of the injury response. In the sections that follow, recent miRNA microarray studies of SCI, TBI and stroke are reviewed, and how our know ledge of injury-associated miRNAs can inform therapeutic strategies for these injuries is also discussed.
Spinal cord injury
SCI has an estimated worldwide annual incidence of 11–60 cases per million people,56 resulting in an estimated financial burden of more than US$1 million per patient over an average lifetime.57 miRNA microarray studies can inform strategies for development of more-effective treatments for SCI, as they provide a broad perspective of the complex molecular and cellular changes that occur after injury.
One microarray study of a contusion model of SCI in rats found that, compared with baseline, over 35% of the miRNAs expressed in the spinal cord were significantly affected within the first 7 days following injury.58 The affected miRNAs were differentially regulated, with some demonstrating a sustained increase in expression after injury and others a sustained decrease. According to the target prediction algorithm miRanda, upregulated miRNAs (such as miR-1 and miR-206) targeted anti-inflammatory and antioxidant mRNAs. Conversely, downregulated miRNAs, including miR-34a and miR-181a, were predicted to target proinflammatory mRNAs such as tumour necrosis factor and IL-1β. Furthermore, antiapoptotic and proapoptotic mRNAs were identified as targets of miRNAs that showed increased and decreased expression, respectively. Similar observations were made in another microarray study of a contusion SCI model in rats,59 which found 343 miRNAs to be modulated following injury, most being downregulated by day 7 post-injury compared with baseline. Interestingly, as the injury response progressed, the number of miRNAs that were downregulated gradually increased, whereas the number of upregulated miRNAs remained constant. These data, together with a separate analysis that assessed changes in mRNAs in similar model of SCI,60 suggest that a negative correlation exists between miRNA and mRNA expression patterns in SCI.
In another SCI model, the number of miRNAs noted to be downregulated compared with baseline was higher at day 14 than at day 4 post-injury.61 Moreover, this study found a direct correlation between the number of miRNAs affected and injury severity. In situ hybridization analyses have confirmed the heterogeneity of temporal expression of miRNAs following SCI, but also revealed spatial hetero geneity of miRNAs after injury. Decreased expression of miR-129-1/2 (a member of the miR-129 family of miRNAs, which prevent post-mitotic cells from re-entering the cell cycle62) was initially confined to perilesion white matter tracts up to day 4 post-injury, but had spread towards the rostrum by day 14.61 Expression of miR-1 (a regulator of transcription, differentiation and cell motility), however, was downregulated at the injury site at both day 4 and day 14 post-injury. Reduction of miR-1 and miR-129 levels could cause cells such as neurons to become aberrantly mitotic, and could explain the increase in apoptosis that is observed at the injury site after SCI.
As well as global miRNA changes, the roles of individual miRNAs in SCI have been elucidated through the use of animal models; some examples are described below.
miR-20a
A role for miR-20a in SCI was implicated following the observation that this miRNA was upregulated in a murine transection model of SCI.63 Infusion of miR-20a into the uninjured spinal cord induced inflammation and neuronal cell death similar to the pathology observed after trauma-induced SCI in patients. Inhibition of miR-20a activity in vivo by use of an miRNA inhibitor ameliorated the effects of SCI, improving hindlimb motor recovery and decreasing neuronal death. Mechanistically, miR-20a was found to target the 3′UTR of the mRNA encoding the basic helix–loop–helix protein neurogenin-1 (NGN-1, or Neurog1), a transcription factor that is involved in neuronal differentiation and specification.64 Importantly, NGN-1 infusion into the spinal cord increased expression of myelin and neuronal markers after transection-induced injury in mice, and markedly improved functional recovery.64 These findings validate the functional relevance of the interaction between miR-20a and NGN-1, but miR-20a has also been shown to target STAT3 (a key mediator in the SCI response),65,66 suggesting that miR-20a can affect the response to SCI via multiple pathways.
miR-486
miR-486 is another miRNA that has a profound role in regulating SCI pathology. Similar to miR-20a, miR-486 expression was increased at day 7 post-injury in a murine contusion model of SCI, and was detected in motor neurons by in situ hybridization.67 Infusion of miR-486 into the spinal cords of uninjured mice produced SCI-like effects, with decreased motor function and increased neuronal death.67 One of the validated targets of miR-486 is NeuroD6, a protein that is important for neuronal differentiation and oxidative stress response.68 As predicted, NeuroD6 levels were suppressed after SCI in the mouse model.67 Infusion of a duplexed small interfering RNA (siRNA) that targeted miR-486 caused an increase in NeuroD6 protein levels, which, in turn, increased expression of the reactive oxygen species (ROS) scaven ger proteins gluthianone peroxidase 3, selenoprotein N and thioredoxin. Such downstream effects caused a decrease in the magnitude of neuronal death and led to a significant improvement in motor recovery. These data suggest that miR-486 is an attractive therapeutic target for SCI, as inhibition of this miRNA leads to a reduction of ROS-mediated cell death, thereby increasing neuronal survival and functional recovery.
miR-21
Studies published in 2011 and 2012 revealed that miR-21 is involved in regulation of the response to SCI,61,69,70 but this findings was somewhat unexpected as miR-21 was first identified as an ‘oncomiR’ (an miRNA related to cancer pathogenesis) owing to its high expression in many cancers including glioblastoma multiforme.71–73 A role for miR-21 has now been identified in other pathologies such as hypertrophic cardiomyopathy.74 In a murine contusion model of SCI, miR-21 levels increased modestly around the lesion site within the first 2 weeks post-injury. In the chronic stages following injury, however, a greater upregulation of miR-21, particularly colocalized with astrocytes, was observed.70
The function of miR-21 in SCI was further elucidated using transgenic mice in which miR-21 was either overexpressed or inhibited specifically in astrocytes.70 Astrocytes normally respond to SCI by becoming hypertrophic and increasing their expression of intermediate filament protein75,76—changes that are thought to be beneficial by facilitating repair of the blood–brain barrier (BBB) and limiting infiltration of inflammatory cells into the CNS.77 Interestingly, the beneficial hypertrophic response to SCI was attenuated in transgenic mice that overexpressed miR-21, whereas inhibition of miR-21 augmented the astrocytic response, even in the chronic stages of SCI recovery when astrocytic hypertrophy has normally diminished.70 Inhibition of miR-21 in astrocytes also had a non-cell-autonomous effect, as the number of axons within the glial scar also increased.
miR-21 is regulated by signalling pathways, such as the bone morphogenetic protein (BMP) pathway, that are pivotal to both the detrimental and recovery responses following SCI.78 Signalling through BMP receptor type 1a (BMPR1a) reduced miR-21 expression in astrocytes, whereas signalling through BMPR1b had the opposite effect.53 Of note, the effects of BMPR1a knockout were similar to those observed in mice when miR-21 was overexpressed, whereas knockout of BMPR1b caused a response similar to that observed following miR-21 inhibition. Collectively, these results suggest that BMP-mediated effects in SCI occur via regulation of miR-21.
