Abstract
Objective
Allergy documentation and exchange are vital to ensuring patient safety. This study aims to analyze and compare various existing standard terminologies for representing allergy information.
Methods
Five terminologies were identified, including the Systemized Nomenclature of Medical Clinical Terms (SNOMED CT), National Drug File–Reference Terminology (NDF-RT), Medication Dictionary for Regulatory Activities (MedDRA), Unique Ingredient Identifier (UNII), and RxNorm. A qualitative analysis was conducted to compare desirable characteristics of each terminology, including content coverage, concept orientation, formal definitions, multiple granularities, vocabulary structure, subset capability, and maintainability. A quantitative analysis was also performed to compare the content coverage of each terminology for (1) common food, drug, and environmental allergens and (2) descriptive concepts for common drug allergies, adverse reactions (AR), and no known allergies.
Results
Our qualitative results show that SNOMED CT fulfilled the greatest number of desirable characteristics, followed by NDF-RT, RxNorm, UNII, and MedDRA. Our quantitative results demonstrate that RxNorm had the highest concept coverage for representing drug allergens, followed by UNII, SNOMED CT, NDF-RT, and MedDRA. For food and environmental allergens, UNII demonstrated the highest concept coverage, followed by SNOMED CT. For representing descriptive allergy concepts and adverse reactions, SNOMED CT and NDF-RT showed the highest coverage. Only SNOMED CT was capable of representing unique concepts for encoding no known allergies.
Conclusions
The proper terminology for encoding a patient's allergy is complex, as multiple elements need to be captured to form a fully structured clinical finding. Our results suggest that while gaps still exist, a combination of SNOMED CT and RxNorm can satisfy most criteria for encoding common allergies and provide sufficient content coverage.
Keywords: Terminology; Standards; Vocabulary, Controlled; Allergy; Hypersensitivity; Drug Intolerance
Introduction
The Asthma and Allergy Foundation states that allergy is the fifth leading chronic disease,1 affecting one in five Americans.2 Each year, allergies account for more than 17 million outpatient office visits.2 The estimated annual cost of allergies is nearly $7 billion.2 Allergies are linked to a host of chronic diseases (eg, asthma) and serious illnesses (eg, angioedema).3 In rare cases, allergic reactions may cause death or severe morbidity.
Proper documentation and exchange of a patient's allergy information is vital to patient care, safety, and education. It is estimated that 12% of medication errors are due to a lack of understanding of drug allergies4 and 8–13% of medication errors are preventable due to an allergy being documented at the time a medication is ordered.5–7 Bates et al8 found a 56% reduction in medication errors due to known allergies after implementing computerized provider order entry with clinical decision support (CDS).
The question of how to structure and encode allergy information to improve documentation as well as data interoperability is being actively researched and has attracted the interest of many national working groups, particularly within the context of meaningful use (MU).9 However, no widely adopted standard terminology for encoding allergy exists.
Our objectives for this study are threefold. First, we define a set of desirable characteristics to assess existing standard terminologies for representing allergy. Second, we conduct a quantitative analysis to examine content coverage of each investigated terminology for representing different types of allergy information. Third, we discuss major challenges and issues, make suggestions for possible solutions, and point out future directions.
Background
Allergy, intolerance, hypersensitivity, and adverse sensitivity
We first clarify the often confused differences between allergy, intolerance, hypersensitivity, and adverse sensitivity, each of which is briefly introduced below along with their immunopathologic mechanisms.3 10 11 13
Hypersensitivity refers to excessive, undesirable reactions initiated by exposure to a defined stimulus at a dose tolerated by normal persons.10 The traditional classification for hypersensitivity reactions is that of Gell and Coombs,12 which divides hypersensitivity into four types based on the mechanisms involved and time taken for the reaction. These types include: (I) immediate hypersensitivity reaction or anaphylaxis; (II) cytotoxic or cytolytic antibody reactions (eg, transfusion reaction); (III) immune-complex reactions (eg, serum sickness); and (IV) delayed T cell mediated reactions (eg, poison ivy). Sell14 and Rajan15 expanded the Gell and Coombs classification and included additional immunopathologic mechanisms: inactivation/activation antibody reactions, T cell cytotoxic reactions, and granulomatous reactions. Drug hypersensitivity is further classified as immediate (occurring within 1 h) and non-immediate (occurring after 1 h).16
An allergy is a type I hypersensitivity reaction initiated by an immune response to a non-self agent resulting in a detrimental, sometimes debilitating, effect on an individual.10 13 The non-self agent or antigen triggers an immune response (IgE antibody mediated or non-IgE mediated) that leads to a cascade of cellular and molecular events resulting in the clinical manifestations of the allergic reaction.10 13 The response can be divided into two phases: an immediate phase, which is primarily IgE mediated, and a late phase, which is mediated by inflammatory markers and cytokines. The term ‘immediate hypersensitivity’ encompasses both of these phases and is clinically referred to as allergy.11
Intolerance is the development of detrimental signs and symptoms from a substance that would not typically produce these symptoms (eg, tinnitus after a single dose of aspirin, or lactose intolerance).17 18 Intolerance symptoms may be due to toxic contaminants in a food or substance, the pharmacologic properties of the substance, metabolic disorders, or idiosyncratic responses from the host.13
Because hypersensitivity confers only an immune-mediated condition, some ongoing efforts adopt the term ‘adverse sensitivity’ to include both allergy and intolerance.19
Allergy documentation and challenges
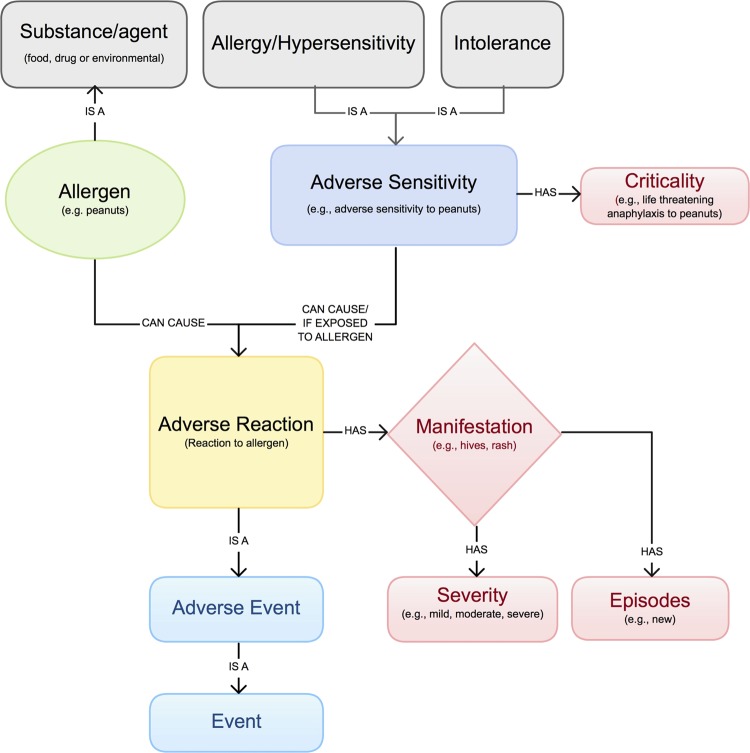
Clinicians routinely elicit allergy information during the medical interview. Key elements (including allergy type, allergen, adverse reaction (AR), severity, episode, and criticality) and their relationships are illustrated in figure 1.19–21 Unfortunately, allergies are often poorly documented in electronic health records (EHRs) and paper charts.22 23 Due to the lack of a comprehensive and standard allergy terminology, most EHRs use a proprietary terminology. Furthermore, when the terminology used by the EHR does not contain the allergy that clinicians are looking for, the allergy is often entered as uncoded free text.24 The resulting allergy information is neither interoperable across clinical information systems nor easily reusable for other applications, such as supporting drug–allergy checking.
