Summary
The intrinsic period of circadian clocks is their defining adaptive property. To identify the biochemical mechanisms whereby casein kinase1 (CK1) determines circadian period in mammals, we created mouse null and tau mutants of Ck1 epsilon. Circadian period lengthened in CK1ε−/−, whereas CK1εtau/tau shortened circadian period of behaviour in vivo and suprachiasmatic nucleus firing rates in vitro, by accelerating PERIOD-dependent molecular feedback loops. CK1εtau/tau also accelerated molecular oscillations in peripheral tissues, revealing its global role in circadian pacemaking. CK1εtau acted by promoting degradation of both nuclear and cytoplasmic PERIOD, but not CRYPTOCHROME, proteins. Together, these whole animal and biochemical studies explain how tau, as a gain-of-function mutation, acts at a specific circadian phase to promote degradation of PERIOD proteins, and thereby accelerate the mammalian clockwork in brain and periphery.
Keywords: Casein kinase 1, Circadian, PERIOD protein, stability
Introduction
In mammals, daily rhythms of sleep and metabolism are driven by the circadian pacemaker within the suprachiasmatic nuclei of the hypothalamus (SCN) which coordinates the activity of sub-ordinate circadian clocks in other brain regions and in peripheral tissues (Reppert and Weaver, 2002) (Hastings et al., 2003) (Lowrey and Takahashi, 2004) (Saper et al., 2005). Precise maintenance of circadian period is the defining adaptive property of these clocks. In both brain and periphery, a central concept is that circadian clocks consist of interlocked auto-regulatory feedback loops in which CLOCK:BMAL1 heterodimers bind to E-box DNA sequences contained within genes encoding the transcriptional repressors PER and CRY. Following their accumulation in the cytoplasm, PER:CRY complexes translocate to the nucleus after a delay of several hours and repress the activity of constitutively bound CLOCK:BMAL1 complexes. These inhibitory complexes are then degraded following a further delay, and the de-repression of CLOCK:BMAL1 activity initiates the next circadian cycle of Per and Cry transcription. Mutations in various elements of this core molecular clock have been described, many of which result in either arrhythmia or alteration in period of rest/activity and sleep cycles.
In addition to periodic transcription, post-translational modifications of clock proteins are crucial for the correct operation of molecular clockworks (Lee et al., 2001) (Gallego and Virshup, 2007). In particular, reversible phosphorylation provides a potential mechanism for the regulated formation of protein complexes, their nuclear entry and ultimate degradation via ubiquitination pathways, each step of which introduces delays into the feedback loop. Current interest has focussed on the role that protein phosphorylation may play in tuning the circadian oscillator to a period of 24 hours. In Drosophila, mutations of CK1 (doubletime, DBT) were the first circadian mutants discovered to involve an enzyme and resulted in either short (dbts) or long (dbtl) rhythms of eclosion and locomotor activity (Kloss et al., 1998) (Price et al., 1998).
The tau mutation of Syrian hamsters was the first mammalian circadian mutation discovered. It causes significant shortening of activity cycles to 20 hours and assorts in a Mendelian fashion (Ralph and Menaker, 1988). Subsequent mapping studies revealed that this mutation resided within casein kinase 1ε as a C to T transversion in position 178 (Lowrey et al., 2000). Protein expression studies suggested a destabilisation of nuclear PER in the tau mutant hamster (Dey et al., 2005), while recent modelling and biochemical studies suggested that tau is a gain of function mutation on specific residues within the PER protein target (Gallego and Virshup, 2007), likely leading to increased degradation, altered stability and accelerated protein turnover. It is unknown whether these latter conclusions, developed in cultured cells and mathematical modelling studies, reflect the real circumstances in living animals. Further, it is unclear whether altered degradation of PER involves cellular compartmentalisation, whether it is tissue specific (neural vs peripheral), and ultimately how it leads to accelerated molecular timing in the SCN.
To define the role of CK1ε in the circadian clockwork, and hence the molecular disturbance underlying advanced sleep phase disorder and accelerated circadian period, we describe here the generation of a mouse model of the hamster tau mutation within CK1 ε. We adapted the loxP-Cre strategy to produce mice carrying the tau allele of the Ck1ε gene and used subsequent cre-mediated in vivo disruption of exon 4, which encodes the catalytic domain of CK1ε (Lowrey et al., 2000) to generate a Ck1ε null allele. This allelic series was assayed by circadian behavioural wheel-running and SCN electrophysiological studies and revealed that tau acts as a gain of function mutation, shortening period in a dose dependent manner. By using PER2 protein reporters (Yoo et al., 2004), we also define, for the first time, the impact of this mutation on the molecular dynamics of both the SCN and peripheral timers, and within the SCN, highlight the role of accelerated protein turnover in shortening circadian period and activity/rest cycles. Finally, western blotting and imaging studies of both transfected cell lines, primary fibroblasts and SCN tissue derived from CK1tau mice reveal that CK1tau specifically targets PERIOD but not CRY proteins during the early nocturnal phase of the circadian cycle, acting on one component of the negative feedback element of the clock, and leading to an acceleration of period by an asymmetric degradation of PER proteins at a specific phase of the circadian cycle.
Results and Discussion
Generation of a tau mutant allele for CK1ε and subsequent disruption
We adapted the loxP-cre recombinase strategy used for creating a conditional knockout to produce a mouse carrying the tau allele of the ck1ε gene. This can be subsequently converted in vivo to a Ck1ε gene knockout by cre-mediated ablation of exon 4, which encodes the catalytic domain of CK1ε (Lowrey et al., 2000) (Figure S1A). Deletion of exon 4 leads to a frame-shift and premature stop codon. The linearised targeting vector was electroporated into ES cells and G418 resistant, homologously recombined ES clones (Figure S1B) subjected to transient transfection with cre recombinase followed by FIAU selection. Resistant colonies that had deleted the selection cassette but retained the mutant tau exon were identified by PCR (Figure S1C) and used to generate germ-line transmitting chimeras. The presence of the tau mutation (the C to T transversion at amino acid position 178) in the founder transgenic mice was confirmed by sequencing of animal-derived genomic DNA through the tau site (Figure S1D). F1 mice carrying the floxed tau allele were crossed with a deletor cre mouse to generate the Ck1ε knockout line. Gene knockout was confirmed genetically by genomic DNA sequencing of the Ck1ε gene as well as by RT-PCR using brain cDNA as templates (Figure S1E). The absence of the CK1ε protein was also confirmed by western blotting in whole-brain extracts using an antibody against the C-terminus of CK1ε (Figure S1F). Mice carrying the tau allele (tau/+ and tau/tau) or the knockout allele (−/+ and −/−) were viable and fertile with no overt deleterious phenotype.
The tau mutation shortens and the null mutation of CK1ε lengthens period
All wild-type and knock-out mice entrained to LD 12hL:12hD exhibited activity in the dark phase, commencing shortly after lights off. In contrast, tau mice exhibited variable responses to LD 12L:12D cycles, and in a sample of 30 animals, 9 exhibited a phase advance of the LD cycle, 5 exhibited masking with onsets coincident with dark onset and 16 animals free-ran across the prevailing LD cycle (Figure S2, A-C). To determine the impact of CK1εtau on free-running behaviour, we monitored wheel-running activity in constant darkness (DD). CK1εtau mice exhibited significant shortening of period in a dose-dependent manner (Figure 1A and 1B): 23.6 ±0.05h for wild-type (WT), 21.8 ±0.08h for heterozygote (CK1εtau/+) and 20.0 ±0.07h for homozygotes (CK1εtau/tau). This is equivalent to 1.80h/copy of CK1εtau and virtually identical to the behavioural phenotype of this mutation in Syrian hamsters (6). In contrast, knock-out mice exhibited a small but significant period lengthening compared to their WT counterparts (24.0 ±0.06 vs 23.6 ±0.05h respectively, p<0.001; Figure 1A and 1B). Importantly, mice carrying a single copy of the knockout allele exhibited a WT-like free-running period of 23.7 ±0.05h, while the tau hemizygote (CK1εtau/−, 21.6 ±0.09h) and heterozygote (CK1εtau/+) mice exhibited virtually identical periods (Figure 1A and 1B). This shows that a single WT copy of Ck1ε provides little protective effect in the face of a tau allele, and thus offers definitive genetic evidence that tau acts as a gain-of-function mutation.
