Abstract
Background
Renin-angiotensin-aldosterone system (RAAS) is the most important endocrine blood pressure control mechanism in our body, genes encoding components of this system have been strong candidates for the investigation of the genetic basis of hypertension. However, previous studies mainly focused on limited polymorphisms, thus we carried out a case-control study in the Han Chinese population to systemically investigate the association between polymorphisms in the RAAS genes and essential hypertension.
Methods
905 essential hypertensive cases and 905 normotensive controls were recruited based on stringent inclusion and exclusion criteria. All 41 tagSNPs within RAAS genes were retrieved from HapMap, and the genotyping was performed using the GenomeLab SNPstream Genotyping System. Logistic regression analysis, Multifactor dimensionality reduction (MDR), stratified analysis and crossover analysis were used to identify and characterize interactions among the SNPs and the non-genetic factors.
Results
Serum levels of total cholesterol (TC) and triglyceride (TG), and body mass index (BMI) were significantly higher in the hypertensive group than in the control group. Of 41 SNPs genotyped, rs3789678 and rs2493132 within AGT, rs4305 within ACE, rs275645 within AGTR1, rs3802230 and rs10086846 within CYP11B2 were shown to associate with hypertension. The MDR analysis demonstrated that the interaction between BMI and rs4305 increased the susceptibility to hypertension. Crossover analysis and stratified analysis further indicated that BMI has a major effect, and rs4305 has a minor effect.
Conclusion
These novel findings indicated that together with non-genetic factors, these genetic variants in the RAAS may play an important role in determining an individual’s susceptibility to hypertension in the Han Chinese.
Introduction
Essential hypertension, defined as high blood pressure (BP) with no identifiable cause, affecting 95% of hypertensive patients [1]. It is considered to be the consequence of an interaction between environmental and genetic factors [2]. Hitherto, many candidate genes in the renin-angiotensin-aldosterone system (RAAS), the sympathetic nervous system, and water-sodium balance system have been widely studied [3]–[6]. Among all these genes which play important roles in the etiology of hypertension, those encoding the main components of the RAAS are deemed the most possible candidate genes since the RAAS plays a fundamental role in the maintenance of blood pressure and cardiovascular homeostasis [7], [8].
RAAS genes encoding renin (REN), angiotensinogen (AGT), angiotensin-converting enzyme (ACE), angiotensin type 1 receptor (AGTR1) and aldosterone synthase gene (CYP11B2) have been widely investigated in different ethnic populations, and dozens of single nucleotide polymorphisms (SNPs) within RAAS genes have been reported to be significantly associated with essential hypertension [9]–[11]. However, previous studies focused on limited SNPs like AGT M235T, AGT A6G, AGT T174M, ACE I/D, AGTR1 A1166C, and CYP11B2 C3344T, and the results are often inconsistent [12]–[15]. Moreover, dozens of genome-wide association studies (GWAS) on hypertension have been carried out, but none of these hot polymorphisms was significantly associated with hypertension [16]–[19]. It is possible that environmental factors, population variation, relatively small sample size, patient selection, and limited genetic alleles may contribute to the conflicting or even contradictory results [20]–[22].
Given these limitations, this study was designed and conducted in a large, homogeneous sample of Han Chinese, which would minimize the potential influence of mixed factors on hypertension. The objective of the present study was to systemically examine the association between polymorphisms in the RAAS candidate genes (REN, AGT, ACE, AGTR, and CYP11B2) and hypertension. Thus, we first conducted a case-control study in the Han Chinese population and genotyped all tagSNPs within RAAS genes. Subsequently, we analyze the interaction among different SNPs and non-genetic risk factors for hypertension, which may give more information on the roles of genetic factors.
Materials and Methods
The protocol of this study was reviewed and approved by the medical ethics committee of Ningbo University. The health records and blood samples of the participants were collected with informed written consent.