Exercise-induced miRNAs
Exercise following SCI has shown promise as a means to improve functional recovery,79 and research suggests that miRNAs can mediate this recovery process. For instance, in rats that were made to exercise after SCI, levels of the proapoptotic miRNA miR-15b were significantly decreased, whereas miR-21 (which targets and blocks the proapoptotic protein programmed cell death protein 4) was upregulated.80 miR-21 also targets PTEN, a negative regulator of the AKT–mTOR (mammalian target of rapamycin) pathway, which governs proliferation and growth. Conditional knockout of PTEN in corticospinal tract neurons enhanced regeneration of axons through the injured tissue.81 An mTOR-targeting miRNA, miR-199a-3p, was also found to be downregulated by exercise.69 These data suggest that the beneficial aspects of exercise after SCI are, at least in part, attributable to changes in miRNAs that have the net effect of promoting cellular plasticity while inhibiting neuronal apoptosis. Interestingly, the findings on miR-21 suggest that the effects of this miRNA on proliferation and growth vary with cell type, with an inhibitory role in astrocytes but a positive effect in neurons. To maximize the therapeutic potential of miRNAs, cell-specific modulation of these molecules may, therefore, be necessary.
Traumatic brain injury
TBI is a leading cause of mortality in the general population in the USA, accounting for over 50,000 deaths annually.1 Within the armed forces, however, post-deployment surveys indicate that up to 20% of soldiers who have returned from Iraq have experienced at least one TBI.82 Similar to their role in SCI, miRNAs may be important mediators of the profound molecular and cellular changes that occur after TBI in both the short and the long term. Microarray analyses in rat models of TBI have revealed dynamic temporal regulation of miRNA expression within the cortex, with the numbers of downregulated and upregulated miRNAs peaking at 24 h and 72 h post-injury, respectively.83
miRNA changes in the hippocampus
Structural changes within the hippocampus are among the most frequent sequelae of TBI; in one MRI study, hippocampal atrophy was observed in 59% of patients with TBI.84 Furthermore, hippocampal pathology is a major predictor of the cognitive and memory deficits in many individuals who have experienced a TBI.85 Microarray studies in animal models of TBI have revealed significant changes in miRNA expression within the hippocampus. Of the 444 verified rodent miRNAs, 35 were found to be upregulated and 50 downregulated after a TBI.86 According to gene-ontology bioinformatics analyses, the predicted targets of these miRNAs are involved in signal transduction, transcription, proliferation and differentiation.
In another study of TBI in rodent models, deep sequencing identified eight upregulated and 13 downregulated miRNAs at 24 h post-injury, whereas three were upregulated and 13 downregulated at day 7 post-injury.87 In this study, the miRNAs that showed changes in expression during the acute post-injury phase were predicted to target genes involved in apoptosis, protein folding, and aerobic respiration—processes that are associated with pathology and stress management in the cell. By contrast, miRNAs that were dysregulated in chronic stages of injury were predicted to regulate genes involved in cytoskeletal organization and intracellular trafficking—processes linked to brain repair mechanisms. As observed in other CNS injuries, miR-21 was upregulated in the TBI-affected hippocampus, particularly in the dentate gyrus and CA3 region.88 Expression of protein cell death protein 4, a target of miR-21, was decreased in these regions, identifying a potential miR-21-regulated survival mechanism in cells that are affected by TBI.
miRNAs as biomarkers
The role of miRNAs in TBI may not be purely mechanistic. When treating a patient with suspected TBI, timely diagnosis is difficult, as cognitive effects of the injury may not manifest for days or weeks, by which point the therapeutic window has narrowed. miRNA-based bio-markers of brain injury in the serum and CSF could serve as tools to quickly identify affected patients after suspected TBI. Studies of CSF from TBI-affected rats found a significant increase in levels of one miRNA, miR-let-7i, as early as 3 h post-injury.89 Prediction analysis revealed that miR-let-7i targets TBI-related proteins such as S100B and UCH-L1, suggesting a possible role for miR-let-7i in regulating TBI pathology.90–92 Studies in patients with TBI have identified three other miRNAs—miR-16, miR-92a, and miR-765—that can serve as diagnostic biomarkers for severe brain injury. Notably, levels of miR-16 and miR-92a are also increased in the plasma of patients with mild TBI.93 Collectively, these studies highlight miRNAs as promising diagnostic tools in TBI.
miRNA changes following treatment
One of the most promising treatments for TBI is hypothermia, a therapy that reduces prolonged neurological damage and improves functional outcomes.94–96 Interestingly, some miRNAs that show altered expression after TBI are also temperature-sensitive. In a rat fluid-percussion model of TBI, the levels of many miRNAs that were upregulated after TBI were reduced under hypothermic conditions.97 One such miRNA, miR-9, is predicted to reduce expression of proteins known to interact with actin-binding proteins and basal plasma membranes, suggesting that an increase in miR-9 levels after TBI would perturb cytoskeleton and cell-adhesion properties, thereby increasing cell death. Reduction in the levels of miR-9, therefore, could contribute to the enhanced cell survival that is observed with hypothermia treatment. Further experiments in animal models of TBI are needed to determine whether modulation of miR-9, either alone or in combination with other miRNAs, can lead to improvements following TBI.