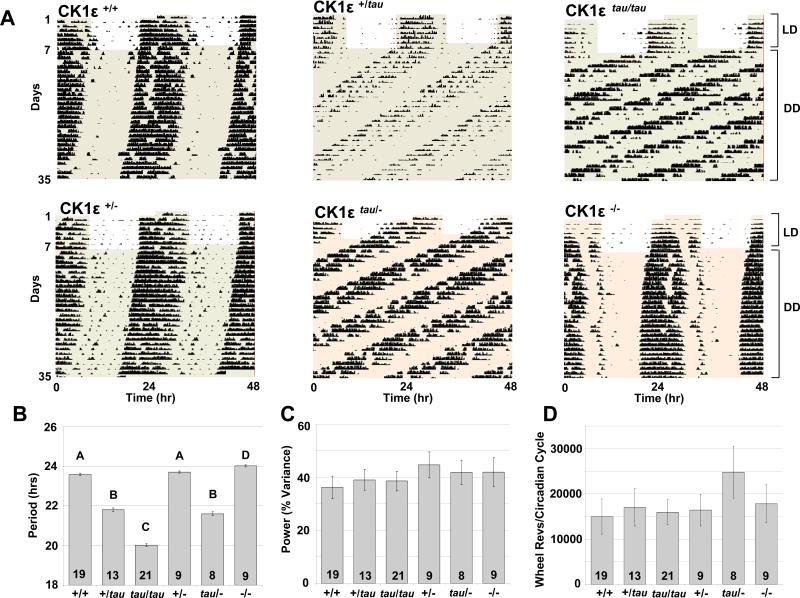
Figure 1. Locomotor activity rhythms for wild type, tau mutant and knock-out mice.
(A) Representative wheel-running activity records (actograms) for wild type (CK1ε +/+; top-left panel), heterozygous (CK1ε +/tau; top-middle panel), homozygous (CK1ε tau/tau; top-right panel), hemizygous (CK1ε +/−; bottom-left panel), hemizygous tau (CK1ε tau/−; bottom-middle panel), and CK1ε knock-out mice (CK1ε −/−; bottom-right panel) are shown in double plotted format. Each horizontal line represents 48 hrs, with the second day plotted to the right and one cycle below the first cycle. Animals were run in 24hr cycles of 12hr Light, 12hr Dark (LD) and then in continuous darkness (DD). The timing of the LD cycles is indicated by the alternating white and grey areas of the actogram.
(B) Periodogram estimates of period for each genotype as mean ± SEM; the number of animals is indicated within each bar. Bars with different letters show a significant difference (p < 0.001 ANOVA and post-hoc Bonferroni compared to wild-type); bars with the same letter are not significantly different.
(C) Circadian amplitude (power from periodogram analyses) for each genotype. There was no significant difference between genotypes (mean±SEM).
(D) Activity levels during the first 4 weeks of DD for each genotype as determined by total wheel-running revolutions per circadian cycle. There were no significant differences between genotypes despite shortened overall circadian period (mean±SEM).
Interestingly, the absolute number of wheel-revolutions per circadian cycle was similar in all genotypes, suggesting period acceleration in the tau mutants may occur due to an asymmetric effect on overall circadian organisation, perhaps sparing nocturnal activity per se (Figure 1C and 1D). Similar effects are reported for circadian mutations in DBT (Price et al., 1998), the CK1 ortholog in Drosophila. The asymmetric acceleration of circadian timing was extended in studies of metabolic rhythms in the CK1εtau and WT mice using indirect calorimetry. Here, we noted that there was a compression of circadian rhythms of oxygen consumption (VO2) in CK1εtau mice which closely matched the periods determined earlier by wheel-running (Figure 2). Remarkably, the pattern of oxygen consumption was highly asymmetric such that the shortening of period was almost entirely associated with a compression of the inactive (subjective day) phase (active phase duration: WT =12.6 ±0.1h; CK1εtau =13.4 ±0.6h; inactive phase duration: WT =11.4 ±0.1h; CK1εtau =6.9 ±0.5h; p < 0.01, Figure 2B and 2C). Hence, our data suggest that the absolute duration of nocturnal physiology and behaviour were unaffected by the CK1εtau mutation, rather, nocturnal events were initiated earlier in CK1εtau than in WT. Such observations are compatible with our earlier demonstration in tau mutant hamsters of an advanced onset of circadian nocturnal melatonin secretion, with the result that overall duration of secretion of this nocturnal hormone is identical to that of wild types (Lucas et al., 1999). Collectively, these data suggest that CK1εtau may accelerate period by compressing a specific phase of the circadian behavioural and physiological cycle. In the case of the knock-out mice, the circadian phenotype was mild (ca 18 minute extension in period). Although we have not undertaken measures of oxygen consumption in knock-out mice, our analysis of the relative proportion of the circadian cycle in which knock-out animals are active (alpha) and the overall structure of the circadian activity cycle revealed that they were not significantly different to wild-types.
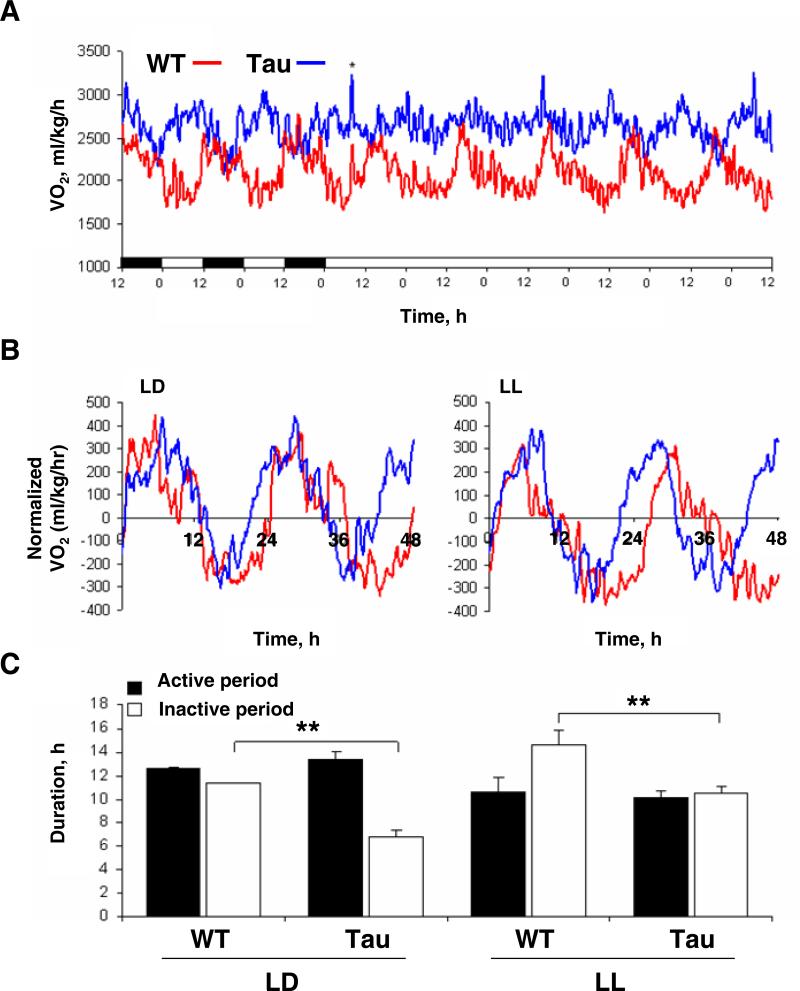
Figure 2. CK1εtau causes asymmetric acceleration of circadian timing in metabolic rhythms.
(A) Representative recordings of oxygen consumption rhythms (VO2) in WT (red) and Tau mutant (blue) mice across a segment of the light-dark (LD) and constant light (LL) cycles, showing an accelerated period in Tau mice. * indicates cages cleaned.
(B) Representative profiles of VO2 for Tau and WT normalized to the midpoint (50%) of the trough to peak and peak to trough amplitudes. Rhythms are double plotted over a 48 hr time-base. The ‘active’ and ‘inactive’ portions of the cycle were defined as the portions of the cycle above or below zero (respectively).