Figure 1.
Allergy and intolerance model (adapted from HL7).19–21 Access the article online to view this figure in colour.
An idealized allergy terminology faces many requirements and challenges. One noteworthy challenge is to facilitate establishment of the connection between medication formulations and drug allergens, which are typically recorded in different EHR modules (ie, medication list and allergy list, respectively). Such connection is critical for triggering CDS algorithms. Another challenge is to facilitate representation of a complete allergy record with all clinically relevant information details and relationships among them. For example, in a reported ‘amoxicillin allergy,’ the substance ‘amoxicillin’ is noted as the causative agent for the observed reaction of ‘severe hives’ and ‘mild shortness of breath.’ In addition, the ideal terminology has to include the right balance of pre-coordination (the representation of a clinical meaning using a single concept identifier) and post-coordination (the representation of a clinical meaning by two or more concepts).25 The underlying terminology model should specify how the members of a post-coordinated collection are related to one another and support logical transformations between different representations which express the same meaning.26
Recent related efforts
Federal Medication Terminologies (FMT) for the allergy domain have been endorsed by the National Committee on Vital and Health Statistics (NCVHS) and the Department of Health and Human Services (HHS) with the aim of creating allergy specifications for interoperable exchange.27 The Health Information Technology Standards Panel (HITSP) has made a preliminary set of terminology recommendations for encoding allergy and drug sensitivity components of either document-based or message-based HITSP constructs (eg, Clinical Document Architecture (CDA) documents, HL7 V2 messages).20 21 These include: Systemized Nomenclature of Medical Clinical Terms (SNOMED CT)28 for allergy/AR, RxNorm29 for medications, National Drug File–Reference Terminology (NDF-RT)30 for drug classes, and the Unique Ingredient Identifier (UNII)31 for food and substance allergens. Task forces from HHS and the National Council for Prescription Drug Programs (NCPDP) are working to define a standardized vocabulary that allows interoperable coding of allergies.32 The NCPDP Allergy Value Set Task Group has created an initial set of recommendations for an idealized interoperable allergy value set that includes RxNorm as the primary source terminology. MU stages 1 and 2 criteria require EHRs to implement drug–allergy checking and maintain active allergy medication lists.9 33 However, HHS has not yet specified which terminologies should be used to encode allergy information.
Review of existing standard terminologies and relevant studies
Based on a systematic literature search and HITSP recommendations, in the following we provide an overview of five existing standard terminologies and relevant studies for encoding allergy concepts. We included the Medication Dictionary for Regulatory Activities (MedDRA) in our analysis because it is an international medical terminology used for classifying adverse event information associated with the use of biopharmaceuticals and other medical products.34 35
SNOMED CT
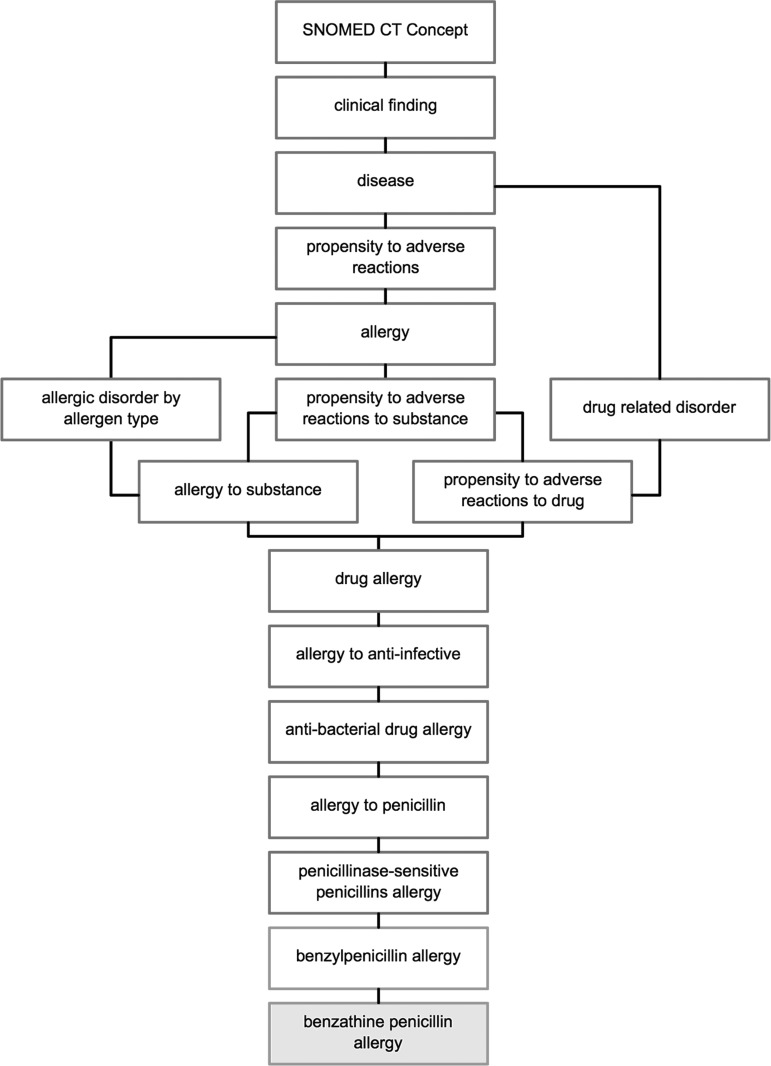
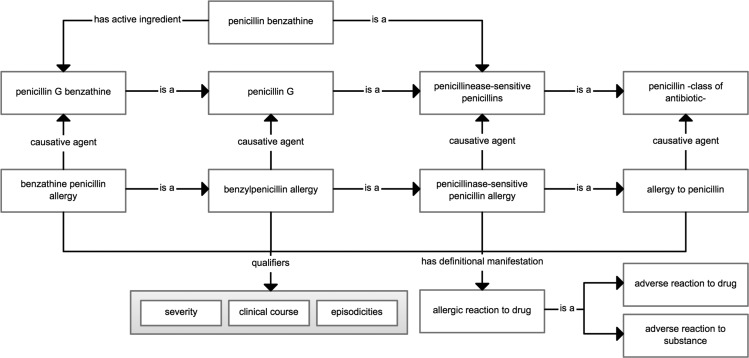
SNOMED CT is a comprehensive, hierarchical terminology used for clinical documentation and reporting.36 Allergies within SNOMED CT are classified as a ‘clinical finding’ with the subordinates ‘disease’ (hypersensitivity disorder) and ‘propensity to adverse reactions.’ Within ‘propensity to adverse reactions’ are three hyponyms: ‘allergy,’ ‘propensity to adverse reactions to a substance,’ and ‘pseudoallergy.’ An allergy example of benzathine penicillin is shown in figure 2 to illustrate the various levels of classification from the root concept level to the specific drug allergy. SNOMED CT describes an allergy concept using a ‘is a’ relationship that links it to its parent concept, a ‘causative agent’ relationship that links it to the substance or ingredient concept, and a ‘has definitional manifestation’ relationship that links it to an allergic reaction concept (figure 3). These semantic relationships facilitate clinical reasoning. For example, through the ‘has a causative agent’ relationship, one could retrieve all the drugs that belong to the penicillin class and that cause a penicillin allergy.