Stroke
One in six people worldwide will have at least one stroke in their lifetime.98 Stroke can result in extensive tissue damage as well as long-term cognitive and motor deficits, which are potentiated by cerebral oedema and neuronal death among other phenomena. Identification of the miRNAs that are involved in regulation of stroke-related cellular and molecular networks could provide insight into new therapeutic avenues. As with TBI, dysregulation of miRNAs following stroke can be assessed in peripheral blood samples. One analysis of whole blood from patients who had experienced a stroke revealed that of the 836 miRNAs tested on the array chip, 157 were differentially regulated: 138 upregulated and 19 down regulated.99 Ontological analyses predicted that the targets of the dysregulated miRNAs were involved in angiogenesis, hypoxia, endothelial cell regulation, and the immune response. Surprisingly, the miRNA profile of large-artery strokes was distinct from that of small-artery strokes, suggesting that the vessel affected by stroke can be identified through analysis of whole-blood miRNA levels.99
miRNA-based differentiation of stroke subtypes
miRNA expression profiles can be used to differentiate between ischaemic and haemorrhagic stroke.100 Analysis of rat blood at day 1 post-injury revealed that 10 and 20 miRNAs were upregulated in ischaemic and haemorrhagic stroke models, respectively. Of these miRNAs, only eight were common to both models. 65 and 21 miRNAs were downregulated in ischaemic and haemorrhagic strokes, respectively, with 14 miRNAs being shared between the stroke subtypes. Analysis of miRNA levels in brain tissue from the animals led to a similar conclusion: miRNA changes in ischaemic or haemorrhagic stroke are largely distinct, but also show some overlap. These studies suggest that a similar set of cellular functions, such as cell cycle and cell death, are affected in both types of stroke; however, as distinct subsets of miRNAs are dysregulated in each stroke subtype, some pathways could be differentially affected depending on stroke aetiology. Further investigation is needed to discern which pathways are unique to each stroke subtype and how they can be exploited to maximize detection and treatment of stroke.
Modulation of cerebral oedema
Cerebral oedema is a common consequence of stroke that contributes to tissue and cellular damage following ischaemia. Levels of aquaporin proteins, which function as cellular water conduits, are known to increase after cerebral ischaemia,101 although their exact role in this process is unclear. Some reports based on rodent models suggest that reduction of aquaporin-4 levels after stroke leads to a reduction in oedema,102 whereas others suggest that upregulation of aquaporin-4 is required to facilitate oedema clearance.103 In a rodent middle cerebral artery occlusion (MCAO) model, levels of miRNAs that are predicted to target aquaporins (such as miR-30a-3p and miR-383) were decreased and aquaporin-4 levels were increased after ischaemia.104 In addition, miR-320a, which is also downregulated in MCAO, has validated binding sites in the 3′ UTR of the aquaporin-1 and aquaporin-4 mRNAs.105 Collectively, these studies suggest that modulation of miRNAs can be used to regulate aquaporins and, therefore, affect oedema following cerebral ischaemia.
miRNAs and neuronal cell death
Neuronal cell death is another prominent feature observed after stroke. In accordance with this phenomenon, levels of miRNAs that regulate apoptosis are altered following cerebral ischaemia. For example, miR-497, which targets the antiapoptotic genes Bcl-2 and Bcl-w, was upregulated in infarcted regions in a mouse model of MCAO.106 Importantly, inhibition of miR-497 in mice reduced infarct size while enhancing neurological function. Bcl-W is an identified target of miR-29b, an miRNA that was found to be upregulated in neurons after MCAO.107 miR-21 is also thought to increase neuronal survival after stroke by targeting and blocking translation of the gene that encodes Fas ligand (a receptor that signals to induce apoptosis).108 Such a mechanism could explain preservation of neurons within the ischaemic boundary zone—a region in which miR-21 levels are increased following ischaemia.
ROS are also a key trigger of neuronal apoptosis. Scavengers of ROS, such as superoxide dismutase 2 (SOD2), help to protect neurons from such stress.109 Interestingly, SOD2 is targeted by miR-145, another miRNA that is upregulated after MCAO. In a mouse model of MCAO, infusion of an miR-145 antagonist (antagomiR-145) caused an increase in neuronal SOD2 expression and a reduction in infarct size.110 This study suggests the existence of a class of miRNAs that can be modulated to improve neuronal survival after ischaemic events.
miRNAs in ischaemic preconditioning
A theoretical strategy for neuroprotection in stroke is ischaemic preconditioning (IPC), which involves brief exposure to nonlethal ischaemia to induce protective mechanisms in the tissue during phases of prolonged ischaemia.111 Microarray studies in animal models have shown that levels of numerous miRNAs are rapidly altered following IPC. Such alterations include upregulation of miRNAs that target members of the mitogen-activated protein kinase (MAPK) and mTOR signalling pathways.112 Conversely, miRNAs that were down regulated seemed to target signalling via Wnt. miRNA-based modulation of these three pathways (MAPK, mTOR and Wnt) can increase cytoprotection and cellular regeneration in the affected tissue. The miR-200 family was also upregulated within 3 h of IPC, with the neuroprotective effect driven by downregulation of prolyl-hydroxylase 2, a negative regulator of the pro-survival protein hypoxiainducible factor-1α.113 Another study identified methyl CpG binding protein 2 (MeCP2)—a global transcriptional regulator and putative mediator of the beneficial effects of IPC—as a target of many of the downregulated miRNAs.114 This observation provided an explanation for the increase in MeCP2 protein expression that was observed after IPC despite the fact that the mRNA levels of MeCP2 remained unchanged.115 These studies serve as guidance for the translation of neuroprotective mechanisms of IPC from bench to bedside.
Common themes of dysregulated miRNAs
Many of the studies reviewed here rely on miRNA microarray data to develop an understanding of the CNS injury response. Although this method of analysis provides a global view of the changes that occur during injury, microarrays have restrictions, such as a limited miRNA expression profile of the microarray chip and need for validation of affected miRNAs. Importantly, many of the predicted mRNA targets of identified miRNAs remain undefined and require further investigation. Analysis of the collective results from microarray studies can, however, provide insights into the characteristics of the CNS injury response. A meta-analysis of micro array studies unmasks the miRNA response kinetics after injury, revealing that many miRNAs are altered as early as day 1 post-injury (Table 1). This observation is consistent with reports from studies that used in vitro models of CNS injuries or synthetic inducible miRNA reporter systems.116,117 The finding of early post-injury modulation of miRNAs suggests that molecules involved in regulation of miRNA expression are at least partially present before injury, and that they do not wholly rely on protein production following activation of signalling components. Furthermore, spatial expression profiles indicate that the affected miRNAs lie within and adjacent to the injury site, suggesting that miRNA modulation involves both autocrine and paracrine signalling. However, as changes in blood miRNAs were also detected, systemic signalling is also likely to be involved in the modulation of miRNA levels following injury.
Table 1.