(C) Duration of the active (black) and inactive (open) phases of the circadian cycle (mean±SEM). Duration of active phase was maintained in Tau mice in both LD and LL, when compared with WT mice. In contrast, the inactive (subjective day) portion of the cycle was reduced in Tau mice under both light conditions. As expected, both genotypes exhibited a lengthening of the inactive period when switched from LD to LL. (**p<0.01 post-hoc Bonferroni).
Individual neurons of the SCN exhibit an accelerated firing rate in CK1εtau mutants
In order to extend our studies to the SCN, we adopted the use of a novel suction electrode method which permits discrimination of firing rate profiles for individual cells for over 48h in vitro (Brown et al., 2006). Using acutely prepared SCN slices, we recorded extra-cellular multi-unit electrical activity in animals in which circadian periods had previously been recorded in running-wheels in DD, permitting comparison of individual behavioural periods to SCN cellular rhythms. In all genotypes (CK1ε+/+, CK1εtau/+, CK1εtau/tau, CK1ε−/−), there was a close concordance with their period of multiunit recording in each slice (Figure 3A). SCN single unit recordings revealed a similar outcome (Figure 3A), with a highly significant correlation (r2 = 0.83, p<0.01) between rhythms of single unit firing rates and behaviour across all 4 genotypes (Figure 3B), indicating that effects of CK1εtau on behavioural period likely arose from corresponding changes in circadian patterns of electrical activity of SCN neurons (Brown et al., 2005). The pattern of electrical activity of the knock-out was normal in terms of amplitude and firing frequency, indicating that gene loss did not alter fundamental aspects of SCN electrophysiological properties. Thus, our generation of CK1εtau in mice and subsequent gene deletion has resulted in a novel allelic series of circadian period, ranging from 20.0 (CK1εtau/tau) to 24.0h (CK1ε−/−), and provides an unprecedented resource to explore the central role of CK1 to circadian rhythm regulation in mammals.
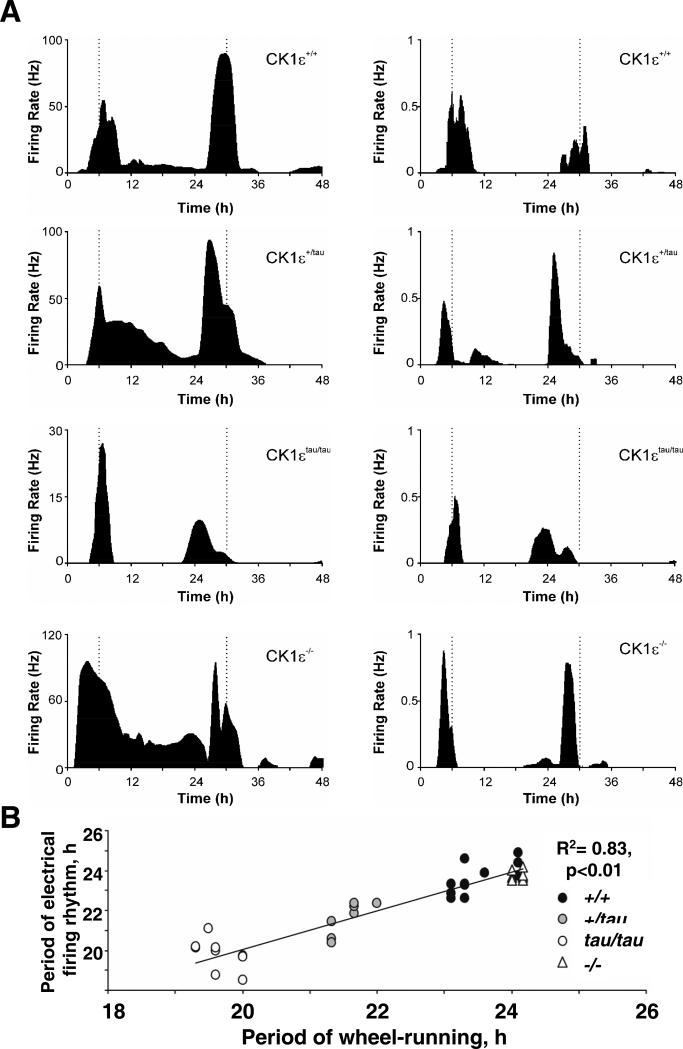
Figure 3. CK1ε mutations alter period of SCN electrical firing rhythm.
(A) Representative SCN Multi-unit (LH) and Single unit (RH) action potential discharge rhythms recorded from wildtype (CK1ε+/+), heterozygote (CK1ε+/tau), homozygote (CK1εtau/tau) and knockout mutant (CK1ε−/−) brain slices.
(B) Correlation between period of wheel-running behavior determined in DD before slice preparation and subsequent single unit SCN firing rates for WT (closed circles), tau heterozygote (grey circles) and homozygote (open circles) and null animals (triangles). Data are derived from 11 out of 13 CK1ε+/+ SCN neurons, 8/9 CK1ε+/tau, 11/12 CK1εtau/tau and 9/10 CK1ε−/− cells that exhibited detectable firing rate rhythms.
Impact of CK1εtau/tau on the molecular pacemaker of the SCN and peripheral tissues
Accelerated circadian periods of activity/rest cycles and also SCN firing rates likely reflect altered dynamics within molecular feedback loops that underpin circadian time keeping within the SCN. To determine the impact of CK1ε on SCN molecular oscillators, CK1εtau mice were crossed into PER2::Luciferase (PER2::LUC) protein fusion reporter mice (Yoo et al., 2004). Organotypic SCN slices were prepared from 5-10 day old pups and luciferase activity recorded using photomultiplier tubes (PMT's) over a minimum of 7-10 cycles. Bioluminescence recorded from PER2::LUC SCN slices closely matched behavioural rhythms for each genotype (Figure 4A and 4B): 24.4 ±0.15h, 21.9 ±0.08h and 20.2 ±0.17h in CK1ε+/+, CK1εtau/+, CK1εtau/tau slices respectively (ANOVA, F=226.5 p<0.01). To extend these studies to the single cell level, we recorded PER2 expression of individual SCN neurons measured using a CCD camera. Single neuron imaging showed that circadian period of single neurons also matched genotype (24.9 ±0.09, 22.1 ±0.04 and 20.5 ±0.05h for CK1ε+/+, CK1εtau/+, CK1εtau/tau respectively), and that CK1εtau had no effect on the inter-cellular synchrony nor regional distribution of circadian gene expression across the SCN (Figure 4C). Therefore, these bioluminescence recordings reveal that both the behavioural and electrophysiological impact of the CK1εtau mutation is underpinned by accelerated molecular time-keeping within individual SCN neurons.
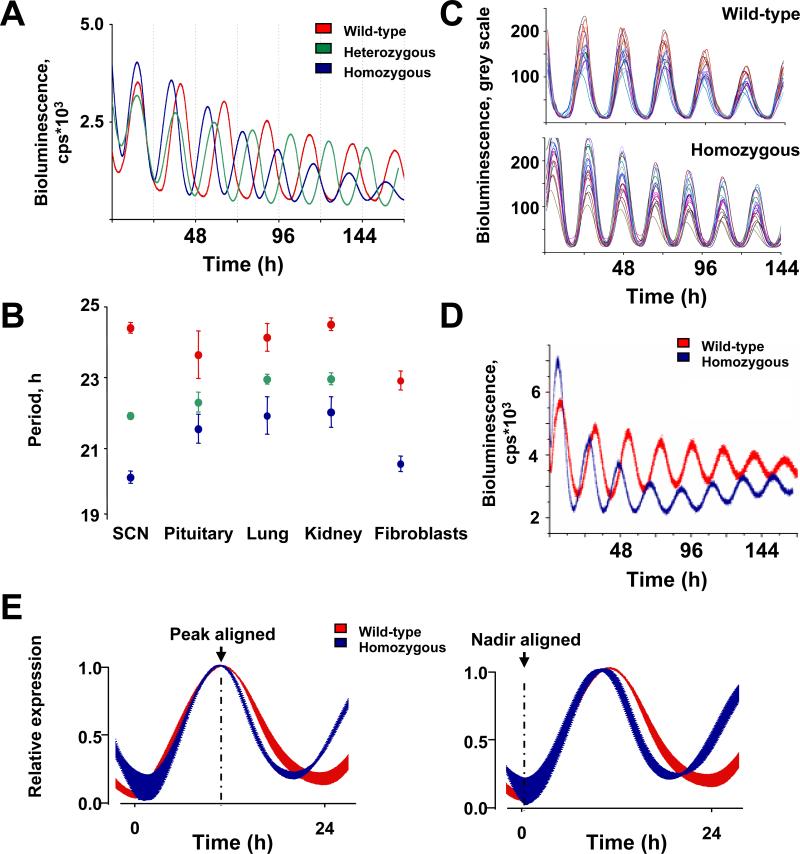
Figure 4. CK1εtau has a global impact on circadian pacemaking, accelerating molecular circadian oscillators in SCN, peripheral tissues and primary fibroblasts.