Study Participants
The participants were chosen from our established community-based epidemiologic study of common diseases. With informed written consent, we collected more than 10,000 health records. Subsequently, participants who fulfilled the following criteria were put into our database: 30 to 75 years old, Han Chinese, living in Ningbo City (East coast of China) for at least three generations without migration history. Finally, 905 essential hypertensive cases and 905 normotensive controls were chosen from this database, which were matched for age and sex. In addition, participants without cardiovascular diseases, diabetes, kidney diseases, or other major chronic illnesses according to their health records were recruited as controls.
Measurement of Blood Pressure and Clinical Parameters
Blood pressure was measured in the morning after the participants had been in sitting position for 10 minutes. Three readings were taken 5 minutes apart using standard mercury sphygmomanometer and the average of last two measurements was recorded. Hypertension in this study was defined as sitting systolic blood pressure (SBP) ≥140 mmHg and/or diastolic blood pressure (DBP) ≥90 mmHg, or self-reported use of anti-hypertensive medication. Patients with secondary hypertension were excluded. Normal blood pressure was defined with SBP≤120 mmHg and DBP≤80 mmHg.
With informed written consent, two milliliter of venous blood was collected with ethylene diamine tetraacetic acid (EDTA) as anticoagulant. Subsequently, serum levels of total cholesterol (TC), high-density lipoprotein (HDL), triglyceride (TG) were measured enzymatically on a Hitachi automatic biochemistry analyzer 7100. Clinical information including body mass index (BMI), and weekly alcohol and cigarettes consumption were also obtained. In this study, who consumed ≥70 g of alcohol per week for more than 1 year were defined as individuals with alcohol abuse. Moreover, who smoked ≥70 cigarettes per week for more than 1 year were defined as individuals with smoking habit.
SNP Genotyping
All 41 tagSNPs were retrieved from HapMap (http://hapmap.ncbi.nlm.nih.gov/), with tagger pairwise method in CHB: R2 cut off = 0.8 and minor allele frequency (MAF) cut off = 0.1 (Table S1). The genomic DNA was extracted from the whole blood using standard phenol-chloroform extraction method. Genotyping was performed using the GenomeLab SNPstream Genotyping System (Beckman Coulter Inc.) according to the manufacturer’s protocol [23].
Statistical Analysis
Continuous variables were presented as the mean ± SD and analyzed by t-test between two groups. Statistical analysis of allele and genotype frequencies between case and control groups were compared by chi-squared test. Effect of confounding variables were identified by logistic regression (SPSS 16.0, SPSS Inc.). Hardy-Weinberg equilibrium (HWE) was calculated in controls by the software PEDSTATS V0.6.8 (http://www.sph.umich.edu/csg/abecasis/). Multifactor dimensionality reduction (MDR), stratified analysis and crossover analysis were used to identify and characterize interactions among the SNPs and the non-genetic factors [24]. The software used for MDR is distributed in a JAVA platform with a graphical user interface and is freely available online (http://www.epistasis.org/mdr.html).
All tests were two-sided, and P values less than 0.05 were considered statistically significant. For chi-squared test, the P values were adjusted for the total number of tested SNPs using the Bonferroni correction method (α = 0.05/41 ≈ 0.0012).
Results
The baseline characteristics of our study participants are summarized in Table 1. The male to female ratio was equal in both groups, and mean age of hypertensive participants and controls were similar, demonstrating that the hypertensive and control groups were well-matched and are appropriate for the following analyses. Serum high-density lipoprotein (HDL) and percentage of regular smoking and alcohol abuse showed no difference between hypertensive and control groups. However, serum levels of TG and TC, and BMI were significantly higher in the hypertensive groups than in the control group (P<0.01).
Table 1. Baseline characters of the investigated participants.
| Variables | Case | Control | P-value |
| Number | 905 | 905 | |
| Male/Female | 392/513 | 392/513 | |
| Age (y) | 56.91±7.37 | 56.60±7.51 | P = 0.38 |
| BMI(Kg/m2) | 24.65±3.24 | 23.21±2.86 | P<0.01 |
| HDL (mM) | 1.41±0.35 | 1.41±0.32 | P = 0.72 |
| TC (mM) | 5.34±1.00 | 5.17±0.93 | P<0.01 |
| TG (mM) | 2.02±1.68 | 1.63±1.12 | P<0.01 |
| Smoking habit | 173 | 147 | P = 0.11 |
| Alcohol abuse | 152 | 148 | P = 0.80 |
BMI, body mass index; HDL, high-density lipoprotein; TC, total cholesterol; TG, triglyceride.
