Abstract
A major mechanism of DNA repair related to homologous recombination is the Fanconi Anemia pathway (FA). FA genes collaborate with BRCA genes to form foci of DNA repair on chromatin following DNA damage, or during S phase of the cell cycle. Our goal was to develop a method capable of evaluating the functional status of the pathway in patients’ tumor tissue, which could also be practically incorporated to large scale screening. In order to develop this method, we first used Western immunoblot to detect FANCD2 protein mono-ubiquitination in fresh tumor specimens of ovarian cancer patients undergoing surgery, and stained formalin fixed paraffin embedded (FFPE) tumor tissue simultaneously with DAPI, FANCD2 and Ki67 antibodies, eventually extending this method to other solid tumors. This triple stain permitted evaluation of the presence, or lack thereof, of FANCD2 subnuclear repair foci in proliferating cells by immunofluorescence microscopy. Overall, we evaluated 156 FFPE tumor samples using the FA triple staining immunofluorescence (FATSI) method. The ratios of FANCD2 foci negative tumors in ovarian, lung, and breast tumor samples were 21%, 20%, and 29.4%, respectively. Our studies have led to the development of a suitable method for screening, capable of identifying tumors with somatic functional defects in the FA pathway. The use of paraffin embedded tissues renders the reported method suitable for large scale screening to select patients for treatment with DNA interstrand crosslinking agents, PARP inhibitors or their combination.
Keywords: patient selection, DNA repair foci
INTRODUCTION
The poly ADP-ribose polymerase (PARP) inhibitors are being developed with the hope of inhibiting base excision repair, a process of prime importance for tumors to survive genotoxic insult. Interestingly, BRCA gene homozygous deficiency has been identified as a potential predictor of response to PARP inhibitors,1,2 and recent clinical trials have demonstrated PARP inhibitors’ antitumor activity in cancer patients with germ line BRCA deficiency.3–5 BRCA2 is involved in homologous recombination (HR), an example of double-strand break repair, thus inhibiting a repair mechanism through PARP inhibition in patients whose tumors are already deficient for another repair mechanism leads to tumor death. The term synthetic lethality is commonly used to describe this phenomenon.
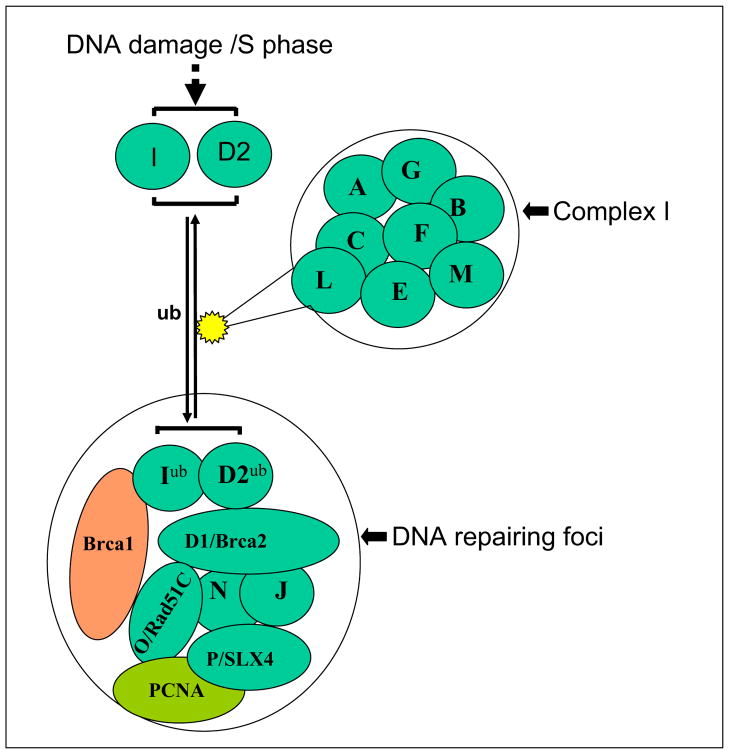
Although the numbers of cancer patients with germline BRCA2 deficiency are low, BRCA2 is just one of many genes that collaborate in the same repair pathway, the Fanconi Anemia (FA) pathway, named for a hereditary condition characterized by developmental abnormalities, progressive bone marrow failure, and cancer predisposition.6–8 At least 15 FA subtypes (A, B, C, D1/BRCA2, D2, E, F, G, I, J, L, M, N, O and P) have been identified to date.9–17 Eight of these proteins (FANC-A, -B, -C, -E, -F, -G, -L and -M) are subunits of an FA core complex (complex I), a nuclear E3 ubiquitin ligase (Fig 1).12,18,19 The FA complex I activates FANCD2 and FANCI by mono-ubiquitinating the proteins as a response to DNA damage.12,20 The activated proteins are subsequently transported to subnuclear foci, thought to be the sites of DNA repair, which also contain BRCA1, FANCD1/BRCA2, FANCJ/BRIP1, FANCN/PALB2, FANCP/SLX4 and FANCO/Rad51C.7,12–17 De-ubiquitination of FANCD2, by the ubiquitin-specific protease 1 results in its inactivation and release from the sites of DNA repair.21 Since the function of many of the FA proteins is to ubiquitinate and to activate FANCD2, this is a key effector protein in the FA pathway. FANCD2 is converted from a short (S) to a long (L) form by mono-ubiquitination, during S phase, or following induction of DNA double-strand breaks or interstrand crosslinks.22 Any mutation or epigenetic change that disrupts components of the core complex also abrogates its E3 ligase function, leading to defective FANCD2 mono-ubiquitination and no nuclear foci formation.7
Figure 1. The Fanconi anemia (FA) pathway and formation of repair foci.