(A) Representative PER2::LUC bioluminescence oscillations in WT (red), heterozygote (green) and homozygote (blue) organotypic SCN slices.
(B) The periodicity of tissues from WT, heterozygote and homozygote tau PER2::LUC mice (mean±SEM; WT, n=4-6; heterozygotes, n=10-11; homozygote, n=5-6).
(C) CCD recordings of SCN PER2::LUC expression at the single cell level for WT and homozygote SCN slices (20 per slice; n=3 slices for each genotype).
(D) Representative traces of PER2::LUC expression in primary fibroblast cultures of WT (red) and tau mutant (blue) mice.
(E) Waveform alignments by nadir or peak for PER2::LUC rhythms from WT (red) and homozygote (blue) organotypic SCN slices. Data were normalised and then plotted as mean±SEM (n=6 per genotype). Note selective reduction in peak to nadir interval regardless of alignment.
It is unknown whether endogenous CK1ε has a global role in setting clock speed in tissues outside the nervous system. We therefore recorded circadian PER2::LUC activity in organotypic slices of pituitary, lung and kidney and primary lung fibroblast cultures. This revealed clear circadian expression of PER2 and, on a group basis, a significant shortening of period by CK1εtau (Figure S3A, 4B and 4D). In contrast to the SCN, however, the magnitude of effect was variable across tissues, indicating partial penetrance of the mutation. Moreover, in lung and kidney the tau allele significantly accelerated the rate of dampening of the molecular cycle (Figure S3B). Thus, our in vitro data show that CK1εtau accelerates pace-making globally, although its detailed impact on SCN and peripheral oscillators is tissue-specific.
We hypothesized from our tau mutant hamster studies that CK1εtau may shorten period by accelerating the loss of nuclear PER proteins specifically in the early nocturnal phase (Dey et al., 2005), leading to earlier de-repression of BMAL1 and CLOCK, and overall circadian acceleration. This “asymmetric acceleration” model of PER protein turnover in CK1εtau mutants was tested in our bioluminescence recordings of SCN slices. By aligning SCN waveforms from WT and CK1εtau (Figure 4E) by either peak or nadir of expression, it is evident that the mutation significantly accelerates the decline of PER2::LUC (time to fall to 50% level WT = 5.3 ±0.3h, CK1εtau/tau = 3.9 ±0.2h, ANOVA F=8.97, p<0.01). Consequently, the cycle in CK1εtau homozygote slices is advanced by ca 3.4h compared to WT over the interval of PER protein decline (hours from peak to trough), whereas over the rising phase CK1εtau/tau has little effect. Additional statistical confirmation of this distorted waveform came from analysis of skewness (Figure S3C), slices with shorter periods having a more negative skew due to asymmetric distortion of waveform. Our data show that most of the accelerated degradation of PER within the SCN occurs at a specific phase and that this acceleration is consistent with, and can account for, the period changes observed in wheel-running behaviour.
CK1εtau selectively accelerates PER degradation independently of sub-cellular localization
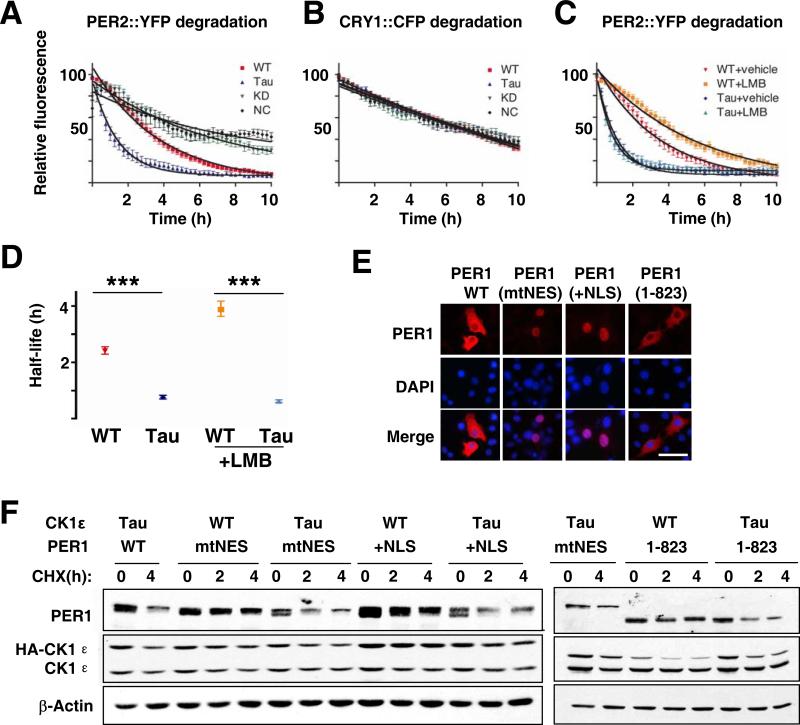
These data indicate that CK1εtau accelerates the onset of nocturnal processes (but not affecting their absolute duration) by accelerating the clearance of endogenous SCN PER2 proteins predominantly in the early circadian night. To explore the biochemical basis of this effect, we used real-time fluorescence videomicroscopy to monitor PER2::YFP degradation in COS-7 cells following blockade of de novo protein synthesis by treatment with cycloheximide (CHX, 20 μg/ml). Co-expression with WT but not kinase-dead CK1ε accelerated PER2::YFP protein degradation, relative to vector control. Degradation was further accelerated by CK1εtau (Figure 5A). In contrast, CRY1::CFP degradation was not affected by any variants of CK1ε, indicating a selective action of CK1ε on PER2 (Figure 5B). Inhibition of nuclear export by leptomycin B (LMB) retained PER2::YFP in the nucleus and slowed its degradation in the presence of WT CK1ε. In contrast LMB did not alter the acceleration of PER2 degradation by CK1εtau (Figure 5C and 5D), confirming a nuclear site of action of this mutation. LMB could alter degradation rates by blocking PER nuclear export, or by altering trafficking of other components required for PER degradation. To differentiate between these possibilities, to extend the range of cell types investigated and to determine the effect of CK1εtau on PER1, we expressed in NIH 3T3 cells WT or mutant forms of PER1 in which either the nuclear export signal was mutated (mtNES) or an additional copy of the nuclear localization signal was added (+NLS). Both mutations trapped most of the PER1 within the nuclear compartment (>65% for mtNES; >88% for +NLS) as determined by immunocytochemisty (Figure 5E). CK1εtau accelerated degradation, not only of wtPER1 as previously reported (Gallego et al., 2006) but also of nuclear-trapped PER1 (residual PER1 at 4 h post-CHX treatment: CK1εtau vs. WT, 45% and 81% for mtNES; 39% and 66% for +NLS, Figure 5F). In addition, CK1εtau accelerated degradation of another mutant form of PER1 (PER1 1-823) which lacked the C-terminus that contains an active NLS as well as the CRY binding site (Vielhaber et al., 2001) and retained PER1 predominantly in the cytoplasm (>87%; Figure 5E and 5F). Together, these in vitro data clearly show that CK1εtau targets both PER1 and PER2 but not CRY1 proteins, and this selective targeting occurs regardless of nuclear or cytoplasmic localisation.
Figure 5. The tau mutation selectively targets PER proteins for accelerated degradation independently of their cellular localisation.