Summary of miRNA microarray studies in CNS injury
| Study | Tissue analysed | Time points analysed | Pathways predicted to be modulated | Number of miRNAs increased at last time point | Number of miRNAs decreased at last time point |
|---|---|---|---|---|---|
| Spinal cord injury | |||||
| Liu et al. (2009)58 | Rat spinal cord | 4 HPI, 1 DPI, 7 DPI | Apopotosis Inflammation Antioxidation |
30 | 16 |
| Strickland et al. (2011)61 | Rat spinal cord | 4 DPI, 14 DPI | Apopotosis Inflammation Cell proliferation Cell differentiation Cell cycle |
3 | 32 |
| Yunta et al. (2012)59 | Rat spinal cord | 1 DPI, 3 DPI, 7 DPI | Apopotosis Inflammation Cellular regeneration | 11 | 192 |
| Traumatic brain injury | |||||
| Lei et al. (2009)83 | Rat cortex | 6 HPI, 24 HPI, 48 HPI, 72 HPI | Not performed | 19 | 5 |
| Redell et al. (2009)86 | Rat hippocampus | 3 HPI, 24 HPI | Cell proliferation Cell differentiation Cell cycle Protein modification |
24 | 28 |
| Hu et al. (2012)87 | Rat hippocampus | 24 HPI, 7 DPI | Apoptosis Transcription Cytoskeleton arrangement Cell adhesion |
3 | 13 |
| Balakathiresan et al. (2012)89 | Rat serum and cerebrospinal fluid | 3 HPI, 24 HPI | Axon guidance Cytoskeleton arrangement Cell adhesion Intracellular signalling |
35 | 4 |
| Redell et al. (2012)93 | Human blood plasma | 3 DPI | Apoptosis Cell proliferation Cell cycle Angiogenesis |
27 | 33 |
| Stroke | |||||
| Jeyaseelan et al. (2008)104 | Rat brain and blood | 24 HPI, 48 HPI | Aquaporins Cellular regeneration Extracellular matrix remodelling |
32 (brain) 10 (blood) |
49 (brain) 15 (blood) |
| Dharap et al. (2009)110 | Rat cortex | 3 HPI, 6 HPI, 12 HPI, 1 DPI, 3 DPI | Antioxidization Inflammation Ion channels Neurotransmitter receptors Transcription factors |
24 | 23 |
| Tan et al. (2009)99 | Human peripheral blood | 6–18 months post-injury | Angiogenesis Cell proliferation Hypoxia Inflammation Intracellular signalling |
176 | 23 |
Abbreviations: DPI, days post-injury; HPI, hours post-injury; miRNA, microRNA.
Analysis of predicted targets of miRNAs that are modu lated following SCI, TBI and stroke indicate that some of the targeted pathways—such as apoptosis, inflammation, cell proliferation and cell differentiation—are shared between these injures (Table 1). However, close inspection reveals that a given cellular function is often regulated by a different set of miRNAs in each injury. Regulation of neuronal differentiation, for example, is affected in the various CNS injuries, but different miRNAs and gene targets have been implicated in each. As detailed above, miR-20a is upregulated after SCI and targets the proneural gene Ngn1.63 In stroke, however, miR-124, which promotes neuronal differentiation of neural progenitor cells by targeting Jag1 mRNA, is downregulated.47 Apoptosis is regulated by miR-15b and miR-486 after SCI,67,80 but is regulated by miR-29b, miR-145 and miR-497 following stroke.106,107,110 Despite such injury-specific miRNAs, certain miRNAs are modulated in all injury types: miR-21 is the most frequently reported of such miRNAs, and may serve as a common cell-death regulator in SCI, TBI and stroke.
The findings of miRNA studies build on our knowledge gained from mRNA-based microarray analysis of CNS injuries. A common theme that has arisen from both mRNA and miRNA studies is the complexity of pathway regulation, as well as recognition of miRNAs as a means to regulate similar pathways through different gene targets. Therapeutic strategies can be designed to target miRNAs that affect common pathways, thereby enabling modulation of mRNA levels to increase cell survival, reduce inflammation, and promote cell proliferation and differentiation in the various injuries. Moreover, as unique sets of miRNAs are altered in each injury type, miRNAs can potentially serve as diagnostic biomarkers for the different types and subtypes of CNS injuries.
miRNA-based therapies
As described above, miRNA levels are altered after CNS injuries, and such changes have profound effects on the downstream targets of these molecules. Restoration or inhibition of miRNA activity, therefore, represents a therapeutic opportunity to modulate cellular process and improve outcomes in patients with CNS in juries. The current state of miRNA-based therapies—including miRNA amplifiers and inhibitors, and artificial miRNAs—are discussed below.
miRNA amplifiers
Few methods that aim to exogenously increase levels of miRNAs after CNS injury have been developed. One such method involves miRNA mimics: modified double-stranded RNA molecules with one strand containing the same sequence as the mature miRNA. Notably, naturally occurring RNA molecules contain uridine bases and uridine–guanine nucleotide pairs that induce a nonspecific immune response by activating Toll-like receptors.118 As the miRNA mimic sequence cannot be changed without its efficacy or specificity being compromised, chemical modifications of these nucleotide bases are neces sary to minimize the immune reaction. For example, a 2′ ribose modification to replace guanine with cyclopentyl-guanine in an miRNA mimic can markedly reduce immune activation while maintaining miRNA activity (Figure 1).119 Modifications to other regions of the molecule, such as the 3′ overhang, have also been shown to increase miRNA-mimic stability by reducing the extent of nuclease degradation of the mimic.120In vivo studies of miRNA mimics have been performed in other fields such as oncology.121 Studies of miRNA mimics in the context of patients with stroke, TBI or SCI are lacking, but these molecules could also be an effective therapy for CNS injuries if they can be deployed in an appropriate manner.
Another strategy for increasing miRNA levels in vivo is to introduce the pri-miRNA or pre-miRNA sequence into the target tissue with the use of a DNA-based plasmid (Figure 1). Successful application of this technology has been demonstrated in a stroke model in which a plasmid encoding pri-miR-181 was introduced into the brain using intraventricular infusion.122 This process led to production of functional miR-181 molecules that success fully targeted heat shock protein 70 family members, and to a reduction in infarct size. One benefit of plasmid-based expression of miRNAs is that the amount of mature miRNA can be amplified by increasing transcription of the plasmid. This method, however, relies on the action of endogenous enzymes such as Drosha and Dicer, the levels and function of which may be reduced in the damaged tissue.123
miRNA inhibitors
Owing to concerted research efforts to block the function of oncomiRs, marked progress has been made in the development of miRNA inhibitors: antisense sequences that bind mature miRNA molecules and block their function.