The FA complex I functions to activate FANCD2 and FANCI by mono-ubiquitinating the proteins following DNA damage. The activated FANCD2 and FANCI proteins are subsequently transported to subnuclear foci, which contain FANCD1/BRCA2, FANCN (PALB2), FANCJ (BRIP1, BACH1), FANCO (Rad51C) and FANCP (SLX4). The FA repair proteins repair DNA damage in conjunction with other associated proteins including BRCA1 and other Proteins.
FA patients have a high incidence of malignancies, and are hypersensitive to DNA interstrand crosslinking agents such as mitomycin C (MMC).23 FANCD2 monoubiquitination is critical for MMC or cisplatin resistance and is required for the FANCD2 protein to form damage-induced foci on chromatin.22–24 Recent evidence links disruption of the FA cascade to sporadic cancers in the general population, which may involve epigenetic silencing of the FA-core complex, mutations of one or several FA genes, or modification of encoded products.25–27 Furthermore, disruption of the FA genes correlates with cisplatin,28 MMC,23 and PARP inhibitors sensitivity.29 Given the number of genes involved in this pathway, and that a number of genetic or epigenetic alterations could interfere with its functionality, we hypothesize that a substantial number of FA pathway deficient patients can be identified across the many histological types of cancer, and that these patients may be more sensitive to cross-link based therapy and to inhibitors (e.g, PARPi) of alternative repair mechanisms.
However, in order to evaluate this hypothesis, an effective method capable of evaluating the functionality of this pathway as a whole, which could be feasibly incorporated in large-scale screening, needs to be developed. Willers et al.30 reported that foci formation of DNA repair proteins BRCA1, FANCD2, and RAD51 was detected in fresh irradiated breast cancer biopsy specimens by immunofluorescence microscopy. Herein we report the development of a FancD2 foci formation triple-stain immunofluorescence based method that can be performed on formalin fixed paraffin embedded (FFPE) tumor specimens.
MATERIALS AND METHODS
Assessment of cellular FANCD2 nuclear foci formation
SV40 transformed empty retroviral vector transduced human fibroblasts PD20 (FANCD2 mutation) and PD220 (FANCA mutation), and their retrovirally corrected (transduced with wild-type FANCD2 to PD20 and wild-type FANCA to PD220 cell) counterparts, were obtained from The Oregon Health and Science University Fanconi Anemia Cell Repository. Human breast cancer cell MCF-7 was obtained from the American Type Culture Collection (ATCC, Rockville, MD). MCF-7 cells were grown in D-MEM medium with 10% fetal bovine serum and 1% penicillin/streptomycin. PD20, PD220 cells and their retrovirally corrected counterparts were tested in our laboratory and grown in D-MEM medium with 10% fetal bovine serum, 1μg/ml puromycin, and 1% penicillin/streptomycin in an incubator (37°C, 5% of CO2). Cells were fixed for 10 min in 4% paraformaldehyde, and permeabilized for 5 min at 4°C in 0.5% Triton X-100. The slides were blocked in 1X phosphate-buffered saline (PBS) containing 2% BSA at room temperature for 1 hour. The cells were incubated in the primary antibody (anti-FANCD2 at concentration of 1:500, Novus Biologicals, Littleton, CO; Catalog #: NB100-182; or anti-gammaH2AX at concentration of 1:500, abCam, Cambridge, MA, Catalog #: ab22551) diluted in blocking solution for 1 h at room temperature, and washed three times for 10 min each with 1X PBS. Fluorescence conjugated secondary antibodies were used for foci detection. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
FANCD2 protein ubiquitination in cells and fresh tumors
After the Ohio State University Institutional Review Board (IRB) approval, and signed informed consent was obtained, fresh tumor specimens from patients undergoing medically required procedures were procured. Western immunoblot analysis was performed as described previously.31 Briefly, cells and tumor tissues were solubilized in lysis buffer. Protein concentrations were evaluated using the Bradford reagent (Bio-Rad, Hercules, CA). 100 μg of total protein was subjected to NuPAGE™ 3–8% Tri-Acetate (Invitrogen, Carlsbad, CA) gel electrophoresis. The separated proteins were electro-blotted onto a nitrocellulose membrane. Membranes were incubated in blocking buffer and with the primary antibody (anti-FANCD2) over night at 4°C. After four 10-minute washes with TBS-T, the membrane was incubated with a horseradish peroxidase-linked secondary antibody at room temperature for one hour. After five 10-minute washes, a chemi-luminescent detection system was used to detect the secondary antibody, and the membranes exposed to autoradiography.