(A, B) Decay of PER2::YFP or CRY1::CFP fluorescence in COS-7 cells co-transfected with WT (red), tau (Tau, blue), kinase-dead (KD, green) CK1ε or empty plasmid (NC, black). Cells were treated with CHX (20 μg/ml) 0.5 hour before recording.
(C) Decay of PER2::YFP fluorescence in COS-7 cells treated with vehicle or leptomycin B (LMB, 10ng/ml) one hour before CHX (20 μg/ml) (red; CK1ε WT+vehicle, yellow; WT+ LMB; blue tau mutant+vehicle, green; tau mutant+LMB). (A-C, individual cell data normalized to maximum and minimum intensity over recording, plotted as mean±SEM, n=14-33, 2-4 independent experiments).
(D) Half lives for PER2::YFP in presence of WT or tau CK1ε, treated with vehicle or LMB (mean±95% confidence limits, ***p < 0.001).
(E) Differential nuclear or cytoplasmic retention of Myc-tagged WT and mutant PER1 in NIH 3T3 cells. Scale bar= 500 Dm.
(F) Western blots of Myc-tagged wild-type mPER1 or with mutated NES (mtNES), with extra NLS (+NLS) or truncated C-terminus removing native NLS (1-823), co-expressed with HA-tagged CK1ε WT or tau mutant in NIH 3T3 cells treated with CHX (40 μg/ml). Data shown are representative of at least three separate experiments.
CK1εtau also selectively accelerates endogenous PER degradation in primary cells and SCN slices
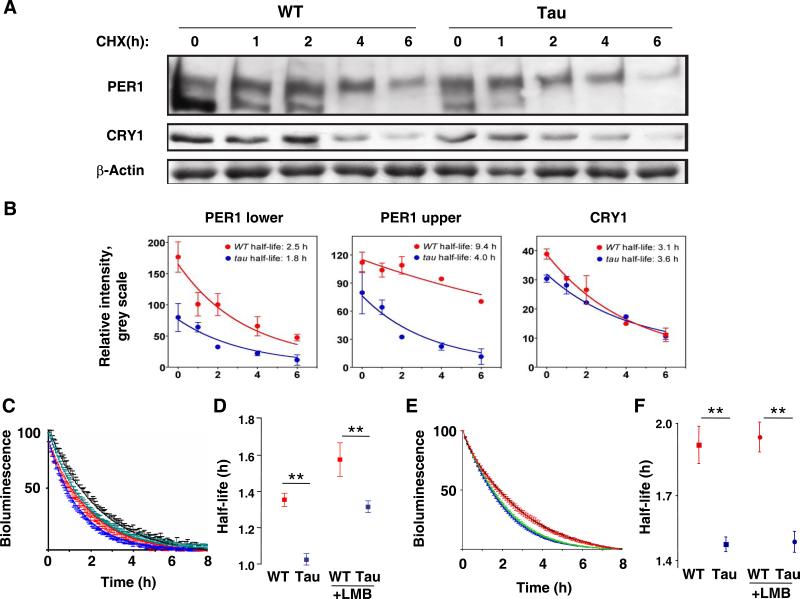
We extended these studies of recombinant protein stability by using Western blots to examine native PER1 and CRY1 expression in WT and mutant primary fibroblast cultures. PER1 proteins typically exhibit two clear bands which represent hypo- (lower band) and hyper-phosphorylated (upper band) forms of PER1 (Lee et al., 2001) (Figure 6A). In tau mutant fibroblasts, the overall levels of PER1 were lower than those seen in WT cells. However, upon treatment with CHX (20 μg/ml), the hyper-phosphorylated forms of PER1 displayed a significantly shorter half-life in tau mutant cells than in WT cells (WT: 9.4 h vs. tau: 4.0 h, p<0.05 using nonlinear exponential decay analysis with GraphPad) (Figure 6B). This is consistent with the idea that hyper-phosphorylated PER1 may be preferentially targeted to the proteasome for degradation (Gallego and Virshup, 2007). In contrast, the overall expression level and CHX-stimulated decline of CRY1 was unaffected by CK1εtau (Figure 6A and 6B). To obtain a more dynamic view of PER degradation we then exploited real-time recording of primary fibroblasts treated with CHX at the peak of PER2::LUC expression. This revealed that CK1εtau significantly shortened (P<0.01) the half-life of PER2::LUC expression, by ca. 25 min. Moreover, even though pre-treatment with LMB extended the half-life in both genotypes, CK1εtau was still effective at accelerating PER2::LUC degradation (P<0.01) (Figure 6C and 6D), consistent with the recombinant protein studies of a nuclear action for the mutation. Finally, we tested PER2::LUC degradation in the physiologically relevant context of the SCN slice, again adding CHX at the peak of circadian expression. As with the fibroblasts, loss of PER2::LUC bioluminescence was more rapid in mutant than WT SCN slices (Figure 6E and 6F), half-life again being shortened by ca. 25 min. Moreover, the effect of CK1εtau remained following pre-treatment with LMB, which had no significant effect on PER2::LUC clearance in either genotype. Together, these data demonstrate that CK1εtau facilitates the degradation of endogenous PER (but not CRY) proteins in SCN and primary cells, and that blockade of nuclear export does not attenuate the tau phenotype.
Figure 6. Degradation of endogenous PER is accelerated in tissue and cells from tau mutant mice.
(A) Representative western blots of endogenous PER1 and CRY1 in lung fibroblasts from WT or tau mice, treated with CHX (20 μg/ml) for 0, 1, 2, 4 and 6 hours. Note two distinct bands for PER1.
(B) Group data (mean±SEM, Red=WT, Blue=mutant) from western blots reveal significant reduction in expression and earlier clearance of PER1 in mutant fibroblasts (F-test, p<0.05 for upper band), but no effect of tau on expression of endogenous CRY1 (F-test, p=0.39). (C) Decay of PER2::LUC bioluminescence from lung fibroblasts, exposed to CHX in presence or absence of LMB. Data are normalized to the peak level of expression (time 0 after CHX) and the minimum after 8 hours, and plotted as mean±SEM; red= WT+vehicle, black= WT+LMB, blue= tau+vehicle, green= tau+LMB).
(D) Half-lives for PER2::LUC bioluminescence from fibroblasts (mean±SEM). CK1Dtau significantly accelerated loss of PER2::LUC signal regardless of LMB treatment. **p<0.01 by t-test.
(E) Decay of PER2::LUC bioluminescence from SCN slices, exposed to CHX in the presence or absence of LMB. Data expressed as in (C).
(F) Half-lives for PER2::LUC bioluminescence from SCN slices (mean±SEM). CK1Dtau significantly accelerated loss of PER2::LUC signal regardless of LMB treatment. **p<0.01 by t-test.
Conclusions
In mammals, different forms of CK1 (CK1ε and CK1δ), act on diverse pathways, including Wnt signalling, neurotransmitter regulation as well as circadian rhythms. Both CK1ε and CK1δ complex with PER and CRY (Lee et al., 2001), while CK1-mediated phosphorylation of BMAL1 increases its transcriptional activity (Eide et al., 2002). In humans, circadian familial advanced sleep disorders (FASPS) are known to be caused by a serine to glycine substitution in the hPER2 gene at position 662 (Toh et al., 2001) and also by a T44A mis-sense mutation in human CK1δ, which causes hypophosphorylation of PER2 (Xu et al., 2005). Transgenic mice over-expressing the hPER2 mutation exhibit shorter free-running periods, mimicking FASPS (Xu et al., 2007), and their phenotype is sensitive to CK1δ: increased dosage of CK1δ shortens their period further. These studies, together with recent in-vitro analyses of the equivalent serine residue on mouse PER2 protein (Vanselow et al., 2006) suggest that expression, degradation, nuclear entry and export of PER2 are modulated by multiple states of phosphorylation and that CK1 activity plays a central role at these various check-points of the circadian cycle (Gallego and Virshup, 2007) (Mignot and Takahashi, 2007).