AntagomiRs
AntagomiRs are first-generation miRNA inhibitors that contain 2′-O-methyl-modified nucleotides, terminal phosphorothioates and 3′-end-conjugated cholesterol groups—features that increase in vivo stability and bio-availability of the inhibitor (Figure 1).124 AntagomiRs have been effectively delivered to many tissues, including the CNS,125 but as high doses of these inhibitors are likely to be needed for in vivo efficacy, more-potent strategies may be required.126
Locked nucleic acids
Locked nucleic acids (LNAs) offer one strategy for high-potency miRNA inhibition (Figure 1). LNAs are modified oligonucleotides that contain locked ribose rings. The modifications to these molecules are designed to minimize endonuclease degradation while markedly enhancing their capacity to bind to target miRNA.127,128 In mice and nonhuman primates, LNA molecules have demonstrated low toxicity and enhanced in vivo function compared with antagomiRs.129,130 In chimpanzees that had been infected with hepatitis C virus, intravenous administration of an LNA that targeted miR-122 (an miRNA that is essential for replication of hepatitis C virus) reduced not only the levels of this miRNA in the liver but also the number of circulating hepatitis C viral particles.131 Following such promising results, a phase II clinical trial of the miR-122-targeting LNA was initiated, and has recently been completed, in patients with chronic hepatitis C.132
miRNA sponges
Although antagomiRs and LNAs are tools that enable knockdown of miRNA functions, their effects are likely to be transient, as the half-life for these molecules in serum is just a few days.133,134 To overcome this problem and obtain persistent inhibition, miRNA-sponge constructs have been developed. An miRNA sponge is an artificial RNA transcript containing multiple repeats that are complementary to the target miRNA (Figure 1).135 Consequently, the sponge serves as a competitive inhibitor, sequestering the miRNA from its endogenous targets. Retroviruses and lentiviruses have been used to integrate plasmid DNA encoding the sponge transcript into the brains of mice, and this approach led to stable in vivo inhibition of the target miRNA.136,137 Sponge transcripts are promising tools for treatment of CNS injuries, as inhibition of the miRNAs that are upregulated during injury is likely to improve patient recovery.
Delivery mechanisms
Development of miRNA mimics and inhibitors has a promising future, but targeted delivery of these molecules, in particular into the CNS, remains a major ob stacle. Direct administration of miRNA therapeutics, for example into the ventricles or parenchyma, is one potential delivery method, but the resulting tissue damage and immune-system activation present some drawbacks. Delivery methods that afford passage across the BBB have, therefore, been explored. One such method involves exosomes—endogenous lipid-membrane ve sicles that are secreted by cells and can serve as carriers for RNA molecules.138,139 On their entry into the systemic circulation, exosomes can cross the BBB and deliver siRNA molecules for gene silencing.140 Although the exosome delivery method has not yet been applied to miRNAs, its use for CNS-targeted miRNA-based therapies is likely to be a natural extension (Figure 1).
Artificial miRNAs
An emerging idea in the field of RNA interference therapy is the use of miRNA-mediated silencing to control gene expression in an artificial manner. After injury to the CNS, many deleterious mRNAs—such as those encoding proteins involved in apoptosis and inflammation—are upregulated.141,142 Artificial miRNAs that target groups of mRNAs involved in such deleterious processes could be a useful therapeutic strategy. Indeed, with the aid of computer algorithms, artificial miRNAs have been developed that can be used to identify miRNA seed sequences in genes of interest.143,144 By mining the vast amount of available mRNA microarray data for transcriptome changes seen after CNS injuries, seed sequences that are common to different mRNAs can be identified and exploited to generate artificial miRNAs. Such an approach could enable precise gene expression control with minimal off-target mRNA binding, as artificial miRNA sequences can be adjusted to bind only to the intended transcript.
Conclusions and future directions
The fact that the cellular and molecular changes that occur after CNS injuries are diverse provides a substantial challenge for single-target therapies. Identification of the miRNAs that are altered following CNS injury provides a means to elucidate global patterns within these changes. An analysis of recent miRNA microarray studies indicates that pathways such as apoptosis, inflammation and cell proliferation are among the most commonly affected path ways in CNS injuries, and that changes in these pathways are conserved across CNS injuries from SCI to stroke. Consequently, therapies aimed at modifying these pathways could be broadly applicable to a variety of CNS injuries, but multifaceted therapies that target many components in multiple pathways are also likely to be required. Exploitation of miRNA-based therapeutics is an appealing means of accomplishing this goal.
Promising advances in synthetic miRNA amplifiers and inhibitors have been made with regard to improved bioavailability, function, and adverse-effect profiles. The success of studies in which known miRNAs were targeted, such as the clinical trial of miR-122 in hepatitis C,132 provide inspiration for development of novel miRNA-based therapies. Furthermore, the power of bioinformatics and target-prediction algorithms can be harnessed for the design of artificial miRNAs that target a diverse set of mRNA transcripts involved in the common pathways of CNS injuries. Full realization of the potential of miRNAs, however, will require validation of findings from animal models and in human tissue; only then can microarray data and rapid miRNA pharmaceutical development be combined to help improve outcomes after CNS injuries.
Key points.
■ MicroRNAs (miRNAs) are critical for normal development but also have a role in disease, particularly of the CNS
■ miRNA microarrays provide the highest-order view of changes that occur following injuries to the CNS, such as stroke, spinal cord injury or traumatic brain injury
■ These CNS injuries affect key cellular pathways such as apoptosis, inflammation and cellular proliferation, all of which are regulated by miRNAs
■ miRNA-based therapies that augment or inhibit miRNA activity, and with enhanced bioavailability and function, are currently being developed
■ Artificial miRNAs can be designed to bind to and modify groups of mRNA transcripts that are not naturally targeted by endogenous miRNAs
Review criteria.
PubMed was searched for articles, including advance online publications published up to 26th January 2013, using the following search terms: “microRNA”, “CNS injury”, “spinal cord injury”, “traumatic brain injury”, “stroke”, “ischaemia”, “microRNA mimics”, “antagomiRs”, “locked nucleic acids”, “microRNA sponge”, and “artificial microRNA”. Full versions of review and original articles were assessed for appropriateness. References were also examined to identify additional articles not captured using the search terms listed. The database at www.ClinicalTrials.gov was searched for miRNA-based therapies.
Acknowledgements
O. G. Bhalala and J. A. Kessler are supported by NIH Grants R01NS20013 and R01NS20778. M. Srikanth is supported by NIH Grant F30NS065590.
Footnotes
Author contributions
O. G. Bhalala researched data for the article. All authors provided substantial contributions to discussion of content, writing the article, and to the review and/or editing of the manuscript before submission.
Competing interests
The authors declare no competing interests.