Figure 2.
Simplified SNOMED CT classification of a ‘benzathine penicillin allergy.’
Figure 3.
Semantic relationships and qualifiers defined within SNOMED CT for an allergy concept.
SNOMED CT uses pre-coordination of the term ‘allergy’ and the substance (eg, ‘penicillin’), allowing discernment between a substance and a clinical finding or disorder (eg, ‘penicillin allergy’). Finer-grained concepts can be represented using a post-coordination mechanism. For example, ‘severity’ (eg, mild) can be post-coordinated with an allergy concept as a qualifier (figure 3).37
RxNorm
Created by the National Library of Medicine, RxNorm is a standardized nomenclature that provides normalized terms for clinical drugs and drug delivery devices, and links its terms to various contributing source vocabularies.29 38 RxNorm's normalized terms for clinical drugs specify drug ingredients, strengths, and/or dose forms.29 Specific term types (TTYs) describe the drug at multiple levels of abstraction that can be important depending on its use (eg, ingredient level information for drug–allergy checking). Drug concepts are related to each other through a set of named relationships (eg, ‘has ingredient’) and drug classes are included by way of Legacy VA Drug Classes from NDF-RT.
Anticipating the need for a drug class hierarchy in RxNorm (eg, for allergy classes), Palchuk et al39 mapped RxNorm to NDF-RT using a set of medication records from two institutions. While feasible, they found the process to be ‘difficult and imperfect,’ citing future needs for a more systematic approach to mapping drug classes within NDF-RT.
NDF-RT
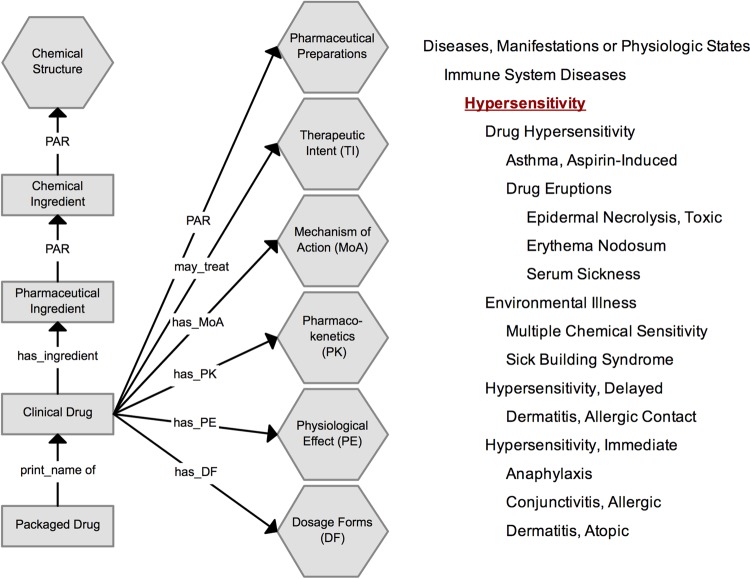
NDF-RT is an extension of the Veterans Health Administration National Drug File (VHA-NDF). It uses a formal multi-axial model that classifies clinical drugs based on pharmaceutical drug class and generic ingredients.30 NDF-RT further specifies drugs and ingredients via defined role relationships to other related concepts by means of mechanism of action, physiologic effect, therapeutic intent, contraindication, pharmacokinetics, and dosage forms (figure 4, left).40 41 Drug products are categorized in a hierarchical fashion through VA Drug Classes. External Pharmacologic Classes (EPC) also exist and are in the process of being integrated into the NDF-RT content model.42 Allergies within NDF-RT are specified using the traditional definition of ‘hypersensitivity’ and assume a hierarchical relationship (figure 4, right).43
Figure 4.
(Left) NDF-RT drug-class hierarchy organization (adapted from Pathak et al45 and Carter et al40). (Right) Hierarchy of NDF-RT for hypersensitivity (adapted from the National Cancer Institute43). NDF-RT, National Drug File–Reference Terminology. Access the article online to view this figure in colour.
For medications, NDF-RT specifies an allergy by assigning a role relationship that defines both the concept and its relationship to other concepts within a domain. For example, when an allergy (hypersensitivity) exists to penicillin, the role relationship ‘contraindicated with’ (CI_with) will be specified denoting a contraindication.