By creating null and tau mutations of CK1ε in mice, we provide behavioural, neurophysiological and cellular evidence showing that CK1εtau acts as a gain-of-function that accelerates the molecular dynamics of circadian time-keeping. These biochemical actions explain the behavioural and physiological phenotype of the mutation in both hamsters and mice. They also provide a new perspective with which to understand the action of PER2 and CK1 mutations which have been linked to circadian period and sleep disturbances in humans (Xu et al., 2005) (Xu et al., 2007). Our data therefore confirm and extend the earlier modelling-based studies, later tested in cell line studies (Gallego et al 2006). Moreover, we show that this acceleration is a consequence of enhanced rates of degradation of PER proteins at specific phases of the circadian cycle i.e. early circadian night, and that such asymmetric effects on PER degradation are translated into altered phasing of physiological and behavioural circuits. Our studies also reveal that CK1εtau can act on both nuclear and cytoplasmic targets, but suggest that within the SCN, CK1-mediated degradation may be primarily a nuclear event, as predicted by our studies of the hamster mutant (Dey et al., 2005). We also show that the knock-out of Ck1ε has a relatively mild phenotype (extends the circadian period by ca 18 minutes a day) and that a single wild-type allele offer no significant protection in the presence of the tau allele. A clear implication therefore is that Ck1δ may phosphorylate target PERIOD proteins, and that wild-type Ck1ε may thus be partially redundant in the circadian timing mechanisms. Resolution of these issues awaits characterization of the phenotypes of Ck1δ knock-outs.
An intriguing feature of our data is that the duration of molecular night is relatively compressed due to the accelerated clearance of PER2 after its peak expression at CT12, but the duration of nocturnal behavioural and physiological processes (CBT, melatonin secretion) is apparently normal, as measured in solar time. Consequently, because the offsets are not governed by the molecular cycle, they occur several hours later than the compressed molecular cycle would predict: nocturnal processes spilling over into the start of the molecular day. Hence the behavioural day (defined by activity offset and onset) is shorter in the mutant, even though the underlying determining change is shortening of the molecular night. These strongly asymmetric effects of CK1εtau on molecular and behavioural timing likely underlie the pronounced disturbances of circadian timekeeping (dampened amplitude, altered phasing) widely observed in the peripheral tissues of tau mutant hamsters (Dey et al., 2005) and may contribute to poor growth and general morbidity associated with the mutation in hamsters (Lucas et al., 2000). The variable responses of peripheral tissues to the tau mutation are intriguing, and may suggest tissue-specific differences in the relative expression of Ck1ε or Ck1δ, or cellular differences in the manner in which PERIOD proteins are trafficked and degraded.
Endogenous PER (but not CRY) proteins (PER1 by western blot, PER2 by bioluminescence recording) are destabilized in the tau mutant background. These effects on protein half-life occur in a context (treatment with cycloheximide) where transcription is irrelevant and thus the observed protein instability is sufficient to explain the behavioural phenotype in full. This point is reinforced by our original findings in the tau mutant hamster (Dey et al., 2005) which showed that the Per mRNA cycle in the SCN mapped to circadian time whereas the PER protein expression cycle was accelerated in vivo due to early clearance. These results from the kinase mutants therefore provide a valuable counterpoint to those Xu et al. (2007) who modeled human FASPS by creating mice carrying transgenes encoding mutated versions of the human PER2 protein. The S662G mutation in hPER2, which leads to one form of FASPS in patients, shortened circadian period but the apparent degradation rate of PER2 protein was unaltered. Rather, expression of hPer2 mRNA was reduced, leading to lower hPER2 protein levels. Our findings with the kinase mutant therefore add further support to the speculation of Xu et al. (2007) that effects mediated via S662 may accelerate the clock by reduced transcription, whereas the tau mutation, as a gain-of-function of CK1ε, targets a separate domain within PER2 which is responsible for accelerated degradation.
The lack of impact of CK1εtau on CRY dynamics complements the recent description of loss of function mutations on F-box proteins, which mediate actions on CRY degradation, resulting in a longer half-life of CRY proteins (Godinho et al., 2007) (Busino et al., 2007) (Siepka et al., 2007). As a result, circadian period is lengthened. These studies, and data reported here further suggest that mechanisms for the degradation of these two core circadian transcription factor families (PER and CRY) operate independently, but must have tightly co-evolved in order to achieve precise overall control on circadian period. Our new mouse model therefore provides new insight into how a circadian mutation accelerates behavioural and cellular circadian rhythms, in both the brain and peripheral tissues, and demonstrates that temporally specific effects on PER degradation can be tracked through to an altered circadian structure of behaviour and physiology.
Experimental procedures
Animal maintenance
All experiments were conducted under the aegis of the 1986 Home Office Animal Procedures Act (UK) and following local ethical review. All animals were reared at 2022 °C, and maintained on standard rodent breeder or maintenance chow under 12hr Light: 12hr Dark lighting schedules.
Antibodies and plasmids
Monoclonal anti-CK1ε antibody was obtained from BD Transduction Laboratories, and anti-Myc (9E10) antibody from Santa Cruz Biotechnology. Anti-PER1 and Anti-CRY1 antibodies were a kind gift from Choogon Lee. Polyclonal anti-Myc antibody (9106) was from ABCAM and anti-α-Tubulin (T6199) and anti-actin (A2066) from Sigma. Myc-PER1 constructs were cloned in PCS2+MT vector and HA-tagged kinases (CK1 ε WT and tau mutant) were cloned in pCEP4 (Invitrogen). The SV40 Tag NLS (PKKKRKVG) was added to the 5' end of Myc-mPER1 by PCR (Gallego et al., 2006). Full-length mPer2 cDNA with C-terminal enhanced yellow fluorescent protein tag derived from pEYFP-N1 (Clontech) was cloned into pcDNA3.1/V5/HisA vector (Invitrogen) (PER2::YFP). Full-length mCry1 cDNA was cloned into enhanced cyan fluorescent protein vector pECFPN1 (Clontech) (CRY1::CFP). Tau mutation plasmid for transfection was prepared by site directed mutagenesis leading to a substitution of arginine 178 (*C*GC) by cysteine (*T*GC). Kinase-dead variant was prepared by substitution of lysine at position 38 (AAG) by alanine (GCC). Ck1ε plasmid was generously donated by Achim Kramer (Humboldt-university, Berlin, Germany). Wild-type, tau or kinase-dead variants of full-length Ck1ε cDNA were cloned into pcDNA-DEST40 vector (Invitrogen) (WT, Tau, KD).
Generation of transgenic mice
The targeting construct contained exons 2-6 of the Ck1ε gene (Figure S1) was prepared using sv129 genomic DNA fragments from a PAC clone. Briefly, an 8.7 kb Sal I – Xba I fragment encompassing exons 2-6 was subcloned into pBluescript (KS+) and verified by end-sequencing. The 1.4 kb Bgl II – Eco RI fragment containing exon 4 was used to introduce the tau mutation (C12555T) by site-directed mutagenesis and verified by sequencing. A floxed NeoTK selection cassette was inserted into the EcoR I site in the intron 4 and a double stranded oligo encoding lox P and Nco I sites ligated into the Bgl II site 5’ of exon 4 (Figure S1A). The targeting construct was linearized with Not l, electroporated into 129/Sv R1 ES cells (http://www.mshri.on.ca/nagy/r1.htm) and genomic DNA from G418 resistant clones isolated as described previously (Talts et al., 1999). Bgl II digested DNA was Southern blotted and hybridized with 32P-labelled external probe, amplified from genomic DNA using the following primers: forward 5’ACTTCCTTCTCTTTACCCACG’3; reverse 5’AGAAGCAAGCATCCATTTCC’3. The floxed selection cassette was deleted from homologouslly recombined ES cell clones by transient transfection with a cre-expressing plasmid followed, 48 hours later, by 5 days of FIAU (1-(2-deoxy-2-fluoro-β-D-arabino-furanosil)-5-iodouracil) selection (Abuin et al., 1996). FIAU-resistant colonies were expanded and genomic DNA analyzed by PCR as described below to determine whether they were tau or knockout alleles. ES cell clones carrying the tau allele (tau/+) were used to create germ-line transmitting chimeras by blastocyst injection. tau/+ mice were crossed with littermates to produce homozygous tau offspring allele or crossed with a deletor cre line to generate the Ck1ε knockout allele in vivo. Wild type (+) and tau alleles were genotyped by PCR using primers ‘a’ (5’CACCTGGGCATTGGTGAGT’3) and ‘b’ (5’GGAGGTCAAGGGGCCAGT’3) (Figure S1A). The presence of the knockout (−) allele was detected using primer ‘c’ (5’GTGGTAGAAGGAGAGAACTGAC’3) and ‘b’ (see above). In all, 11 chimeric mice derived from 3 clones were used to generate a founder line of Ck1ε mice, which were then introgressed over 6 generations into a C57/B6 background.