To evaluate the response of FANCD2 protein to irradiation, fresh ovarian tumors were cut into 1 to 2 mm pieces and treated with a 137Cesium γ-source at a dose of 5Gy. Irradiated samples were incubated in D-MEM medium with 10% of fetal bovine serum for 15 to 16 hours and subsequently snap frozen for Western blot analysis and embedded in paraffin for FANCD2 foci evaluation.
FA triple staining immunofluorescence (FATSI) method using FFPE tissues
Human tumor tissue samples were obtained from The Tissue Procurement Shared Resources of the Ohio State University Comprehensive Cancer Center and The Cooperative Human Tissue Network, Midwestern Division at The Ohio State University, after IRB approval. FFPE tumor tissue was cut at 4 microns, placed on positively charged slides and stained with hematoxylin and eosin. Additional sections for immunofluorescence staining were placed in a 60°C oven for 1 h, cooled, deparaffinized and rehydrated through xylenes and graded ethanol solutions to water in standard fashion. Antigen retrieval was performed by placing slides in Dako’s TRS antigen retrieval solution (Dako, Carpenteria, CA) in a calibrated vegetable steamer (Black and Decker) at 94°C for 25 minutes. Slides then were placed on a Dako Autostainer for automated staining. The tissue sections were incubated with a primary antibody cocktail of rabbit polyclonal FANC-D2 antibody (Novus Biologicals, Littleton, CO) at a dilution of 1:1000 and a monoclonal anti-Ki67 mouse antibody (Dako, Carpenteria, CA) at a dilution of 1:150, for 1 hour at room temperature. Sections then were co-incubated with a secondary antibody (FITC conjugated to anti-rabbit IgG and Alexa fluor 594 donkey anti-mouse IgG, Invitrogen, Carlsbad, CA) at 1:1000 for 1 hour at room temperature. All rinses were performed on the autostainer with TBST. The sections were mounted on glass slides using a 4′ 6-diamidino-2-phenylindole (DAPI)–containing embedding medium (Vysis Dapi 1, Abbott Laboratories, Downers Grove, IL). Formalin fixed paraffin embedded (FFPE) FANCD2 foci negative cells (PD220 or PD20) and foci positive cells (MCF-7 or FA corrected PD220 or FA corrected PD20) were used as controls on the sample slide during the procedure. The slides were analyzed under a Nikon E-400 fluorescence microscope or an Olympus BX51 equipped with fluorescent detection for designated immunofluorescence. The FANCD2 foci were analyzed under a 50X or 100 X oil objectives.
FFPE control cells
5 × 106 cells were fixed in 10% neutral buffered formalin (NBF) for 2–4 hours prior to procedure. The cells were spun down at 400g for 5 min at room temperature on a bench-top Beckman centrifuge. The formalin was decanted and rinsed carefully with 1X PBS to remove residual formalin. The cells were re-suspended gently by flicking the tube tips, and then transferred to a new centrifuge tube (2ml). The cells were washed again with 1x PBS and decanted as much as possible. One drop of undiluted eosin (Richard-Allan Scientific, Kalamazoo, MI) was added to the tube, and mixed by flicking the tube tip. Ten drops of human plasma were added and mixed followed by addition of 15 drops of thrombin solution (Thrombin Topical Bovine Gentrac, Inc. Middleton, WI). The mixed solution with the control cells were inverted gently to mix and form clot. The tube was then centrifuged at 400g for 5 min and the clot was pulled out with a wooded tip, placed and wrapped in tissue bag and placed in a cassette. The tissue was then embedded in paraffin using conventional tissue embedding procedures. The paper conforms to the ethical guidelines for human research.