References
- 1.Coronado VG, et al. Surveillance for traumatic brain injury-related deaths—United States, 1997–2007. MMWR Surveill. Summ. 2011;60:1–32. [PubMed] [Google Scholar]
- 2.Paralysis facts & figures. Christopher & Dana Reeve Foundation Paralysis Resource Center. 2012 [online], http://www.christopherreeve.org/site/c.mtKZKgMWKwG/b.5184189/k.5587/Paralysis_Facts__Figures.htm.
- 3.Stroke 101 fact sheet. National Stroke Association. 2012 [online] http://www.stroke.org/site/DocServer/STROKE101_2009.pdf?docID=4541.
- 4.Munro KM, Perreau VM. Current and future applications of transcriptomics for discovery in CNS disease and injury. Neurosignals. 2009;17:311–327. doi: 10.1159/000231897. [DOI] [PubMed] [Google Scholar]
- 5.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012;11:189–200. doi: 10.1016/S1474-4422(11)70286-1. [DOI] [PubMed] [Google Scholar]
- 7.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bian S, Sun T. Functions of noncoding RNAs in neural development and neurological diseases. Mol. Neurobiol. 2011;44:359–373. doi: 10.1007/s12035-011-8211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meza-Sosa KF, Valle-García D, Pedraza-Alva G, Pérez-Martínez L. Role of microRNAs in central nervous system development and pathology. J. Neurosci. Res. 2012;90:1–12. doi: 10.1002/jnr.22701. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basyuk E, Suavet F, Doglio A, Bordonné R, Bertrand E. Human let-7 stem-loop precursors harbor features of RNase III cleavage products. Nucleic Acids Res. 2003;31:6593–6597. doi: 10.1093/nar/gkg855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han J, et al. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 16.Hutvágner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 17.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long JM, Lahiri DK. Advances in microRNA experimental approaches to study physiological regulation of gene products implicated in CNS disorders. Exp. Neurol. 2012;235:402–418. doi: 10.1016/j.expneurol.2011.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nass D, et al. miR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 2009;19:375–383. doi: 10.1111/j.1750-3639.2008.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 21.Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 22.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 24.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 25.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 28.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 29.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal microRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 30.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 32.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 34.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 35.Bernstein E, et al. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 36.De Pietri Tonelli D, et al. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis TH, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 39.Bak M, et al. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miska EA, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith B, et al. Large-scale expression analysis reveals distinct microRNA profiles at different stages of human neurodevelopment. PLoS ONE. 2010;5:e11109. doi: 10.1371/journal.pone.0011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng K, Li H, Zhu Y, Zhu Q, Qiu M. MicroRNAs are essential for the developmental switch from neurogenesis to gliogenesis in the developing spinal cord. J. Neurosci. 2010;30:8245–8250. doi: 10.1523/JNEUROSCI.1169-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Wang F, Lee JA, Gao FB. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu XS, et al. MicroRNA profiling in subventricular zone after stroke: miR-124a regulates proliferation of neural progenitor cells through Notch signaling pathway. PLoS ONE. 2011;6:e23461. doi: 10.1371/journal.pone.0023461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He X, Yu Y, Awatramani R, Lu QR. Unwrapping myelination by microRNAs. Neuroscientist. 2012;18:45–55. doi: 10.1177/1073858410392382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dugas JC, et al. Dicer1 and miR-219 are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X, et al. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–626. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Budde H, et al. Control of oligodendroglial cell number by the miR-17-92 cluster. Development. 2010;137:2127–2132. doi: 10.1242/dev.050633. [DOI] [PubMed] [Google Scholar]
- 52.Smirnova L, et al. Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 53.Sahni V, et al. BMPR1a and BMPR1b signaling exert opposing effects on gliosis after spinal cord injury. J. Neurosci. 2010;30:1839–1855. doi: 10.1523/JNEUROSCI.4459-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng K, Li H, Huang H, Qiu M. MicroRNAs and glial cell development. Neuroscientist. 2012;18:114–118. doi: 10.1177/1073858411398322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tao J, et al. Deletion of astroglial Dicer causes non-cell-autonomous neuronal dysfunction and degeneration. J. Neurosci. 2011;31:8306–8319. doi: 10.1523/JNEUROSCI.0567-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ackery A, Tator C, Krassioukov A. A global perspective on spinal cord injury epidemiology. J. Neurotrauma. 2004;21:1355–1370. doi: 10.1089/neu.2004.21.1355. [DOI] [PubMed] [Google Scholar]
- 57.Spinal cord injury facts and figures at a glance. National Spinal Cord Injury Statistical Center. 2012 [online], https://www.nscisc.uab.edu/PublicDocuments/fact_figures_docs/Facts%202012%20Feb%20Final.pdf.
- 58.Liu NK, Wang XF, Lu QB, Xu XM. Altered microRNA expression following traumatic spinal cord injury. Exp. Neurol. 2009;219:424–429. doi: 10.1016/j.expneurol.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yunta M, et al. MicroRNA dysregulation in the spinal cord following traumatic injury. PLoS ONE. 2012;7:e34534. doi: 10.1371/journal.pone.0034534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Biase A, et al. Gene expression profiling of experimental traumatic spinal cord injury as a function of distance from impact site and injury severity. Physiol. Genomics. 2005;22:368–381. doi: 10.1152/physiolgenomics.00081.2005. [DOI] [PubMed] [Google Scholar]
- 61.Strickland ER, et al. MicroRNA dysregulation following spinal cord contusion: implications for neural plasticity and repair. Neuroscience. 2011;186:146–160. doi: 10.1016/j.neuroscience.2011.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu J, et al. miR-129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle. 2010;9:1809–1818. doi: 10.4161/cc.9.9.11535. [DOI] [PubMed] [Google Scholar]
- 63.Jee MK, Jung JS, Im YB, Jung SJ, Kang SK. Silencing of miR20a is crucial for Ngn1-mediated neuroprotection in injured spinal cord. Hum. Gene Ther. 2012;23:508–520. doi: 10.1089/hum.2011.121. [DOI] [PubMed] [Google Scholar]