RT-PCR
Total RNA from brain of CK1ε knockout mice and wild-type siblings was isolated using Trizol (Invitrogen). To exclude genomic DNA contamination, total RNA was digested with DNase I (Roche, Germany). 1 μg of total RNA was reverse transcribed using OligodT oligonucleotides by the SuperScript™ II RNase H-Reverse Transcription kit (Invitrogen). In parallel, negative control samples lacked Reverse Transcriptase. 2μl of the resultant first strand cDNA was PCR-amplified with appropriate primer pairs using a hot-master PCR kit (Eppendorf) as follows: 1) 96 °C for 5 mins. 2) 30 × 94 °C for 20 s, 55 °C for 10 s, 72 °C for 50 s. 3) 72 °C for 7 min. PCR products were visualized on 1.5 % agarose gel. Primer pairs were based on mouse sequences as follows (all 5’-3’): Ck1ε forward, GCCTCTGGTGAGGAAGTAG; reverse, CGGTAGGGAATATGCTGGTG. Gapdh forward, CCTTCATTGACCTCAACTAC; reverse,GGAAGGCCATGCCAGTGAGC.
Western Blotting of CK1ε
Protein extracts were prepared from whole brain of wild type, CK1εtau/tau knockout and CK1ε mice. Mice were euthanized by cervical dislocation, whole brains removed and frozen on dry ice. Tissues were mechanically homogenised at 4 °C in 3 volumes of extraction buffer (0.1 M KCl, 20 mM HEPES [pH 7.5], 5 mM EDTA, 1 mM dithiothreitol, 0.1% Triton X-100, 5% glycerol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM MgCl2, 10 μg/ml of aprotinin, 5 μg/ml of leupeptin, 1 μg/ml of pepstatin A). Homogenates were cleared by centrifugation (2x 10 mins, 13000 × g at 4 °C). Supernatants were mixed with 2•protein loading buffer, boiled and spun at 13000 × g for 1 min at room temperature. Total protein (70 μg) was resolved on 10% denaturing PAGE gels and transferred to nitrocellulose membrane (Bio-Rad). Membranes were blocked with 5% skimmed milk in Tris-buffered saline containing 0.05% Tween-20 and incubated with relevant antibodies. ECL plus detection was used (Amersham Biosciences).
Behavioural Analysis
6-16 week old mice were single-housed in cages equipped with running wheels and ad libitum food and water, contained within a light-tight chamber at constant temperature (°C) and humidity (% H2O). Mice were entrained to a 12hr light and 12hr dark (LD) cycle for 7 days and released into constant darkness (DD) for ~28 days. Wheel revolutions were recorded in 10 mins time bins by DataQuest III acquisition program (Data Sciences Inc., St. Paul, MN), transferred to the TauActo 1.2 analysis software (developed in house by T. Brown) to produce double-plotted actograms, periodogram, intensity of running(alpha/rho) and power of rhythm. χ2 periodogram analyses were performed using 28 days of data in DD.
Electrophysiological recordings
Electrophysiological recordings were conducted and analyzed as previously described (Brown et al., 2006). Mice (housed with running wheels in DD) were killed at the beginning of the inactive phase and coronal brain sections (350 μm thick) were cut using a vibroslicer (Campden Instruments, Leicester, UK) and transferred to an interface style brain slice chamber continuously perfused (~1.5 ml/min) with oxygenated (95% O2 / 5% CO2) aCSF supplemented with 0.0005% gentamicin (Sigma, UK) and warmed to 36 ± 1 °C. Extracellular multiunit activity (MUA: signal-to-noise > 2:1) was recorded from the SCN using aCSF-filled suction electrodes, differentially amplified (x20,000) and bandpass filtered (300-3000Hz) via a Neurolog system (Digitimer, Welwyn Garden City, UK), digitized (25,000 Hz) using a micro 1401 mkII interface (Cambridge Electronic Design (CED), Cambridge, UK) and recorded on a PC running Spike2 version 5 software (CED). Single unit activity was discriminated from MUA recordings offline on the basis of waveform shape and validated by measurement-based clustering and the presence of a clear refractory period in an interspike interval histogram. Firing rate data were moderately smoothed using a 1h moving average and presented as the mean firing rate each min. The circadian period of single cell and MUA rhythms, were analyzed by curve-fitting developed in house (Clockwise; Dr T. Brown). Initially data were normalized such that data spanned a range of values between 100 and −100. The normalized data were then fit with the equation Y=Asin(B(x+C)) using the Newton-Raphson iterative method, where A equaled the amplitude of the rhythm, B equaled the period in radians/h and C determined the phase. Initial values of A, B and C were estimated from the best fitting curve of a series of > 3000 standard curves that spanned the period range 3-34h and exhibited a range of different amplitudes and phasing.. Peak widths were defined as the duration over which firing rate was above the mean for that cell.
Metabolic rhythm measurement and analysis
Indirect calorimetry: Tau and WT mice (n=6 per genotype) were single-housed in indirect calorimetric cages (Columbus Instruments, Columbus, OH, USA). Mice were monitored for a minimum of 10 days in LD followed by a minimum of 5 days in constant light (LL), during which time oxygen consumption (VO2) was measured every 10 min. Illustrations of VO2 traces in Figure 2 depict group (genotype) averages taken from a representative experiment. Analysis of metabolic rhythms: Circadian rhythms in VO2 were analyzed using curve-fitting software (Clockwise) as described above. Recordings from individual animals during LD and LL portions of the experiment were analyzed separately. Once period length was determined, a composite circadian cycle of metabolic rate was generated for each animal from the entire test period (a separate composite was generated for each lighting condition). From these representative circadian profiles, ‘active’ and ‘inactive’ portions of the cycle were determined by normalizing metabolic rate to the midpoint (50%) of the peak to trough amplitude, calculated separately for the rise and fall of the rhythm. Duration of active and inactive phases in LD and LL were analyzed using repeated measures Two-way ANOVA, with a Bonferonni's post hoc test.
Bioluminescence recording and imaging
Whole slice bioluminescence emission and individual cellular emission were recorded as described (Maywood et al., 2006) (Prosser et al., 2007) using photomultiplier assemblies and CCD cameras supplied by Hamamatsu (UK). Waveforms of rhythmic bioluminescence emission from whole slices and individual cells were analyzed in BRASS software and RAP algorithm.
Cell isolation, culture and transfection
For culture of primary lung fibroblast, lungs were removed from euthanized adult mice for peripheral fibroblast preparation. Briefly, lung was chopped and minced in sterile conditions and washed twice in chilled modified Hank's Balanced Salt Solution (Sigma). Cells were then dissociated by means of shaking at 37 °C for 2 hrs in 100 u/ml of Collagenase IA (Sigma) (dissolved in PBS) and 3mM CaCl2. Once dissociated, cells were filtered through a sterile nylon mesh followed by 2 washes and centrifugation in chilled Hank's solution. Pellets were re-suspended in culture medium (DMEM with 4.5 g/L glucose, Glutamax and pyruvate, supplemented with 10% FBS, 100 U/ml Penicillin and 100 μg/ml streptomycin) and plated on a 10cm culture dish. Cultures were maintained at 37 °C (5% CO2) for 2-3 days until confluent and ready for splitting. Confluent cells in 35 mm dishes were synchronized with Dexamethasone (100 nM) for 1 hr prior to PMT recording of PER2::LUC bioluminescence (Izumo et al., 2003).