RESULTS
Mono-ubiquitinated FANCD2 protein and FANCD2 foci are absent in cells with an inactive FA pathway
Human fibroblasts PD20 and PD220 are two of the most frequently used FA defective cell lines in investigating of biological function of FA pathway. To demonstrate the feasibility in detecting FA foci formation in our laboratory, SV40-transformed fibroblasts derived from patients with FA group D2 (PD20 cells, FANCD2 mutation) or group A (PD220, FANCA mutation), and their retrovirally corrected counterparts were used for the Western blot analysis. The supplementary Figure A (Fig. SA) demonstrated that the FA pathway corrected PD220 (transduced with wildtype FANCA) and PD20 (transduced with wild type FANCD2) cells were able to produce the ubiquitinated form of FANCD2 protein, which appeared as a protein with larger molecular weight, or large band (FANCD2-L) on Western film. In contrast, the FA pathway defective PD20 and PD220 cells were lacking the FANCD2-L band, and produced only one (FANCD2 in PD220 cell) or no (PD20 cell) product. Using immunofluorescence staining, we were able to detect FANCD2 foci formation in both PD20-corrected and PD220-corrected cells, whereas, FANCD2 foci were absent from the FA defective PD20 and PD220 cells (Fig. SB).
To further demonstrate that FANCD2 foci are markers related to DNA repair, we investigated co-localization of FANCD2 foci and gamma-H2AX foci using the human breast cancer cell MCF-7. The results showed that FANCD2 foci were co-localized with gamma-H2AX foci (Fig. SC).
Effect of irradiation on FANCD2 protein monoubiquitination and foci formation in fresh tumors
It has been reported that irradiation promote FANCD2 protein monoubiquitination and foci formation. We analyzed five fresh ovarian tumor samples following exposure to 5 Gy of radiation. Irradiated samples were incubated in a cell culture incubator for 15 to 16 hours and were subsequently snap-frozen for Western blot analysis and embedded in paraffin for FANCD2 foci evaluation. The amount of FACND2-L was increased in three tumors post irradiation, whereas FANCD2-L was absent in two other tumors, both pre and post irradiation, suggesting that these two tumors are FA defective (Fig. 2A). Immunofluorescence staining showed that the number of FANCD2 foci was 2.6 fold increased post irradiation as compared to no irradiation in the FA intact tumors. As expected, FANCD2 foci were absent from both pre and post irradiated tissues in the FA defective tumors (Fig. 2B).
Figure 2. Irradiation increases FANCD2 monoubiquitination and foci formation in ovarian cancers.
The FA intact and FA defective tumor were analyzed with Western blotting and immunofluorescent staining pre and post irradiation (5Gy). (A) The FA deficient tumor lost the monoubiquitinated FANCD2 protein both pre and post irradiation. In contrast, monoubiquitinated FANCD2 (FANCD2-L) protein was increased post irradiation in FA intact tumor. FANCD2: FANCD2 protein without modification, FANCD2-L: monoubiquitinated FANCD2 protein, T: ovarian cancer, T-IR: ovarian cancer treated by irradiation (5Gy). (B) FANCD2 foci were evaluated with FFPE tumor samples. Number of FANCD2 foci were increased post irradiation as compared with the tissue from the same tumor without irradiation in the FA intact tumor. However, in the FA defective tumor, FANCD2 foci were absent regardless of irradiation.
FANCD2 protein monoubiqutination and foci formation in patient’s tumor specimens without stimulation
A clinical protocol (OSU2007C0087) for collecting tumor and normal tissue of cancer patients undergoing surgical procedures was activated and tumor tissue samples were obtained. Western blotting analysis of 25 ovarian tumors showed that the FANCD2-L band was absent in four of these tumors, reflecting no FANCD2 monoubiquitination.
In order to evaluate for FANCD2 repair foci formation in tumor samples, and to test the feasibility of using paraffin embedded tissue blocks for this purpose, a small section of fresh ovarian tumor was embedded in Tissue Freezing Medium and stored in a −80 freezer, while another section from the same tumor was fixed in 10% formalin for 12 hours and embedded in paraffin. Among 10 ovarian tumors assessed for FANCD2 foci formation in both frozen and FFPE tissue, there was full concordance and the foci were morphologically similar (Fig. 3). Considering the availability and simplicity to obtain FFPE samples, we subsequently used the FFPE tumor samples for immunofluorescence evaluation in the rest of the tumors. FANCD2 foci were absent in six of the 25 ovarian tumor samples. However, further analysis revealed that one of the foci negative specimens contained mainly non-proliferating necrotic tissue, thus it was considered a false negative.
Figure 3. Comparison of FANCD2 foci formation between frozen sections and FFPE samples.