- 64.Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 65.Carraro G, et al. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev. Biol. 2009;333:238–250. doi: 10.1016/j.ydbio.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan H, et al. MicroRNA-20a overexpression inhibited proliferation and metastasis of pancreatic carcinoma cells. Hum. Gene Ther. 2010;21:1723–1734. doi: 10.1089/hum.2010.061. [DOI] [PubMed] [Google Scholar]
- 67.Jee MK, et al. MicroRNA 486 is a potentially novel target for the treatment of spinal cord injury. Brain. 2012;135:1237–1252. doi: 10.1093/brain/aws047. [DOI] [PubMed] [Google Scholar]
- 68.Uittenbogaard M, Baxter KK, Chiaramello A. The neurogenic basic helix-loop-helix transcription factor NeuroD6 confers tolerance to oxidative stress by triggering an antioxidant response and sustaining the mitochondrial biomass. ASN Neuro. 2010;2:e00034. doi: 10.1042/AN20100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu G, Detloff MR, Miller KN, Santi L, Houlé JD. Exercise modulates microRNAs that affect the PTEN/mTOR pathway in rats after spinal cord injury. Exp. Neurol. 2012;233:447–456. doi: 10.1016/j.expneurol.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhalala OG, et al. microRNA-21 regulates astrocytic response following spinal cord injury. J. Neurosci. 2012;32:17935–17947. doi: 10.1523/JNEUROSCI.3860-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 72.Ciafrè SA, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 73.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Rooij E, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl Acad. Sci. USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fawcett JW, Asher RA. The glial scar and central nervous system repair. Brain Res. Bull. 1999;49:377–391. doi: 10.1016/s0361-9230(99)00072-6. [DOI] [PubMed] [Google Scholar]
- 76.Barnabé-Heider F, Frisén J. Stem cells for spinal cord repair. Cell Stem Cell. 2008;3:16–24. doi: 10.1016/j.stem.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 77.Sofroniew MV. Reactive astrocytes in neural repair and protection. Neuroscientist. 2005;11:400–407. doi: 10.1177/1073858405278321. [DOI] [PubMed] [Google Scholar]
- 78.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van den Brand R, et al. Restoring voluntary control of locomotion after paralyzing spinal cord injury. Science. 2012;336:1182–1185. doi: 10.1126/science.1217416. [DOI] [PubMed] [Google Scholar]
- 80.Liu G, Keeler BE, Zhukareva V, Houlé JD. Cycling exercise affects the expression of apoptosis-associated microRNAs after spinal cord injury in rats. Exp. Neurol. 2010;226:200–206. doi: 10.1016/j.expneurol.2010.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu K, et al. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat. Neurosci. 2010;13:1075–1081. doi: 10.1038/nn.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shively SB, Perl DP. Traumatic brain injury, shell shock, and posttraumatic stress disorder in the military—past, present, and future. J. Head Trauma Rehabil. 2012;27:234–239. doi: 10.1097/HTR.0b013e318250e9dd. [DOI] [PubMed] [Google Scholar]
- 83.Lei P, Li Y, Chen X, Yang S, Zhang J. Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Res. 2009;1284:191–201. doi: 10.1016/j.brainres.2009.05.074. [DOI] [PubMed] [Google Scholar]
- 84.Orrison WW, et al. Traumatic brain injury: a review and high-field MRI findings in 100 unarmed combatants using a literature-based checklist approach. J. Neurotrauma. 2009;26:689–701. doi: 10.1089/neu.2008.0636. [DOI] [PubMed] [Google Scholar]
- 85.Cohen AS, et al. Injury-induced alterations in CNS electrophysiology. Prog. Brain Res. 2007;161:143–169. doi: 10.1016/S0079-6123(06)61010-8. [DOI] [PubMed] [Google Scholar]
- 86.Redell JB, Liu Y, Dash PK. Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J. Neurosci. Res. 2009;87:1435–1448. doi: 10.1002/jnr.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hu Z, et al. Expression of miRNAs and their cooperative regulation of the pathophysiology in traumatic brain injury. PLoS ONE. 2012;7:e39357. doi: 10.1371/journal.pone.0039357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Redell JB, Zhao J, Dash PK. Altered expression of miRNA-21 and its targets in the hippocampus after traumatic brain injury. J. Neurosci. Res. 2011;89:212–221. doi: 10.1002/jnr.22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Balakathiresan N, et al. MicroRNA let-7i is a promising serum biomarker for blast-induced traumatic brain injury. J. Neurotrauma. 2012;29:1379–1387. doi: 10.1089/neu.2011.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Geyer C, Ulrich A, Gräfe G, Stach B, Till H. Diagnostic value of S100B and neuron-specific enolase in mild pediatric traumatic brain injury. J. Neurosurg. Pediatr. 2009;4:339–344. doi: 10.3171/2009.5.PEDS08481. [DOI] [PubMed] [Google Scholar]
- 91.Papa L, et al. Ubiquitin C-terminal hydrolase is a novel biomarker in humans for severe traumatic brain injury. Crit. Care Med. 2010;38:138–144. doi: 10.1097/CCM.0b013e3181b788ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Svetlov SI, et al. Morphologic and biochemical characterization of brain injury in a model of controlled blast overpressure exposure. J. Trauma. 2010;69:795–804. doi: 10.1097/TA.0b013e3181bbd885. [DOI] [PubMed] [Google Scholar]
- 93.Redell JB, Moore AN, Ward NH, 3rd, Hergenroeder GW, Dash PK. Human traumatic brain injury alters plasma microRNA levels. J. Neurotrauma. 2010;27:2147–2156. doi: 10.1089/neu.2010.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marion D, Bullock MR. Current and future role of therapeutic hypothermia. J. Neurotrauma. 2009;26:455–467. doi: 10.1089/neu.2008.0582. [DOI] [PubMed] [Google Scholar]
- 95.Dietrich WD, Atkins CM, Bramlett HM. Protection in animal models of brain and spinal cord injury with mild to moderate hypothermia. J. Neurotrauma. 2009;26:301–312. doi: 10.1089/neu.2008.0806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Choi HA, Badjatia N, Mayer SA. Hypothermia for acute brain injury—mechanisms and practical aspects. Nat. Rev. Neurol. 2012;8:214–222. doi: 10.1038/nrneurol.2012.21. [DOI] [PubMed] [Google Scholar]
- 97.Truettner JS, Alonso OF, Bramlett HM, Dietrich WD. Therapeutic hypothermia alters microRNA responses to traumatic brain injury in rats. J. Cereb. Blood Flow Metab. 2011;31:1897–1907. doi: 10.1038/jcbfm.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bushnell CD. Stroke and the female brain. Nat. Clin. Pract. Neurol. 2008;4:22–33. doi: 10.1038/ncpneuro0686. [DOI] [PubMed] [Google Scholar]
- 99.Tan KS, et al. Expression profile of MicroRNAs in young stroke patients. PLoS ONE. 2009;4:e7689. doi: 10.1371/journal.pone.0007689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu DZ, et al. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J. Cereb. Blood Flow Metab. 2010;30:92–101. doi: 10.1038/jcbfm.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Badaut J, Lasbennes F, Magistretti PJ, Regli L. Aquaporins in brain: distribution, physiology, and pathophysiology. J. Cereb. Blood Flow Metab. 2002;22:367–378. doi: 10.1097/00004647-200204000-00001. [DOI] [PubMed] [Google Scholar]