At present, NDF-RT is the terminology recommended by HITSP for representing medication drug classes.20 However, these recommendations constrain its use to concepts that span chemical structure, mechanisms of action, and physiologic effect. To date, adoption of specified subsets of NDF-RT for the purpose of allergy documentation has been limited, as little implementation guidance on the use of NDF-RT in the context of translating class-based medication allergies to defined subsets has been provided. As a means to mitigate these limitations, recent work by the NCPDP Allergy Value Set Task Group has identified a starter set of NDF-RT drug classes (that span attribute values of Ingredient_Kind, Mechanism_of_Action_Kind, or Physiological_Effect_Kind) for use within interoperable exchanges.32
MedDRA
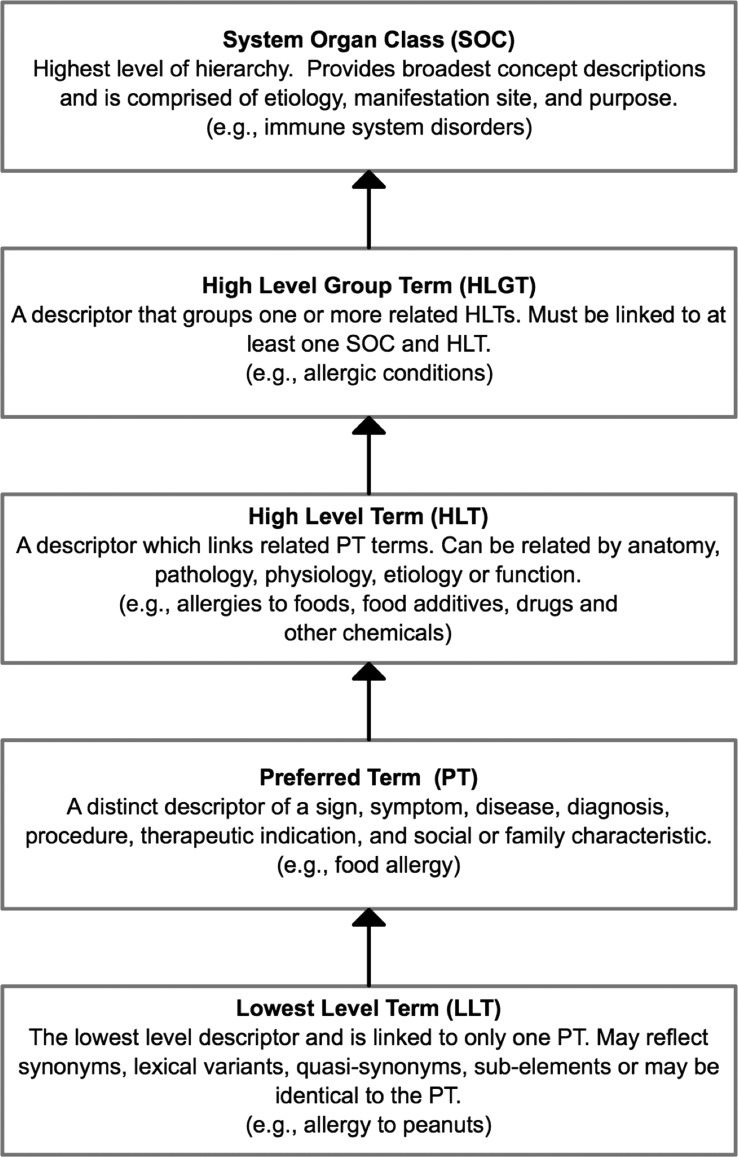
MedDRA is developed by the International Conference on Harmonization (ICH)44 to monitor AR from both medications and devices.34 35 By virtue of being primarily designed for regulatory purposes and health effects in individual patients, MedDRA excludes information on drug/product terminology, equipment/device terminology, and descriptors of severity.34 35 MedDRA is a hierarchical terminology divided into five tiers ranging in granularity from system organ class to lowest level terms (LLT) (figure 5). LLTs are linked to only one preferred term (PT) and typically reflect synonyms (SY). Allergies are classified within the high-level group term (HLGT) ‘Allergic Conditions,’ a descendent of ‘Immune System Disorders.’
Figure 5.
Hierarchical representation of the Medication Dictionary for Regulatory Activities (MedDRA).35
Several studies have evaluated desiderata and mapping AR with MedDRA. Bousquet et al46 found MedDRA had better completeness for adverse drug reactions than other adverse drug reaction dictionaries but did not fulfill many of Cimino's desiderata and formal definitions for its terms. Bodenreider found that 58% of PTs in MedDRA have mappings to SNOMED CT. However, mapping of HLGT and HLT terms yielded lower results (below 30%).47 Differences in the level of granularity were identified for the two terminologies. In MedDRA, PT terms representing a terminal node may have multiple decedents in SNOMED CT that further specify the allergy and its attributes. Nadkarni and Darer also found limitations in mapping MedDRA to SNOMED CT through the Unified Medical Language System (UMLS) due to duplication of terms and no semantic consistency to distinguish LLT and PT terms.48
UNII
UNII is developed and maintained by the US Food and Drug Administration and provides UNIIs for drugs, biologics, foods, and devices.31 49 For allergens, UNII provides the chemical structure, chemical formula, molecular weight, chemical abstract service number, PT, and SY.49
UNII has high concept granularity by virtue of being based on chemical structure, but it has several limitations in representing patient allergies. There is no hierarchical structure to organize substances into classes, and substances are enumerated as a simple list. For example, the IS A relationship between ‘blue shrimp’ and ‘shrimp’ is not specified. An EHR user could conceivably have to filter through 272 types of shrimp to encode a shrimp allergy, making it difficult for clinical use. Further, SY and brand names (TR), which often represent semantically different concepts, share the same unique identifier as the ‘preferred substance name.’ For example, ‘Moxifloxacin hydrochloride,’ ‘Avelox,’ and ‘Vigamox’ share the same unique identifier ‘C53598599T.’
While prior studies have evaluated allergy information exchange and terminology mapping, our intent is to compare the desirable characteristics and content coverage of reference terminologies applicable to allergies. Our goal is to identify the terminologies best suited for documenting and encoding allergies.
Methods
Our analyses of these five terminologies include both a qualitative analysis that assesses the capability of each terminology to support documentation of an allergy observation and a quantitative analysis that measures the breadth of coverage within specific domains. Analyses were performed using the 2011 versions of RxNorm (April), SNOMED CT (January), NDF-RT (July), UNII (December), and MedDRA (March). Each were downloaded and imported into Microsoft SQL Server 2008.
As part of our qualitative analysis, we defined a set of 16 desirable terminology characteristics (tables 1 and 2) based on generic desiderata proposed by Cimino50 and Elkin et al51 and justified the relevant desiderata (ie, content coverage, concept orientation, formal definitions, multiple granularities, vocabulary structure, and maintainability) in the specific context of the allergy domain. A specific criterion, subset capability, is added, which refers to a terminology's capability in defining a relatively compact set of allergy concepts for ease of implementation and to facilitate allergy documentation and data entry. Appraisal of each are graded using a five-point Likert scale with four stars being excellent; three, good; two, fair; one, poor; and zero, indicating no fulfillment. Ratings were assigned by study authors (FRG, LZ, JMP, GR, RR) and consensus was achieved via an iterative process. The majority of the desiderata can be fairly judged due to the intrinsic characteristics of the terminologies (eg, concept orientation, formal definition, vocabulary structure, etc). In terms of content coverage and multiple granularities, results from our quantitative analysis were used as a reference for the ratings.
Table 1.