A small section of fresh ovarian tumor was embedded in Tissue Freezing Medium and stored in a −80 freezer, while another section from the same tumor was fixed in 10% formalin for 12 hours and embedded in paraffin. Both tissue sections were cut and incubated with rabbit polyclonal FANC-D2 primary antibody (Novus Biologicals, Littleton, CO) at a dilution of 1:1000 for 1 hour at room temperature. Sections then were incubated with a FITC conjugated secondary antibody for 1 hour at room temperature. The sections were mounted on glass slides using a 4′ 6-diamidino-2-phenylindole (DAPI)–containing embedding medium (Vysis Dapi 1, Abbott Laboratories, Downers Grove, IL). The slides were analyzed under a fluorescence microscope. Magnification: 1000X
Comparisons of the Western data with FANCD2 foci formation in the fresh ovarian specimens showed that four of five evaluable tumors with negative foci formation produced no FANCD2-L, whereas one foci negative tumor was able to produce the FANCD2-L protein. Nineteen foci positive ovarian tumors were evaluated for FANCD2 protein. In three of these specimens, the very low level of FANCD2 protein found precluded the determination of FANCD2 protein monoubiquitination status. FANCD2-L was present in the remaining 16 cases. No FANCD2 protein was detected in the one necrotic tumor tissue, discussed above.
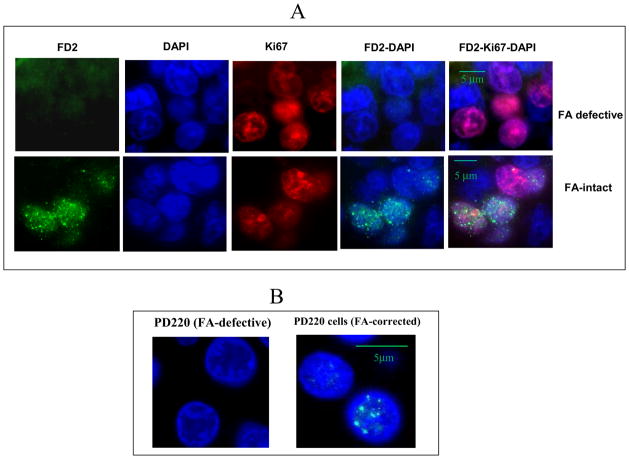
In the absence of DNA damage insult, the FANCD2 sub-nuclear foci are likely to be observed mainly during the S phase of the cell cycle. Thus, if a tumor sample lacks proliferating cells, FANCD2 repair foci may not be observed, leading to potential false FANCD2 foci negative cases. Because some cancer samples or sections contain mainly non-proliferating or necrotic cells, in order to circumvent the potential problem with false negative cases, we incorporated to the immunofluorescence evaluation the additional evaluation of Ki67 protein, as an indicator of adequate cell proliferation. Figure 4 shows the use of FA triple (FANCD2, Ki-67 and DAPI) staining immunoflorescence (FATSI) method to identify FANCD2 foci positive and foci negative cancer samples in breast cancers (Fig. 4A). Formalin fixed paraffin embedded (FFPE) FANCD2 foci positive and negative cells (Fig. 4B) were used as controls on the sample slide during the procedure.
Fig. 4. Detection of FANCD2 foci formation in human FFPE samples by the immunofluorescent triple staining analysis.
Paraffin embedded tissues sections were deparaffinized and rehydrated. Tissue sections were incubated with a primary antibody cocktail of rabbit polyclonal FANC-D2 antibody (Novus Biologicals, Littleton, CO) at a dilution of 1:1000 and a monoclonal anti-Ki67 mouse antibody (Dako, Carpenteria, CA) at a dilution of 1: 150 for 1 hour at room temperature. Sections then were incubated with a secondary antibody cocktail containing FITC conjugated anti-rabbit IgG and Alexafluor 594 donkey anti-mouse secondary for 1 hour at room temperature. The sections were mounted on glass slides using a 4′ 6-diamidino-2-phenylindole (DAPI)–containing embedding medium (Vysis Dapi 1, Abbott Laboratories, Downers Grove, IL). The slides were analyzed under a fluorescence microscope. (A) Breast cancer, and (B) FFPE PD220 (FA defective) and corrected PD220 (FA positive) cells. Magnification: 1000X.
Ki-67 nuclear positivity of greater than 10% (Ki-67/DAPI) was subsequently required to consider a sample evaluable. Evaluation of a total of 300 cells (300 DAPI stained nuclei) with less than one foci positive nucleus (a foci positive nucleus is defined by at least three foci) per 100 cells was arbitrarily set as the benchmark to define FA pathway deficiency. We evaluated 156 FFPE breast, lung and ovarian cancer samples (Table 1). According to Ki67 staining, 141 of the 156 tumors were evaluable. The rate of FANCD2 foci negative ovarian tumors was 21%, 20% for lung and 29.4% for breast (Table 1).
Table 1.