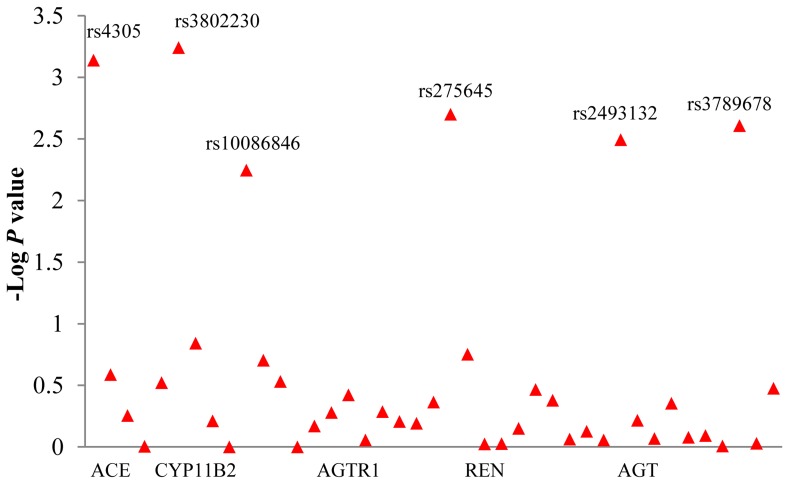
The P value of 41 tagSNPs within RAAS genes were shown in Figure 1. The rs10935724 within AGTR1 and rs6414 within CYP11B2 were failed in genotyping, and the genotyping success rate of other 39 SNPs was 99%. Two of 39 SNPs, rs3789678 and rs10086846, deviated from Hardy-Weinberg equilibrium (P<0.05). However, the genotype distribution and MAF of these two SNPs were similar to those of HapMap CHB. To exclude the genotyping error, we randomly regenotyped 20% of the samples for these two SNPs by Tm-shift genotyping method [25], the results were same. Therefore, they were still included in the following analyses. In addition, with the prevalence, odds ratio (OR), and MAF in this study, the Genetic Power Calculator (available online http://pngu.mgh.harvard.edu/~purcell/gpc/) indicated that the sample size is big enough to do case-control analysis with 80% power [26]. According to the chi-square test P values (P<0.05) and odds ratios, rs3789678 and rs2493132 within AGT, rs4305 within ACE, rs275645 within AGTR1, rs3802230 and rs10086846 within CYP11B2 were shown to associate with hypertension (Table 2). No significant association was found between polymorphisms within REN and hypertension. The genotype information for the remaining 35 SNPs that did not reach significance in the association analyses were shown in Table S2. After Bonferroni correction, only rs4305 and rs3802230 were still significant, the other 4 SNPs were marginally significant.
Figure 1. Negative Log P values for the association of 41 single-nucleotide polymorphisms in 5 candidate genes of renin-angiotensin-aldosterone system with hypertension.
The P values were obtained from the comparison of two allele frequencies. Labeled SNPs had a P value less than 0.01.
Table 2. Genotype distributions of those SNPs significantly associated with hypertension.