Desiderata with descriptions and justification
| Desiderata | Description | Justification (examples) |
|---|---|---|
| Content coverage | Domain specific content coverage | Content necessary for encoding of allergen, reaction, severities, and episode |
| Concept orientation | Concepts correspond to no more than one meaning/non-ambiguous | Non-ambiguous representation of allergens and synonyms |
| Formal definitions | Definitions, attributes, and relationships between concepts | Defined relationships for allergies based on drug class or ingredient |
| Multiple granularities | Fine to coarse-grained descriptions of concepts | Representation of both ‘coarse’ and ‘fine’ details of allergic reactions and substances |
| Vocabulary structure | Support of hierarchy and composite concepts | Hierarchical representation of allergy content and pre- and post-coordination |
| Subset capability | A relatively compact collection of related concepts in a terminology | Facilitate documentation/encoding of key elements of a patient's allergy |
| Maintainability | Capacity to evolve, remain current, and useable over time | Frequency of updates, change history, and adherence to editorial policies |
Table 2.
Appraisal of terminologies for allergy using desiderata
| Criteria | SNOMED CT | NDF-RT | RxNorm* | UNII | MedDRA |
|---|---|---|---|---|---|
| Content coverage | |||||
| Food allergens | *** | ** | * | ** | * |
| Drug allergens | *** | *** | **** | *** | * |
| Environmental allergens | *** | ** | * | ** | * |
| Reaction to allergen | **** | ** | *** | ||
| Severity of reaction | **** | * | |||
| Episode of reaction | **** | ||||
| No known allergies | *** | ||||
| Concept orientation | **** | **** | **** | **** | **** |
| Formal definitions | **** | **** | **** | ||
| Multiple granularities | |||||
| Substance | *** | *** | **** | **** | * |
| Reaction | **** | * | ** | ||
| Vocabulary structure | |||||
| Polyhierarchy | **** | **** | * | ||
| Pre-coordinated terms† | **** | ** | |||
| Support for post-coordination | *** | ||||
| Subset capability | **** | ** | ** | * | * |
| Maintainability | **** | **** | **** | **** | **** |
*RxNorm contains multiple source terminologies. For this analysis, only RxNorm itself is considered (ie, SAB=RxNorm).
†Capability of terminology in specifying pre-coordinated descriptive terms for allergy (ie, penicillin allergy).
Likert Scale: four stars, excellent; three stars, good; two stars, fair; one star, poor; zero stars, no fulfillment.
MedDRA, Medication Dictionary for Regulatory Activities; NDF-RT, National Drug File–Reference Terminology; SNOMED CT, Systemized Nomenclature of Medical Clinical Terms; UNII, Unique Ingredient Identifier.
We further conducted a quantitative analysis using methods based on string and pattern matching followed by a manual review to compare each terminology's coverage of (1) common allergen concepts and (2) descriptive concepts specifying allergy, AR, and no known allergies.
Allergen concepts
Where applicable, we calculated the number of allergen/substance concepts (including drug, food, and environmental) in each identified terminology. For drug allergens, we used the NCPDP Allergy Value Set Task Group ‘starter set’ to obtain the top 10 most frequently observed drug allergy classes.32 Members of each drug group (eg, amoxicillin is a type of penicillin) were obtained from NDF-RT, SNOMED CT, and First Databank (FDB).52 Specifically, we used the owl version of NDF-RT and Protégé to look up each drug class and its members. For SNOMED CT we used the terminology browser CliniClue to look up each drug class and its members. Lastly, we used our local drug class relationships from FDB. These initial lists were then compiled to create one single list (with duplicates removed). For food and environmental allergens, we used the eight major food allergen categories stated by the Food Allergen Labeling and Consumer Protection Act (FALCPA),53 as well as those specified by NDF-RT and SNOMED CT.
Descriptive concepts for drug allergy and adverse reaction
We analyzed and calculated the total number of concepts specifying allergy or AR for each member in the above drug, food, and environmental classes within the applicable terminologies (SNOMED CT, NDF-RT, and MedDRA). These concepts do not represent substances but rather allergy descriptions, descriptions of AR, drug activity, or drug toxicity or poisoning. Additionally, we evaluated which terminologies contained concepts to represent ‘no known allergies.’
In order to reduce false negatives, the keywords used for searching against each terminology also included brand names, SY, and other lexical variants. For example, lexical variants for each allergen class and its members were obtained from the National Cancer Institute (NCI) Metathesaurus and Term Browser.43 Additionally, we used RxNorm to identify brand names, SY, and lexical variants by searching for each drug class and its members by RxCUI. The keyword lists were then collated, manually reviewed, and normalized if necessary. The final set was used to search relevant allergy concepts against each terminology. We used the SQL pattern match commands, ‘like’ and ‘equal’ with the keyword bounded by ‘%’ (eg, WHERE [STR] like ‘%amoxicillin%’), to include allergens that contained the keyword. All results were manually reviewed by three reviewers (FRG, LZ, JMP).
Results
Qualitative analysis
The results of our qualitative analysis are shown in table 2. Overall, SNOMED CT consistently met most criteria in all areas, followed by NDF-RT, RxNorm, UNII, and MedDRA. Only SNOMED CT had the capability to encode ‘no known allergies.’ RxNorm exceeded other terminologies in representing drug allergens, but lacked coverage for food and environmental allergens, reactions, severities, and episodes. MedDRA had good coverage for reactions; poor coverage of food, drug, and environmental allergens; and no coverage for severity or episodes. Content coverage will be demonstrated in detail in the following quantitative analysis. UNII and MedDRA had no formal definitions for their concepts. Granularity was highest in UNII and RxNorm for substances, and in SNOMED CT for reactions. Use of polyhierarchy was available in SNOMED CT and NDF-RT, and only SNOMED CT supported both pre- and post-coordination for allergy concepts. Allergen subset creation was possible by most terminologies except MedDRA and UNII. In RxNorm, while subset creation is possible by ingredient, it is very difficult to subset by therapeutic class or medication type. All terminologies have excellent maintenance mechanisms.
Quantitative analysis
The results of our quantitative comparison are presented in tables 3 and 4. Table 3 represents the quantity of allergen concepts. The first two columns indicate the allergen class and number of keywords used in the search. The following columns show the number of concepts returned from each terminology. Our results show that among drug allergens, RxNorm provided the most comprehensive coverage with 16 619 concepts followed by UNII at 7053, SNOMED CT at 4068, NDF-RT at 2236, and MedDRA with 1 concept. For food and environmental allergens, UNII provided the most coverage with 1836 concepts, followed by SNOMED CT at 848, RxNorm at 420, NDF-RT at 283, and MedDRA with 22 concepts.
Table 4 represents the quantity of descriptive concepts, containing allergy (A) and AR. Only a small number of allergens have corresponding descriptive concepts (eg, there is not a pre-coordinated concept ‘penicillin G allergy’ in SNOMED CT as shown in figure 3). SNOMED CT and NDF-RT had the most drug allergy concepts with 169 and 163, respectively, followed by MedDRA with 3. For concepts specifying AR, SNOMED CT returned the largest number at 271 and MedDRA had 11. For food and environmental allergy concepts, SNOMED CT had the largest number at 80. Of note, NDF-RT did not specify a concept to represent AR. Only SNOMED CT contained concepts for specifying ‘no known allergies’ (five concepts).
Table 4.