Status of FANCD2 foci in the FFPE tumors
| Tumor type | Total No of tumor tested | Ki-67 positive | FANCD2 foci presence | FANCD2 foci absence | % FANCD2 foci negative tumor |
|---|---|---|---|---|---|
| Ovarian | 25 | 24 | 19 | 5 | 21% |
| Breast | 111 | 102 | 72 | 30 | 29.4% |
| Lung | 20 | 15 | 12 | 3 | 20% |
Among the 24 evaluable epithelial ovarian cancers (21 primary and 3 recurrent cancers) 19 were serous (16 stage IIIC, 1 stage IV and 2 recurrent) and 5 were non-serous histology (1 stage IC, 2 stage IIC, 1 stage IIIC and 1 recurrent). Five tumors had absence of FANCD2 subnuclear foci and the remaining 19 had foci formation. Lack of foci formation was significantly associated with non-serous histology (3/5 non-serous with a defective pathway vs. 2/19 serous with a defective pathway, P=0.04, 2-tail Fisher’s exact test). Lack of foci formation was also significantly associated with recurrent stage (2/3 recurrent with a defective pathway vs. 3/21 non-recurrent ovarian tumor with a defective pathway, p= 0.0988, 2-tail Fisher’s exact test). Among 20 lung tumors (3 large cell carcinomas, 4 squamous cell carcinomas and 13 adenocarcinomas) five were non-evaluable because of lacking enough ki67 positive cells. Three of 15 evaluable (20%) were FANCD2 foci negative, with no significant association for histological type. Among the 111 breast tumors 102 were evaluable (92 were invasive ductal carcinoma [IDC] and 10 were non-IDC). There was no significant difference (Fisher’s exact test) in FANCD2 foci deficiency between the histology types (29.35% for IDC and 30% for non-IDC). Among the 102 evaluable samples, 62 ER, PR and Her2neu negative. Seventeen (27.4%) of these triple negative tumors were FANCD2 foci negative. Among the 40 non- triple negative tumors, thirteen (32.5%) were foci negative. There was no significant difference (Fisher’s exact test) in FANCD2 foci deficiency among the molecular subtypes.
DISCUSSION
BRCA gene homozygous deficiency has been identified as a potential predictor of tumor response to PARP inhibitors, a phenomenon believed to be due to PARP inhibitor induced deficiency of an alternative repair pathway in tumors already incapable of HR repair. Given the large number of proteins needing to interact in the FA pathway for efficient HR repair, we have hypothesized that a substantial number of cancer patients will have somatic FA repair deficiency in their tumors, and that these patients would also be suitable candidates for treatment with DNA interstrand crosslinking agents and for therapeutic studies of PARP inhibition.
Previous data support a somatic disruption of the FA cascade in sporadic cancers.25–27 These disruptions are caused by either epigenetic silencing of the FA genes, or by mutations of one or several FA genes. Because of the large number of genes, the variety of possible alterations, and collaboration with other proteins, functional assessment of the pathway, rather than searching for individual genetic or epigenetic abnormalities, ought to be the most practical course of action to develop a predictive biomarker. This is best assessed either at the FancD2 monoubiquitination step, or by evaluation of subnuclear repair protein foci formation.
Under normal stress conditions FANCD2 foci are formed in the S phase of the cell cycle, and because the rate of cell proliferation is increased in cancer tissues, it is possible to evaluate FANCD2 foci formation in tumor tissues without stimulation. In this manuscript, we demonstrate the ability to evaluate FANCD2 foci formation in paraffin embedded tissue, including archived FFPE blocks that have been stored for several years. However, since the tumors of interest would be FA functionally deficient tumors (i.e., foci negative), it is of paramount importance to assure that tumors are not falsely assessed as negative solely due to low proliferation or necrosis. Given these considerations, and in order to reduce the rate of false FANCD2 foci negative case, we added Ki67 staining to the test, to clearly identify the formation or lack of foci in proliferating cells.
An unintended potential consequence of this additional staining, while substantially decreasing the number of false negatives, is that it may bias the selection towards patients with highly proliferative tumors. It is possible, on the other hand, as has been postulated by others, that repair deficiency increases tumor mutagenicity, and that higher proliferation is an effect on itself of the repair deficiency. Another aspect to be considered is tumor heterogeneity. For this reason we recommend the use of FFPE blocks rather than tissue microarray (TMA) for the FANCD2 foci evaluation.
An issue for debate is how comprehensive of an assessment of the functionality of the whole pathway is provided by the FATSI test. Is it only able to pick up deficiencies of genes interacting in the proximal part of the pathway? (i.e., Complex I activation), or by virtue of requirement of the action of other genes for foci formation, would it also pick up downstream defects? We tested the ovarian cancer cell line UWB1.289 which contains a mutant BRCA1 (BRCA1-2594delC, truncated BRCA1 protein) for FANCD2 foci formation. We found the UWB1.289 cell was defective in FANCD2 foci formation. In addition, we evaluated Hcc1937, a BRCA1 defective breast cancer cell line which contains BRCA1 mutation, (insertion C at nucleotide 5382) for foci formation for both FANCD2 and RAD51 (antibodies: Novus NB100-182 for FANCD2 and Santa Cruz H-92 for RAD51). Compared to RAD51 staining, the FANCD2 foci are diminished in the Hcc1937 cells (results not shown).