| SNP | Gene | Group | Genotype | MAF | P Value | OR | 95% CI | ||
| rs3789678 | AGT | Case | 690 (CC) | 159 (CT) | 45 (TT) | 0.14 | 0.002 | 1.32 | 1.10–1.58 |
| Control | 626 (CC) | 236 (CT) | 41 (TT) | 0.18 | |||||
| rs2493132 | AGT | Case | 373 (CC) | 410 (CT) | 111 (TT) | 0.35 | 0.003 | 1.23 | 1.07–1.40 |
| Control | 316 (CC) | 452 (CT) | 137 (TT) | 0.40 | |||||
| rs4305 | ACE | Case | 155 (AA) | 441 (AG) | 300 (GG) | 0.36 | 0.001 | 1.26 | 1.10–1.44 |
| Control | 103 (AA) | 450 (AG) | 348 (GG) | 0.42 | |||||
| rs275645 | AGTR1 | Case | 455 (AA) | 395 (AG) | 43 (GG) | 0.27 | 0.002 | 1.27 | 1.09–1.48 |
| Control | 535 (AA) | 333 (AG) | 37 (GG) | 0.22 | |||||
| rs3802230 | CYP11B2 | Case | 80 (AA) | 348 (AC) | 470 (CC) | 0.28 | 0.001 | 1.28 | 1.13–1.48 |
| Control | 89 (AA) | 428 (AC) | 385 (CC) | 0.34 | |||||
| rs10086846 | CYP11B2 | Case | 469 (CC) | 277 (CT) | 151 (TT) | 0.32 | 0.006 | 1.21 | 1.06–1.39 |
| Control | 413(CC) | 318 (CT) | 172 (TT) | 0.37 | |||||
The P values were obtained from the comparison of two allele frequencies. OR, odds ratio; CI, confidence interval.
Considering the effect of confounding variables, we further carried out logistic regression analysis with genetic and non-genetic factors. The result showed that rs2493132, rs10086846, and TC were no longer associated with hypertension (P>0.05, Table 3).
Table 3. Logistic regression for genetic and non-genetic factors.
| Variables | B | P value | Exp (B) | 95% CI |
| rs3789678 | 0.216 | 0.033 * | 1.241 | 1.02–1.51 |
| rs2493132 | 0.131 | 0.108 | 1.140 | 0.97–1.34 |
| rs4305 | 0.242 | 0.001 * | 1.273 | 1.10–1.47 |
| rs275645 | 0.276 | 0.001 * | 1.318 | 1.11–1.56 |
| rs3802230 | 0.313 | 0.011 * | 1.368 | 1.08–1.74 |
| rs10086846 | 0.063 | 0.551 | 1.065 | 0.87–1.31 |
| BMI | 0.143 | 0.000 * | 1.154 | 1.12–1.19 |
| TC | 0.077 | 0.149 | 1.080 | 0.97–1.20 |
| TG | 0.138 | 0.001 * | 1.148 | 1.06–1.25 |
| Constant | −5.432 | 0.000 * | 0.004 |
The P values of 6 SNPs were obtained from the comparison of two allele frequencies. BMI, body mass index; TC, total cholesterol; TG, triglyceride.
P value was less than 0.05.
Moreover, MDR was used to analyze the interaction among ‘significant’ SNPs and non-genetic risk factors for hypertension. After input the genotypes of 6 SNPs together with information about TG, TC, and BMI, the software output the best model for ‘BMI and rs4305’ with 10/10 Cross-validation Consistency (Table 4). In order to delineate how BMI and rs4305 interacts to cause hypertension, we carried out crossover analysis. The result showed that both BMI and the A allele of rs4305 increased the susceptibility to hypertension, but BMI had the main effect (Table 5). The stratified analysis further showed that when BMI ≥25, the A allele of rs4305 has no association with hypertension [P = 0.85, OR = 1.02, 95% confidence interval (CI) = 0.81–1.30] (Table 6). However, when BMI <25, the A allele showed significant association with hypertension (P<0.001, OR = 1.41, 95% CI = 1.19–1.66) (Table 6), which also indicates that BMI has the major effect, and rs4305 had a minor effect.