Number of descriptive concepts specifying allergies or adverse reactions for the most frequently observed allergies by terminology*
| Drug allergies and adverse reactions | SNOMED CT | MedDRA | NDF-RT | |||
|---|---|---|---|---|---|---|
| A | AR | A | AR | A | AR | |
| Penicillin | 36 | 56 | 1 | 2 | 14 | – |
| Sulfonamides | 31 | 49 | 1 | 1 | 28 | – |
| Macrolides | 4 | 14 | 0 | 1 | 5 | – |
| Iodine containing | 12 | 17 | 1 | 0 | 26 | – |
| Cephalosporins | 25 | 40 | 0 | 1 | 24 | – |
| ACE inhibitors | 9 | 12 | 0 | 2 | 10 | – |
| NSAIDS | 32 | 47 | 0 | 3 | 37 | – |
| Tetracyclines | 11 | 22 | 0 | 1 | 6 | – |
| Heparin | 6 | 7 | 0 | 0 | 7 | – |
| Statins | 3 | 7 | 0 | 0 | 6 | – |
| Subtotal | 169 | 271 | 3 | 11 | 163 | – |
| Non-drug allergies and adverse reactions | ||||||
| Food | 38 | 10 | 6 | 6 | 5 | – |
| Environmental | 42 | 7 | 6 | 1 | 1 | – |
| Subtotal | 80 | 17 | 12 | 7 | 6 | – |
| Total | 249 | 288 | 15 | 18 | 169 | – |
*For NDF-RT, ‘contraindicated with’ role relationship to hypersensitivity was used to specify drug allergy.
A, allergy concepts (eg, allergy to penicillin); AR, adverse reaction concepts (eg, adverse reaction to penicillin); MedDRA, Medication Dictionary for Regulatory Activities; NDF-RT, National Drug File–Reference Terminology; SNOMED CT, Systemized Nomenclature of Medical Clinical Terms.
Below we use the statin drug class to illustrate how SY and lexical variants were obtained and how the search was conducted. More detailed descriptions can be found in the online supplement. We initially identified eight drugs (eg, simvastatin, atorvastatin, lovastatin, etc) in this class through NDF-RT, SNOMED CT, and FDB. Using the NCI Metathesaurus and RxNorm to obtain brand names, SY, and lexical variants, this number was expanded to 47 drugs (eg, Lipitor, Zocor, Crestor, etc). Searching across each terminology for drug allergens we found RxNorm had the most at 419, followed by UNII at 152, SNOMED CT at 113, NDF-RT at 93, and MedDRA with zero (table 3). These differences are likely due to the presence of multiple dose forms within RxNorm and finer-grained representation of drug substances by way of chemical and molecular descriptions in UNII. SNOMED CT and NDF-RT, however, do not have this level of granularity, explaining why their numbers are lower. When evaluating descriptive allergy concepts for statins (table 4), SNOMED CT had three allergy concepts (eg, simvastatin allergy) and seven AR concepts (eg, simvastatin adverse reaction). NDF-RT had six allergy concepts and MedDRA had none.
Table 3.
Number of substances found in each terminology for encoding the most frequently observed drug, food, and environment allergens*
| Number of keywords | SNOMED CT | NDF-RT | MedDRA | RxNorm | UNII | |
|---|---|---|---|---|---|---|
| Drug allergens | ||||||
| Penicillin | 283 | 363 | 253 | 0 | 1769 | 773 |
| Sulfonamides | 151 | 983 | 270 | 0 | 1253 | 586 |
| Macrolides | 98 | 174 | 96 | 0 | 777 | 357 |
| Iodine containing | 502 | 635 | 384 | 0 | 1627 | 1182 |
| Cephalosporins | 283 | 411 | 249 | 0 | 1200 | 882 |
| ACE inhibitors | 81 | 177 | 108 | 0 | 727 | 319 |
| NSAIDs | 545 | 875 | 567 | 1 | 7317 | 2391 |
| Tetracyclines | 78 | 177 | 98 | 0 | 979 | 299 |
| Heparin | 90 | 160 | 118 | 0 | 551 | 112 |
| Statins | 47 | 113 | 93 | 0 | 419 | 152 |
| Subtotal | 2168 | 4068 | 2236 | 1 | 16619 | 7053 |
| Non-drug allergens | ||||||
| Food allergens | 42 | 496 | 155 | 11 | 146 | 1493 |
| Environmental allergens | 24 | 352 | 128 | 11 | 274 | 343 |
| Subtotal | 66 | 848 | 283 | 22 | 420 | 1836 |
| Total | 2234 | 4916 | 2519 | 23 | 17039 | 8889 |
*An example of queries used is provided in online supplement section 1.
MedDRA, Medication Dictionary for Regulatory Activities; NDF-RT, National Drug File–Reference Terminology; SNOMED CT, Systemized Nomenclature of Medical Clinical Terms; UNII, Unique Ingredient Identifier.
An example of queries used is provided in online supplement section 2.
Discussion
Encoding allergy content is inherently complex by virtue of needing to represent the allergen and the resulting allergic reaction with its clinical manifestations and severity. We found significant variability in how reference terminologies can be used to represent allergy information, particularly given their intended purpose (eg, MedDRA for regulatory use). No single terminology is, by itself, a complete solution.
Our results indicate that some terminologies may be better suited than others in encoding aspects of an allergy. SNOMED CT, NDF-RT, and RxNorm fulfilled most of the desired criteria we evaluated. SNOMED CT, however, exceeded the others with regard to being able to finely describe an allergic reaction through the use of post-coordination. It also provided a sufficiently large number of pre-coordinated terms for allergies that will facilitate clinicians’ finding of allergy concepts. Relationships between concepts in SNOMED CT are clearly defined, making it desirable for drug–allergy checking and other forms of CDS. Quantitatively, we found RxNorm particularly strong with regard to medication allergens, given its intended use as a medication terminology. SNOMED CT provided very good coverage of descriptive drug allergies, food allergies, and environmental allergies. NDF-RT exhibited well-developed relationships for drug classes, yet did not have sufficient content for encoding allergy severity, and efficient searching of allergens by clinicians may be limited given its compositional structure. UNII contained an impressive number of substances, however its simple structure does not support hierarchical or other semantic relationships among substance concepts. MedDRA, given its intended use for encoding AR, does contain a large number of concepts that could be used to describe the manifestations (eg, signs and symptoms) of an AR. However, it lacks formal definitions to relate a manifestation to its causative agent or severity. Additionally, granularity is restricted to five levels which limits classification beyond the LLT (eg, allergy to penicillin cannot be further specified). Considering the desirable characteristics and content coverage, our findings suggest that for common allergens, SNOMED CT combined with RxNorm would fulfill most criteria with sufficient content coverage.