Moreover, it has been reported that S-phase RAD51 foci form normally in CAPAN-1 cells expressing truncated BRCA2.32 It is plausible, that whether a BRCA2 alteration impedes FA foci formation depends on the position of the alteration and the conformation change of the BRCA2 protein. To test our hypothesis, cells and tissues containing various BRCA2 mutation/deletion are currently under investigation in our laboratory. We are also evaluating FATSI in clinically annotated specimens on archives at the OSU Clinical Cancer Genetics tumor bank, from cancer patients in which the BRCA germ-line mutation status is known.
To demonstrate the presence of an underlying FA genetic or epigenetic defect in FANCD2 foci-negative tumor samples, we analyzed two foci negative lung tumors and matched non-tumor lung tissues by RNAseq. Results showed that one of the tumor samples contained a nonsynonymous substitution at codon 595 (TTA to GTA, L595V) in the FANC-I gene. This mutation was absent from the matched non-tumor tissue, suggesting a somatic (non germ-line) defect. It has been reported that mutation in FANCI is responsible for loss of a functional FA pathway in a patient with Fanconi anemia complementation group I12.
No mutation was detected FA genes in the second tumor by RNAseq. However SNPs analysis showed loss of heterozygosity (LOH) in the FANCD1 locus. FANCF mRNA expression was reduced (−1.6 fold) as compared to matched non-tumor tissue. Furthermore, FANCF promoter methylation was also detected by MS-PCR (data not shown). The primers and methods have been previously described.28
Overall, with the limitations posed by the small sample size, our data suggest that 20–30% of tumors have inactivation of the FA pathway. If as hypothesized, FA defective tumors are sensitive to DNA interstrand crosslinking agents and PARP inhibitors, a test with such simplicity can have widespread application. The identification of patients with somatic functional deficiency of the FA pathway may also lead to a better understanding of the specific genetic/epigenetic events that drive the cancer in these patients.
Supplementary Material
(SA) Western blot analysis of SV40-transformed fibroblasts derived from patients with FA group D2 (PD20, FANCD2 mutation) and group A (PD220, FANCA) mutations. The right panel demonstrates absence of both FANCD2 bands in PD20, and restoration of both bands in corrected cells. The left panel demonstrates absence of the mono-ubiquitinated band (FANCD2-L) in PD220 and restoration of this band in corrected cells. (SB) FANCD2 foci formation in the FA defective cells PD220, PD20 and their corrected counterparts. The PD20 and PD220 cells and their retrovirally corrected counterparts were stained with a rabbit polyclonal anti-FANCD2 antibody. FITC conjugated secondary antibody was used for the foci detection. Cell nuclei were counterstained with DAPI. Representative images illustrate the absence of foci formation in FA defective PD20 and PD220 cells. In contrast, subnuclear FANCD2 foci were formed in retrovirally corrected (transduced with wild-type FANCD2 for PD20 or wild-type FANCA for PD220) PD20 and PD220 cells. (SC) Co-localization of FANCD2 foci and gamma-H2AX foci in the human breast cancer cell MCF-7.
Acknowledgments
Human fibroblasts PD20 and PD220 and their retrovirally corrected counterparts were provided by the The Oregon Health and Science University Fanconi Anemia Cell Repository. We thank the Pathology Core Lab and Tissue Procurement facilities of the OSUCCC, and The Cooperative Human Tissue Network Midwestern Division at The Ohio State University, for their assistance. This work was supported by The Ohio State University Comprehensive Cancer Center Cancer Fund (to M.V), NCI R01CA152101 (to M. V); and American Cancer Society Institutional Seed Grant (IRG-67-003-44 to W.D), and NIH U01 grant GM092655 (W.S.).