Table 4. MDR analysis of gene-environment interaction.
| Best model | TestingAccuracy | Testing Sensitivity | Testing Odds Ratio | Testing X2 | Cross-validation Consistency |
| BMI | 0.59 | 0.43 | 2.26 (95%CI: 1.20–4.27) | 6.50 (P = 0.011) | 10/10 |
| BMI, rs4305 | 0.60 | 0.53 | 2.32 (95%CI: 1.27–4.23) | 7.62 (P = 0.006) | 10/10 |
| BMI, rs275645, rs3802230 | 0.57 | 0.53 | 1.82 (95%CI: 1.01–3.29) | 3.94 (P = 0.047) | 5/10 |
Table 5. Crossover analysis of interaction between BMI and rs4305.
| BMI | Allele | Case | Control | P Value | OR | 95% CI |
| <25 | G | 581 | 876 | 1 | 1 | NA |
| ≥25 | A | 300 | 172 | <0.001 | 2.63 | 2.12–3.26 |
| ≥25 | G | 460 | 270 | <0.001 | 2.57 | 2.14–3.09 |
| <25 | A | 451 | 484 | <0.001 | 1.41 | 1.19–1.66 |
The P values were obtained from the comparison of two allele frequencies. OR, odds ratio; CI, confidence interval.
Table 6. Stratified analysis of interaction between BMI and rs4305.
| BMI | Group | GG | GA | AA | G | A | P Value | OR | 95% CI |
| <25 | Case | 160 | 261 | 95 | 581 | 451 | P<0.001 | 1.41 | 1.19–1.66 |
| Control | 271 | 334 | 75 | 876 | 484 | ||||
| ≥25 | Case | 140 | 180 | 60 | 460 | 300 | P = 0.85 | 1.02 | 0.81–1.30 |
| Control | 77 | 116 | 28 | 270 | 172 |
The P values were obtained from the comparison of two allele frequencies. OR, odds ratio; CI, confidence interval.
Discussion
Since RAAS is the most important mechanism regulates blood pressure in our body [27], genes encoding components of this system have been strong candidates for the investigation of the genetic basis of hypertension and major targets for antihypertensive drugs [28]. However, previous studies mainly focused on limited polymorphisms, thus we carried out a case-control study to systemically investigate the association between polymorphisms in the RAAS genes and essential hypertension. The present study identified several novel genetic variants in the RAAS genes that may play critical roles in BP regulation and susceptibility for hypertension. According to Chi-square test and logistic regression analysis, rs3789678 within AGT, rs4305 within ACE, rs275645 within AGTR1, rs3802230 within CYP11B2 were shown to associate with hypertension.
Similar to this study, some of the susceptibility SNPs were also found to be associated with hypertensive traits in previous studies. The rs4305 has been related to the risk of hypertension (P = 3.0×10−5), and associated with SBP (P = 4.6×10−4) and DBP (P = 6.0×10−5) in a study compromising 23 cohorts of three independent studies [Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE); Global BPgen Consortium; and Women’s Genome Health Study] and a total of 86,588 participants [29]. Furthermore, a recent GWAS for ACE enzyme activity found strong association of rs4343 with increased activity in the Han Chinese (P = 3.0×10−25), and the mean ACE activity among subjects with G allele of rs4343 increased by 3.5 IU/L per copy of the allele [30]. The G allele of rs4343 is in strong linkage disequilibrium (LD) with A allele of rs4305 (HapMap JPT/CHB D’ = 0.97, r2 = 0.80), suggesting a potential common link between the studies. In the present study, A allele of rs4305 increases the susceptibility to hypertension, which might be associated with increased ACE activity.
Prior to our study, Chen et al. also studied the association of CYP11B2 gene and essential hypertension in southwest Han Chinese population [31]. Four tag SNPs (rs4536, rs4545, rs3097, and 3802230) within the CYP11B2 gene were selected through HapMap. In addition, C344T (rs1799998) and K173R (rs4539) polymorphisms that previous studies were mostly interested, were also selected for the study. The result showed that among the six SNPs, only the C allele of rs3802230 was significantly more prevalent in the hypertension subjects than in the control subjects (P = 0.006, OR = 1.28, 95% CI: 1.07–1.52). Since the results of both studies were similar, we further calculated the pooled P value and OR. The combined P = 0.001, OR = 1.20, 95% CI = 1.08–1.34, I2 = 0.0% (P = 0.39), which means no heterogeneity existed between two studies, and the C allele of rs3802230 might be a risk factor for essential hypertension in the Han Chinese population. Chen et al. also analyzed these SNPs in Yi and Hani Minorities of China, and found rs4536 was significantly associated with hypertension in the Hani minority, however, no association was found in the Yi minority [32].