There are significant challenges in trying to use and integrate multiple reference terminologies to encode allergies (as per HITSP recommendations), in essence a ‘best of breed’ approach. The question remains of how to create a common model that allows each of these terminologies to be combined with reconciliation of overlapping concepts and terms. Current EHR modules for allergies do not specify how different reference terminologies can be used in combination. Similarly, CDS applications that rely upon hierarchical associations between classes and members require that substances, related medication concepts, and classifications be compiled within a common terminology framework. Within the NCPDP and National Quality Forum Data Module, use of RxCUIs from RxNorm has been suggested as a means to allow interoperable exchange of medication allergies. While this would remove the complexity of different codes (eg, NDF-RT codes for drug classes, UNII codes for substances, etc) and help achieve MU stage 2 objectives for mediation reconciliation,33 the challenge of establishing hierarchical associations between drug classes and members for CDS still persists.
Future directions
Future research is needed to refine the complex interactions of terminologies by developing a common terminology model for allergies. Additionally, if RxNorm is to be used as the source terminology for medications and SNOMED CT for reactions, further research is needed to determine how content and class-based relationships will be represented in RxNorm. The inclusion of NDF-RT in RxNorm is an attractive option, as semantic relationships could be developed to represent drug classes and members by RxCUI. Given the recent release of the Convergent Medical Terminology (CMT) by the National Library of Medicine54 for documenting problems, development of a similar subset for allergen classes, common reactions (subset of problems), and food and environmental allergies may be of interest. Benefits for end-users would be the compilation of a number of frequently observed allergy terms across multiple institutions to facilitate documentation and encoding of patient allergies using recommended standard terminologies.
Another important direction for future research is to study how to represent negative allergy findings. Documenting pertinent negative findings is no less important than documenting positive ones. Failure to do so may cause legal, malpractice, and compliance issues and can ultimately jeopardize patient safety. Our results showed that only SNOMED CT contains concepts to represent no known allergies, signifying an imperative challenge in the domain of terminology and ontology. As pointed out by previous studies,55 56 the current concept-based terminology paradigm suffers from various problems including misclassifications of terms containing negation.55 Ontology languages based on description logics provide insufficient expressive power to represent axiom negations.56 Therefore, more research is needed to explore general frameworks and mechanisms to represent negative findings in EHRs.
Limitations
Several limitations exist with the approach we used. The drug-based classes from the NCPDP represent a subset of the most frequently observed drug allergies and do not represent all allergens. Similarly, we chose to evaluate only a subset of common environmental and food allergens. For the quantitative comparison, although we applied multiple resources to identify additional SY and lexical variants, false negatives may exist due to missing certain terms. Manual review of all the results was necessary to remove false positives as a small number of keywords were contained within the middle of a word (ie, ‘corn’ is contained in the word ‘cornea’). Future studies may apply other methods beyond string matching (eg, ontology alignment).
Conclusion
The proper terminology for encoding a patient's allergy is complex, as multiple elements need to be captured to form a fully structured clinical finding. Our results suggest that while gaps still exist, a combination of SNOMED CT and RxNorm can satisfy most criteria for encoding common allergies and provide sufficient content coverage.
Footnotes
Contributors: FRG and LZ conceived of the topic and wrote the manuscript. JMP, GR, and RAR contributed to the manuscript. CB and BM reviewed the manuscript.
Funding: This project was supported by a National Library of Medicine training grant T15- LM007092 (FRG).
Competing interests: GR is employed by First Databank. No funding was received from GR or First Databank.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.National Academy on an Aging Society. 2000. Chronic Conditions: A challenge for the 21st Century.
- 2.Centers for Disease Control “CDC Fast Facts A-Z,” Vital Health Statistics. 2003. [cited 28 Oct 2011]. http://www.cdc.gov/nchs/fastats/allergies.htm, http://www.aafa.org/display.cfm?id=9&sub=30 (accessed 5 Sept 2011)
- 3.World Allergy Organization. [cited 28 Oct 2011]. http://www.worldallergy.org (accessed 5 Sept 2011)
- 4.Lesar TS, Briceland L, Stein DS. Factors related to errors in medication prescribing. JAMA 1997;277:312–17 [PubMed] [Google Scholar]
- 5.Gandhi TK, Burstin HR, Cook EF, et al. Drug complications in outpatients. J Gen Intern Med 2000;15:149–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bates DW, Cullen DJ, Laird N, et al. Incidence of adverse drug events and potential adverse drug events. Implications for prevention. ADE Prevention Study Group. JAMA 1995;274:29–34 [PubMed] [Google Scholar]
- 7.Kuperman GJ, Marston E, Paterno M, et al. Creating an enterprise-wide allergy repository at Partners HealthCare System. AMIA Annu Symp Proc 2003:376–80 [PMC free article] [PubMed] [Google Scholar]
- 8.Bates DW, Leape LL, Cullen DJ, et al. Effect of computerized physician order entry and a team intervention on prevention of serious medication errors. JAMA 1998;280:1311–16 [DOI] [PubMed] [Google Scholar]
- 9.Office of the National Coordinator for Health Information Technology Department of Health and Human Services Health information technology: initial set of standards, implementation specifications, and certification criteria for electronic health record technology. Interim final rule. Fed Regist 2010;75:2013–47 [PubMed] [Google Scholar]
- 10.Johansson SG, Bieber T, Dahl R, et al. Revised nomenclature for allergy for global use: report of the nomenclature review committee of the World Allergy Organization, October 2003. J Allergy Clin Immunol 2004;113:832–6 [DOI] [PubMed] [Google Scholar]
- 11.Abbas AK, Lichtman AH, Pillai S. Cellular and molecular immunology. 7th edn. Philadelphia, PA: W.B. Saunders Company, 2011 [Google Scholar]
- 12.Gell PGH, Coombs RRA. Clinical aspects of immunology. 1st edn. Oxford, England: Blackwell, 1963 [Google Scholar]
- 13.Adkinson N, Yunginger J, Busse W, et al. Middleton's allergy: principles and practice. 7th edn. Philadelphia, PA: Elsevier Inc, 2008 [Google Scholar]
- 14.Sell S. Immunopathology. In: Rich RR, Fleisher TA, Schwartz BD, et al., eds. Clinical immunology: principles and practice. St Louis: Mosby, 1996:449–77 [Google Scholar]
- 15.Rajan TV. The Gell-Coombs classification of hypersensitivity reactions: a re-interpretation. Trends Immunol 2003;24:376–9 [DOI] [PubMed] [Google Scholar]
- 16.Romano A, Torres MJ, Castells M, et al. Diagnosis and management of drug hypersensitivity reactions. J Allergy Clin Immunol 2011;127(3 Suppl):S67–73 [DOI] [PubMed] [Google Scholar]
- 17.Hogan WR. Towards an ontological theory of substance intolerance and hypersensitivity. J Biomed Inform 2011;44:26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gruchalla RS. Drug allergy. J Allergy Clin Immunol 2003;111(2 Suppl):S548–59 [DOI] [PubMed] [Google Scholar]
- 19.HL7 Working Group Allergy and Intolerance. 2012. [cited 28 Jun 2012]. http://wiki.hl7.org/index.php?title=Allergy_%26_Intolerance (accessed 20 March 2012)
- 20.Health Information Technology Standards Panel C80 HITSP/C80 Clinical Document and Message Terminology Component v 2.0, Section 2.2.3.3.9 “Medication Drug Class” and Section 2.2.3.1.1 “Problem”. January 25 2010. [cited 6 Sep 2011]. http://www.hitsp.org/ConstructSet_Details.aspx?&PrefixAlpha=4&PrefixNumeric=80 (accessed 5 Sept 2011)
- 21.Health Information Technology Standards Panel C83 HITSP/C83 CDA Content Modules Component v 2.0, 2.2.2.6 Allergy/Drug Sensitivity. January 25 2010. [cited 6 Sep 2011]. http://www.hitsp.org/ConstructSet_Details.aspx?&PrefixAlpha=4&PrefixNumeric=83 (accessed 5 Sept 2011)
- 22.Hsieh TC, Kuperman GJ, Jaggi T, et al. Characteristics and consequences of drug allergy alert overrides in a computerized physician order entry system. J Am Med Inform Assoc 2004;11:482–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khalil H, Leversha A, Khalil V. Drug allergy documentation–time for a change? Int J Clin Pharm 2011;33:610–13: [DOI] [PubMed] [Google Scholar]
- 24.Zimmerman CR, Chaffee BW, Lazarou J, et al. Maintaining the enterprisewide continuity and interoperability of patient allergy data. Am J Health Syst Pharm 2009;66:671–9 [DOI] [PubMed] [Google Scholar]
- 25.Rosenbloom ST, Miller RA, Johnson KB, et al. Interface terminologies: facilitating direct entry of clinical data into electronic health record systems. J Am Med Inform Assoc 2006;13:277–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Health Terminology Standards Development Organization. SNOMED CT® Technical Implementation Guide, International Release (US English) January 2012.