NIH 3T3 cells were grown in DMEM supplemented with 10% calf serum and antibiotics. At 90% confluence in 6-well plates, cells were transfected with relevant plasmid combinations using Lipofectamine 2000 (Invitrogen). The amount of transfected DNA was 0.8 μg of Myc-Per1 and 0.4 μg encoding HA-tagged kinases for each well. COS-7 cells were grown in DMEM supplemented with 10% calf serum and antibiotics. At 8090% confluence in glass-bottomed dishes (MatTek), cells were transfected with combinations of relevant plasmids using 3 μl of GeneJuice (Novagen). The total amount of transfected DNA was 1μg (0.6 μg of Per2::YFP + 0.4 μg of CK1 ε or 0.5 μg of Cry1::CFP + 0.5 μg of CK1ε) for each dish. 24 hours after transfection, DMEM was exchanged for air medium and subjected to live cell fluorescent imaging (Maywood et al., 2006).
Protein degradation assay
For real-time monitoring of PER2::YFP and CRY1::CFP degradation, COS-7 cells were treated with 20 μg/ml cycloheximide (Sigma), with or without LMB pre-treatment (10 ng/ml for 1 hr, Calbiochem). Fluorescence was recorded every 15 minutes for at least 17 hours by time-lapse microscopy using Openlab software (Improvision Single cell fluorescent data were analyzed by IPlab software (Scanalytics) and one-phase exponential decay curve fitting. Half-life calculations and statistics were performed in Prism (GraphPad Software). Data were plotted as mean± SEM with fitted exponential decay curve. Half-lives are plotted as mean± 95% confidence interval of exponential decay curve fitting.
For detection of Myc-tagged PER1 degradation by western blotting, 20 h after transfection, cells were treated with CHX (40 μg/ml) for 0, 2 and 4 hours and then lysed in lysis buffer (150 mM NaCl/20 mM Hepes, pH 7.5/0.1% Nonidet P-40/1 mM EDTA/2 mM DTT) supplemented with 1× Complete protease inhibitor mixture (Roche Applied Science). The cells were then mechanically sheared, and lysates were centrifuged at 16,000 × g for 10 min at 4°C. Total protein (100 μg) was resolved in SDS/PAGE, and then subject to Western blotting. For endogenous PER1 and CRY1 degradation, primary lung fibroblasts were treated with CHX (20 μg/ml) for 0, 1, 2, 4 and 6 hours, then subjected to Western blotting using anti-PER1 and anti-CRY1 antibodies. These experiments were duplicated. For real-time monitoring of PER2::LUC degradation by PMT, SCN slices or lung fibroblasts were treated with CHX (40 μg/ml) with or without LMB pre-treatment (10 ng/ml for 1 hr). For control experiments, DMSO (for CHX) or ethanol (for LMB) was used as vehicle.
Immuno-stainning of PER1 and mutants in NIH 3T3 cells
Twenty hours after transfection of different PER1 constructs, cells were re-plated on glass cover slips. Twenty four hours later cells were washed three times with phosphate-buffered saline (PBS), and fixed with 3.7% paraformaldehyde for 15 min. After three more washes, cells were permeabilized with 0.1% Triton X-100 in PBS for 5 min and washed with 3% BSA/0.1% TX-100/PBS for 30 min. Cells were incubated with an anti-Myc polyclonal antibody for 1 h in 3% BSA/0.1% TX-100/PBS, washed three times and incubated with a secondary Alexa Fluor 594 goat anti-rabbit IgG (Molecular Probes, Inc.) for 1 h. Cells were washed three and incubated with DAPI (1 μg/ml) in 0.1% TX100/PBS. Cells were visualized using an Olympus AX70 fluorescence microscope (magnification • 60), and images were digitally recorded via an AxioCam (Carl Zeiss MicroImaging, Inc.) and saved using the AxioVision 3.1 program. Overlays of the immunofluorescence images were performed in Photoshop (Adobe).
Supplementary Material
Acknowledgements
We thank Tracey Butcher, Graham Morrissey, Emma Owens, Sarah Atkinson, Helen Lydon, Jian Li, Julie Gibbs and Shin Yamazaki for technical assistance. We thank Russell Foster, Owen Jones and Rob Lucas for comments on the manuscript, Simon Luckman for access to the metabolic monitoring equipment (CLAMS) and Andrew Millar for BRASS software package, Masahiro Ishiura for RAP software package. We thank A. Kramer for kind provision of research materials. The project was supported by the Biotechnology and Biological Sciences Research Council and Medical Research Council of the UK for funding.
References
- Abuin A, Bradley A. Recycling selectable markers in mouse embryonic stem cells. Mol Cell Biol. 1996;16:1851–1856. doi: 10.1128/mcb.16.4.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Banks JR, Piggins HD. A novel suction electrode recording technique for monitoring circadian rhythms in single and multiunit discharge from brain slices. J Neurosci Methods. 2006;156:173–181. doi: 10.1016/j.jneumeth.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Brown TM, Hughes AT, Piggins HD. Gastrin-releasing peptide promotes suprachiasmatic nuclei cellular rhythmicity in the absence of vasoactive intestinal polypeptide-VPAC2 receptor signaling. J Neurosci. 2005;25:11155–11164. doi: 10.1523/JNEUROSCI.3821-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Dey J, Carr AJ, Cagampang FR, Semikhodskii AS, Loudon AS, Hastings MH, Maywood ES. The tau mutation in the Syrian hamster differentially reprograms the circadian clock in the SCN and peripheral tissues. J Biol Rhythms. 2005;20:99–110. doi: 10.1177/0748730404274264. [DOI] [PubMed] [Google Scholar]
- Eide EJ, Vielhaber EL, Hinz WA, Virshup DM. The circadian regulatory proteins BMAL1 and cryptochromes are substrates of casein kinase Iepsilon. J Biol Chem. 2002;277:17248–17254. doi: 10.1074/jbc.M111466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Eide EJ, Woolf MF, Virshup DM, Forger DB. An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc Natl Acad Sci U S A. 2006;103:10618–10623. doi: 10.1073/pnas.0604511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- Izumo M, Johnson CH, Yamazaki S. Circadian gene expression in mammalian fibroblasts revealed by real-time luminescence reporting: Temperature compensation and damping. Proc Natl Acad Sci USA. 2003;100:16089–16094. doi: 10.1073/pnas.2536313100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase 1e. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas RJ, Stirland JA, Mohammad YN, Loudon AS. Postnatal growth rate and gonadal development in circadian tau mutant hamsters reared in constant dim red light. J Reprod Fertil. 2000;118:327–330. doi: 10.1530/jrf.0.1180327. [DOI] [PubMed] [Google Scholar]
- Lucas RJ, Stirland JA, Darrow JM, Menaker M, Loudon AS. Free running circadian rhythms of melatonin, luteinizing hormone, and cortisol in Syrian hamsters bearing the circadian tau mutation. Endocrinology. 1999;140:758–764. doi: 10.1210/endo.140.2.6538. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GKY, O'Neil J, O'Brien JA, McMahon DG, Harmar AJ, Hastings MH. Synchronization and maintenance of timekeeping in superchiasmatic circadian clock cells by neuropeptidergic signalling. Current Biology. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Mignot E, Takahashi JS. A circadian sleep disorder reveals a complex clock. Cell. 2007;128:22–23. doi: 10.1016/j.cell.2006.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- Prosser HM, Bradley A, Chesham JE, Ebling FJP, Hastings MH, Maywood ES. Prokineticin receptor 2 (Prokr2) is essential for the regulation of circadian behavior by the suprachiasmatic nuclei. Proc Natl Acad Sci U S A. 2007;104:648653. doi: 10.1073/pnas.0606884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Menaker M. A mutation of the circadian system in golden hamsters. Science. 1988;241:1225–1227. doi: 10.1126/science.3413487. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Saper CB, Scammell TE, Lu J. Hypothalamic regulation of sleep and circadian rhythms. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian Mutant Overtime Reveals F-box Protein FBXL3 Regulation of Cryptochrome and Period Gene Expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talts JF, Brakebusch C, Fassler R. Integrin gene targeting. Methods Mol Biol. 1999;129:153–187. doi: 10.1385/1-59259-249-X:153. [DOI] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielhaber EL, Duricka D, Ullman KS, Virshup DM. Nuclear export of mammalian PERIOD proteins. J Biol Chem. 2001;276:45921–45927. doi: 10.1074/jbc.M107726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.