Pickering and colleagues have initially suggested that hypertension and blood pressure are complex traits [33], and previous epidemiologic studies have found dozens of risk factors, such as obesity, high-fat diets, smoking, alcohol abuse, excessive salt intake, mental stress, and others to associate with high blood pressure [34]–[36]. There is growing evidence that interactions among multiple genes and environmental factors may play an important role in determining the susceptibility to various common diseases including hypertension [37]. Our previous study indicated that interaction analysis might give a little more information than the single genetic study [38]. In the present study, high BMI and serum TG level were confirmed as risk factors for hypertension by logistic regression analysis. The MDR analysis further demonstrated that the interaction between BMI and rs4305 was associated with hypertension. Since BMI represents the internal metabolic and physiological environment that plays a key role in development of high blood pressure [39], and ACE is one of the most important target for design of anti-hypertensive drugs, it’s not surprising that the interaction of them may play an important role in the susceptibility to hypertension. Previous genetic epidemiologic study also found the interactions between MMP3 gene polymorphism rs679620 and BMI in predicting blood pressure in African-American women with hypertension [40]. The recent important genetic studies are mainly carried out in well-organized cohorts like Global BPgen, CHARGE, and GenSalt Study, which means the epidemiologic data are readily available [19], [41]–[43]. With the development of statistic methods for evaluation of gene-environment interaction, we can expect more missing inheritability to be found [44], [45].
In conclusion, we identified several genetic variants in the RAAS genes that were significantly associated with hypertension in the Han Chinese population. Most notable, the interaction between BMI and rs4305 increased the susceptibility to hypertension, meanwhile BMI has a major effect, and rs4305 has a minor effect.
Supporting Information
All 41 tagSNPs within genes coding for RAAS.
(DOC)
Genotype distributions of 35 SNPs not associated with hypertension.
(DOC)
Acknowledgments
The authors acknowledge Ms. Shifang Wu, Chunlin Zhu, and Jieqiong Jin from Kunming Institute of Zoology, Chinese Academy of Sciences, for helpful technical support.
Funding Statement
This study was financially supported by Zhejiang Natural Science Foundation (LQ13C060001), Ningbo Natural Science Foundation (2012A610237), Research Project of Zhejiang Education Department (Y201224146), as well as the KC Wong Magna Fund in Ningbo University. The funders had no role in the study design, data collection, analysis, decision to publish or preparation of the manuscript.
References
- 1. Carretero OA, Oparil S (2000) Essential hypertension. Part I: definition and etiology. Circulation 101: 329–335. [DOI] [PubMed] [Google Scholar]
- 2. Joseph PG, Pare G, Anand SS (2013) Exploring gene-environment relationships in cardiovascular disease. Can J Cardiol 29: 37–45. [DOI] [PubMed] [Google Scholar]
- 3. Simino J, Rao DC, Freedman BI (2012) Novel findings and future directions on the genetics of hypertension. Curr Opin Nephrol Hypertens 21: 500–507. [DOI] [PubMed] [Google Scholar]
- 4. Brand SM (2012) Genetics, genomics and other molecular approaches: example of salt-sensitive hypertension. J Hypertens 30: 877–879. [DOI] [PubMed] [Google Scholar]
- 5. Basson J, Simino J, Rao DC (2012) Between candidate genes and whole genomes: time for alternative approaches in blood pressure genetics. Curr Hypertens Rep 14: 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Franceschini N, Reiner AP, Heiss G (2011) Recent findings in the genetics of blood pressure and hypertension traits. Am J Hypertens 24: 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schmieder RE, Hilgers KF, Schlaich MP, Schmidt BM (2007) Renin-angiotensin system and cardiovascular risk. Lancet 369: 1208–1219. [DOI] [PubMed] [Google Scholar]