Footnotes
All authors have read the journal’s policy on disclosure of potential conflicts of interest, and the authors have none to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 2.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 3.Tutt A, Robson M, Garber J, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 4.Audeh MW, Carmichael J, Penson RT, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 5.Fong P, Boss D, Yap T, et al. Inhibition of poly(ADP-ribose) polymerase in tumors fromBRCA mutation carriers. N Engl J Med. 2009;361:23–34. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 6.Bagby GC., Jr Genetic basis of Fanconi anemia. Curr Opin Hematol. 2003;10:68–76. doi: 10.1097/00062752-200301000-00011. [DOI] [PubMed] [Google Scholar]
- 7.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 8.Auerbach AD, Allen RG. Leukemia and preleukemia in Fanconi anemia patients. A review of the literature and report of the International Fanconi Anemia Registry. Cancer Genet Cytogenet. 1991;51:1–12. doi: 10.1016/0165-4608(91)90002-c. [DOI] [PubMed] [Google Scholar]
- 9.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 10.Xia B, Dorsman JC, Ameziane N, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 11.Xia B, Sheng Q, Nakanishi K, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Smogorzewska A, Matsuoka S, Vinciguerra P, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto KN, Kobayashi S, Tsuda M, et al. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci U S A. 2011;108:6492–6496. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim Y, Lach FP, Desetty R, Hanenberg H, Auerbach AD, Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoepker C, Hain K, Schuster B, et al. SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat Genet. 2011;43:138–141. doi: 10.1038/ng.751. [DOI] [PubMed] [Google Scholar]
- 16.Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 17.Somyajit K, Subramanya S, Nagaraju G. RAD51C: a novel cancer susceptibility gene is linked to Fanconi anemia and breast cancer. Carcinogenesis. 2010;31:2031–2038. doi: 10.1093/carcin/bgq210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machida YJ, Machida Y, Chen Y, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 19.Meetei AR, Yan Z, Wang W. FANCL replaces BRCA1 as the likely ubiquitin ligase responsible for FANCD2 monoubiquitination. Cell Cycle. 2004;3:179–181. [PubMed] [Google Scholar]
- 20.Meetei AR, de Winter JP, Medhurst AL, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 21.Nijman SM, Huang TT, Dirac AM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Higuera I, Taniguchi T, Ganesan S, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 23.Clarke AA, Gibson FM, Scott J, Myatt N, Rutherford TR. Fanconi’s anemia cell lines show distinct mechanisms of cell death in response to mitomycin C or agonistic anti-Fas antibodies. Haematologica. 2004;89:11–20. [PubMed] [Google Scholar]
- 24.Wang Y, Wiltshire T, Senft J, Wenger SL, Reed E, Wang W. Fanconi anemia D2 protein confers chemoresistance in response to the anticancer agent, irofulven. Mol Cancer Ther. 2006;5:3153–3161. doi: 10.1158/1535-7163.MCT-06-0427. [DOI] [PubMed] [Google Scholar]
- 25.Lyakhovich A, Surralles J. Disruption of the Fanconi anemia/BRCA pathway in sporadic cancer. Cancer Lett. 2006;232:99–106. doi: 10.1016/j.canlet.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 26.Neveling K, Kalb R, Florl AR, et al. Disruption of the FA/BRCA pathway in bladder cancer. Cytogenet Genome Res. 2007;118:166–176. doi: 10.1159/000108297. [DOI] [PubMed] [Google Scholar]
- 27.Tischkowitz M, Xia B, Sabbaghian N, et al. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci U S A. 2007;104:6788–6793. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taniguchi T, Tischkowitz M, Ameziane N, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 29.McCabe N, Turner NC, Lord CJ. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 30.Willers H, Taghian AG, Luo CM, Treszezamsky A, Sgroi DC, Powell SN. Utility of DNA repair protein foci for the detection of putative BRCA1 pathway defects in breast cancer biopsies. Mol Cancer Res. 2009;7:1304–1309. doi: 10.1158/1541-7786.MCR-09-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duan W, Gao L, Wu X, et al. MicroRNA-34a is an important component of PRIMA-1-induced apoptotic network in human lung cancer cells. Int J Cancer. 2010;172:313–320. doi: 10.1002/ijc.25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarsounas M, Davis D, West S. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22(8):1115–23. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(SA) Western blot analysis of SV40-transformed fibroblasts derived from patients with FA group D2 (PD20, FANCD2 mutation) and group A (PD220, FANCA) mutations. The right panel demonstrates absence of both FANCD2 bands in PD20, and restoration of both bands in corrected cells. The left panel demonstrates absence of the mono-ubiquitinated band (FANCD2-L) in PD220 and restoration of this band in corrected cells. (SB) FANCD2 foci formation in the FA defective cells PD220, PD20 and their corrected counterparts. The PD20 and PD220 cells and their retrovirally corrected counterparts were stained with a rabbit polyclonal anti-FANCD2 antibody. FITC conjugated secondary antibody was used for the foci detection. Cell nuclei were counterstained with DAPI. Representative images illustrate the absence of foci formation in FA defective PD20 and PD220 cells. In contrast, subnuclear FANCD2 foci were formed in retrovirally corrected (transduced with wild-type FANCD2 for PD20 or wild-type FANCA for PD220) PD20 and PD220 cells. (SC) Co-localization of FANCD2 foci and gamma-H2AX foci in the human breast cancer cell MCF-7.