- 102.Manley GT, et al. Aquaporin-4 deletion in mice reduces brain edema after acute water intoxication and ischemic stroke. Nat. Med. 2000;6:159–163. doi: 10.1038/72256. [DOI] [PubMed] [Google Scholar]
- 103.Papadopoulos MC, Manley GT, Krishna S, Verkman AS. Aquaporin-4 facilitates reabsorption of excess fluid in vasogenic brain edema. FASEB J. 2004;18:1291–1293. doi: 10.1096/fj.04-1723fje. [DOI] [PubMed] [Google Scholar]
- 104.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 105.Sepramaniam S, et al. MicroRNA 320a functions as a novel endogenous modulator of aquaporins 1 and 4 as well as a potential therapeutic target in cerebral ischemia. J. Biol. Chem. 2010;285:29223–29230. doi: 10.1074/jbc.M110.144576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yin KJ, et al. miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol. Dis. 2010;38:17–26. doi: 10.1016/j.nbd.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shi G, et al. Upregulated miR-29b promotes neuronal cell death by inhibiting Bcl2L2 after ischemic brain injury. Exp. Brain Res. 2012;216:225–230. doi: 10.1007/s00221-011-2925-3. [DOI] [PubMed] [Google Scholar]
- 108.Buller B, et al. MicroRNA-21 protects neurons from ischemic death. FEBS J. 2010;277:4299–4307. doi: 10.1111/j.1742-4658.2010.07818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raha S, Robinson BH. Mitochondria, oxygen free radicals, and apoptosis. Am. J. Med. Genet. 2001;106:62–70. doi: 10.1002/ajmg.1398. [DOI] [PubMed] [Google Scholar]
- 110.Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J. Cereb. Blood Flow Metab. 2009;29:675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eltzschig HK, Eckle T. Ischemia and reperfusion—from mechanism to translation. Nat. Med. 2011;17:1391–1401. doi: 10.1038/nm.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dharap A, Vemuganti R. Ischemic preconditioning alters cerebral microRNAs that are upstream to neuroprotective signaling pathways. J. Neurochem. 2010;113:1685–1691. doi: 10.1111/j.1471-4159.2010.06735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee ST, et al. MicroRNAs induced during ischemic preconditioning. Stroke. 2010;41:1646–1651. doi: 10.1161/STROKEAHA.110.579649. [DOI] [PubMed] [Google Scholar]
- 114.Lusardi TA, et al. Ischemic preconditioning regulates expression of microRNAs and a predicted target, MeCP2, in mouse cortex. J. Cereb. Blood Flow Metab. 2010;30:744–756. doi: 10.1038/jcbfm.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stenzel-Poore MP, et al. Effect of ischaemic preconditioning on genomic response to cerebral ischaemia: similarity to neuroprotective strategies in hibernation and hypoxia-tolerant states. Lancet. 2003;362:1028–1037. doi: 10.1016/S0140-6736(03)14412-1. [DOI] [PubMed] [Google Scholar]
- 116.Ziu M, Fletcher L, Rana S, Jimenez DF, Digicaylioglu M. Temporal differences in microRNA expression patterns in astrocytes and neurons after ischemic injury. PLoS ONE. 2011;6:e14724. doi: 10.1371/journal.pone.0014724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shih JD, Waks Z, Kedersha N, Silver PA. Visualization of single mRNAs reveals temporal association of proteins with microRNA-regulated mRNA. Nucleic Acids Res. 2011;39:7740–7749. doi: 10.1093/nar/gkr456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Judge AD, et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 119.Peacock H, et al. Nucleobase and ribose modifications control immunostimulation by a microRNA-122-mimetic RNA. J. Am. Chem. Soc. 2011;133:9200–9203. doi: 10.1021/ja202492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Akao Y, et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther. 2010;17:398–408. doi: 10.1038/cgt.2009.88. [DOI] [PubMed] [Google Scholar]
- 121.Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov. Today. 2012;18:282–289. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 122.Ouyang YB, et al. miR-181 regulates GRP78 and influences outcome from cerebral ischemia in vitro and in vivo. Neurobiol. Dis. 2012;45:555–563. doi: 10.1016/j.nbd.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee ST, et al. Altered microRNA regulation in Huntington's disease models. Exp. Neurol. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 124.Krützfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 125.Krützfeldt J, et al. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Broderick JA, Zamore PD. MicroRNA therapeutics. Gene Ther. 2011;18:1104–1110. doi: 10.1038/gt.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Grünweller A, Hartmann RK. Locked nucleic acid oligonucleotides: the next generation of antisense agents? BioDrugs. 2007;21:235–243. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]
- 128.Stenvang J, Silahtaroglu AN, Lindow M, Elmen J, Kauppinen S. The utility of LNA in microRNA-based cancer diagnostics and therapeutics. Semin. Cancer Biol. 2008;18:89–102. doi: 10.1016/j.semcancer.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 129.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 130.Elmen J, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.US National Library of Medicine ClinicalTrials.gov. 2012 [online], http://www.clinicaltrials.gov/ct2/show/NCT01200420?term=NCT01200420&rank=1.
- 133.Elmen J, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang H, et al. Synthetic microRNA cassette dosing: pharmacokinetics, tissue distribution and bioactivity. Mol. Pharm. 2012;9:1638–1644. doi: 10.1021/mp2006483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Luikart BW, et al. miR-132 mediates the integration of newborn neurons into the adult dentate gyrus. PLoS ONE. 2011;6:e19077. doi: 10.1371/journal.pone.0019077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gentner B, et al. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat. Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 138.Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 140.Alvarez-Erviti L, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 141.Kotipatruni RR, et al. p53- and Bax-mediated apoptosis in injured rat spinal cord. Neurochem. Res. 2011;36:2063–2074. doi: 10.1007/s11064-011-0530-2. [DOI] [PubMed] [Google Scholar]
- 142.Nagel S, et al. Microarray analysis of the global gene expression profile following hypothermia and transient focal cerebral ischemia. Neuroscience. 2012;208:109–122. doi: 10.1016/j.neuroscience.2012.01.048. [DOI] [PubMed] [Google Scholar]
- 143.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 144.De Guire V, et al. Designing small multiple-target artificial RNAs. Nucleic Acids Res. 2010;38:e140. doi: 10.1093/nar/gkq354. [DOI] [PMC free article] [PubMed] [Google Scholar]