- 9. Konoshita T, Genomic Disease Outcome Consortium Study I (2011) Do genetic variants of the Renin-Angiotensin system predict blood pressure response to Renin-Angiotensin system-blocking drugs?: a systematic review of pharmacogenomics in the Renin-Angiotensin system. Curr Hypertens Rep 13: 356–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He J, Gu D, Kelly TN, Hixson JE, Rao DC, et al. (2011) Genetic variants in the renin-angiotensin-aldosterone system and blood pressure responses to potassium intake. J Hypertens 29: 1719–1730. [DOI] [PubMed] [Google Scholar]
- 11. Wang L, Hou L, Li H, Chen J, Kelly TN, et al. (2010) Genetic variants in the renin-angiotensin system and blood pressure reactions to the cold pressor test. J Hypertens 28: 2422–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ji LD, Zhang LN, Shen P, Wang P, Zhang YM, et al. (2010) Association of angiotensinogen gene M235T and angiotensin-converting enzyme gene I/D polymorphisms with essential hypertension in Han Chinese population: a meta-analysis. J Hypertens 28: 419–428. [DOI] [PubMed] [Google Scholar]
- 13. Pereira TV, Nunes AC, Rudnicki M, Yamada Y, Pereira AC, et al. (2008) Meta-analysis of the association of 4 angiotensinogen polymorphisms with essential hypertension: a role beyond M235T? Hypertension 51: 778–783. [DOI] [PubMed] [Google Scholar]
- 14. Mottl AK, Shoham DA, North KE (2008) Angiotensin II type 1 receptor polymorphisms and susceptibility to hypertension: a HuGE review. Genet Med 10: 560–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sookoian S, Gianotti TF, Gonzalez CD, Pirola CJ (2007) Association of the C-344T aldosterone synthase gene variant with essential hypertension: a meta-analysis. J Hypertens 25: 5–13. [DOI] [PubMed] [Google Scholar]
- 16. Kurtz TW (2010) Genome-wide association studies will unlock the genetic basis of hypertension.: con side of the argument. Hypertension 56: 1021–1025. [DOI] [PubMed] [Google Scholar]
- 17.Dominiczak AF, Munroe PB (2010) Genome-wide association studies will unlock the genetic basis of hypertension: pro side of the argument. Hypertension 56: 1017–1020; discussion 1025. [DOI] [PubMed]
- 18. Ehret GB (2010) Genome-wide association studies: contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep 12: 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. International Consortium for Blood Pressure Genome-Wide Association S, Ehret GB, Munroe PB, Rice KM, Bochud M, et al (2011) Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature 478: 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Farahani P, Dolovich L, Levine M (2007) Exploring design-related bias in clinical studies on receptor genetic polymorphism of hypertension. J Clin Epidemiol 60: 1–7. [DOI] [PubMed] [Google Scholar]
- 21. Beeks E, Kessels AG, Kroon AA, van der Klauw MM, de Leeuw PW (2004) Genetic predisposition to salt-sensitivity: a systematic review. J Hypertens 22: 1243–1249. [DOI] [PubMed] [Google Scholar]
- 22. Staessen JA, Li Y, Thijs L (2007) Meta-analysis of blood pressure and the CYP11B2 polymorphism highlights the need for better designed studies. J Hypertens 25: 37–39. [DOI] [PubMed] [Google Scholar]
- 23.Bell PA, Chaturvedi S, Gelfand CA, Huang CY, Kochersperger M, et al.. (2002) SNPstream UHT: ultra-high throughput SNP genotyping for pharmacogenomics and drug discovery. Biotechniques Suppl: 70–72, 74, 76–77. [PubMed]
- 24. Motsinger AA, Ritchie MD (2006) The effect of reduction in cross-validation intervals on the performance of multifactor dimensionality reduction. Genet Epidemiol 30: 546–555. [DOI] [PubMed] [Google Scholar]
- 25. Wang J, Chuang K, Ahluwalia M, Patel S, Umblas N, et al. (2005) High-throughput SNP genotyping by single-tube PCR with Tm-shift primers. Biotechniques 39: 885–893. [DOI] [PubMed] [Google Scholar]
- 26. Purcell S, Cherny SS, Sham PC (2003) Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19: 149–150. [DOI] [PubMed] [Google Scholar]
- 27. Marteau JB, Zaiou M, Siest G, Visvikis-Siest S (2005) Genetic determinants of blood pressure regulation. J Hypertens 23: 2127–2143. [DOI] [PubMed] [Google Scholar]
- 28. Nebert DW (1997) Polymorphisms in drug-metabolizing enzymes: what is their clinical relevance and why do they exist? Am J Hum Genet 60: 265–271. [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson AD, Newton-Cheh C, Chasman DI, Ehret GB, Johnson T, et al. (2011) Association of hypertension drug target genes with blood pressure and hypertension in 86,588 individuals. Hypertension 57: 903–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chung CM, Wang RY, Chen JW, Fann CS, Leu HB, et al. (2010) A genome-wide association study identifies new loci for ACE activity: potential implications for response to ACE inhibitor. Pharmacogenomics J 10: 537–544. [DOI] [PubMed] [Google Scholar]
- 31. Chen B, Nie S, Luo S, Zhang W, Xiao C (2011) Association of the human CYP11B2 gene and essential hypertension in southwest Han Chinese population: a haplotype-based case-control study. Clin Exp Hypertens 33: 106–112. [DOI] [PubMed] [Google Scholar]
- 32. Chen B, Nie S, Yue Z, Shou W, Xiao C (2011) Haplotype-based case-control study of the human CYP11B2 gene and essential hypertension in Yi and Hani minorities of China. Biochem Genet 49: 122–137. [DOI] [PubMed] [Google Scholar]
- 33. Pickering GW, Keen H, Rose G, Smith A (1959) The nature of essential hypertension. Lancet 2: 1027–1030. [PubMed] [Google Scholar]
- 34. Kotchen TA (2010) Obesity-related hypertension: epidemiology, pathophysiology, and clinical management. Am J Hypertens 23: 1170–1178. [DOI] [PubMed] [Google Scholar]
- 35. Wang TJ, Vasan RS (2005) Epidemiology of uncontrolled hypertension in the United States. Circulation 112: 1651–1662. [DOI] [PubMed] [Google Scholar]
- 36. Horan MJ, Lenfant C (1990) Epidemiology of blood pressure and predictors of hypertension. Hypertension 15: I20–24. [DOI] [PubMed] [Google Scholar]
- 37. Kunes J, Zicha J (2006) Developmental windows and environment as important factors in the expression of genetic information: a cardiovascular physiologist’s view. Clin Sci (Lond) 111: 295–305. [DOI] [PubMed] [Google Scholar]
- 38.Xu J, Ji LD, Zhang LN, Dong CZ, Fei LJ, et al.. (2012) Lack of association between STK39 and hypertension in the Chinese population. J Hum Hypertens. [DOI] [PubMed]
- 39. Feng RN, Zhao C, Wang C, Niu YC, Li K, et al. (2012) BMI is strongly associated with hypertension, and waist circumference is strongly associated with type 2 diabetes and dyslipidemia, in northern Chinese adults. J Epidemiol 22: 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Taylor J, Sun YV, Chu J, Mosley TH, Kardia SL (2008) Interactions between metallopeptidase 3 polymorphism rs679620 and BMI in predicting blood pressure in African-American women with hypertension. J Hypertens 26: 2312–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, et al. (2009) Genome-wide association study of blood pressure and hypertension. Nat Genet 41: 677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, et al. (2009) Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet 41: 666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. GenSalt Collaborative Research G (2007) GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens 21: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mukherjee B, Ahn J, Gruber SB, Chatterjee N (2012) Testing gene-environment interaction in large-scale case-control association studies: possible choices and comparisons. Am J Epidemiol 175: 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kazma R, Babron MC, Genin E (2011) Genetic association and gene-environment interaction: a new method for overcoming the lack of exposure information in controls. Am J Epidemiol 173: 225–235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All 41 tagSNPs within genes coding for RAAS.
(DOC)
Genotype distributions of 35 SNPs not associated with hypertension.
(DOC)