- 27.NCVHS allergy recommendations to Secretary of U.S Department of Health and Human Services 2006. [cited 6 Sep 2011]. http://ncvhs.hhs.gov/060913lt.htm (accessed 5 Sept 2011)
- 28.SNOMED CT (Systematized Nomenclature of Medicine-Clinical Terms). [cited 28 Oct 2011]. http://www.ihtsdo.org/snomed-ct/ (accessed 5 Sept 2011)
- 29.RxNorm [cited 20 Aug 2011]. http://www.nlm.nih.gov/research/umls/rxnorm/ (accessed 5 Sept 2011)
- 30.NDF-RT National Drug File—Reference Terminology (NDF-RT) Documentation. 2010. [cited 5 Sep 2011]. http://evs.nci.nih.gov/ftp1/NDF-RT/NDF-RTDocumentation.pdf (accessed 5 Sept 2011)
- 31.Substance Registration System-Unique Ingredient Identifier (UNII). [cited 27 Oct 2011]. http://www.fda.gov/ForIndustry/DataStandards/SubstanceRegistrationSystem-UniqueIngredientIdentifierUNII/default.htm (accessed 5 Sept 2011)
- 32.National Council for Prescription Drug Programs Interoperable Medication Allergy Vocabulary Recommendations. 2011. May 24, 2011 [cited 5 Sep 2011]. healthit.hhs.gov/...pt/.../ncpdp-recommendation-06–24–11.pdf (accessed 5 Sept 2011)
- 33.Office of the National Coordinator (ONC) for Health Information Technology Department of Health and Human Services Health Information Technology: standards, implementation specifications, and certification criteria for electronic health record technology, 2014 Edition; Revisions to the permanent certification program for health information technology. Fed Regist 2012;77:54163–292 [PubMed] [Google Scholar]
- 34.Medical Dictionary of Regulatory Activities website. [cited 5 Sep 2011]. http://www.meddramsso.com/ (accessed 5 Sept 2011)
- 35.Medical Dictionary of Regulatory Activities Introductory Guide to MedDRA Version 14.0. MedDRA Maintenance and Support Services Organization. Chantilly, Virginia: MedDRA Maintenance and Support Services Organization, 2011 [Google Scholar]
- 36.Donnelly K. SNOMED-CT: the advanced terminology and coding system for eHealth. Stud Health Technol Inform 2006;121:279–90 [PubMed] [Google Scholar]
- 37.International Health Terminology Standards Development Organization. SNOMED CT® User Guide, International Release (US English) January 2012.
- 38.Nelson SJ, Zeng K, Kilbourne J, et al. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc 2011;18:441–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palchuk MB, Klumpenaar M, Jatkar T, et al. Enabling hierarchical view of RxNorm with NDF-RT drug classes. AMIA Annu Symp Proc 2010;577–81 [PMC free article] [PubMed] [Google Scholar]
- 40.Carter JS, Brown SH, Erlbaum MS, et al. Initializing the VA medication reference terminology using UMLS metathesaurus co-occurrences. AMIA Annu Symp Proc 2002;116–20 [PMC free article] [PubMed] [Google Scholar]
- 41.Brown SH, Elkin PL, Rosenbloom ST, et al. VA national drug file reference terminology: a cross-institutional content coverage study. Stud Health Technol Inform 2004;107(Pt 1):477–81 [PubMed] [Google Scholar]
- 42.NDF-RT National Drug File—Reference Terminology (NDF-RT) Documentation. 2012. [cited 5 Sep 2011]. http://evs.nci.nih.gov/ftp1/NDF-RT/NDF-RTDocumentation.pdf (accessed 5 Sept 2011)
- 43.National Cancer Institute NCI Term Browser and Metathesaurus. [cited 22 Sep 2011]. http://nciterms.nci.nih.gov/ncitbrowser/pages/multiple_search.jsf (accessed 5 Sept 2011)
- 44.International Conference on Harmonization [cited 30 Sep 2011]. http://www.ich.org/ (accessed 5 Sept 2011)
- 45.Pathak J, Richesson RL. Use of standard drug vocabularies in clinical research: a case study in pediatrics. AMIA Annu Symp Proc 2010;607–11 [PMC free article] [PubMed] [Google Scholar]
- 46.Bousquet C, Lagier G, Lillo-Le Louet A, et al. Appraisal of the MedDRA conceptual structure for describing and grouping adverse drug reactions. Drug Saf 2005;28:19–34 [DOI] [PubMed] [Google Scholar]
- 47.Bodenreider O. Using SNOMED CT in combination with MedDRA for reporting signal detection and adverse drug reactions reporting. AMIA Annu Symp Proc 2009;45–9 [PMC free article] [PubMed] [Google Scholar]
- 48.Nadkarni PM, Darer JD. Determining correspondences between high-frequency MedDRA concepts and SNOMED: a case study. BMC Med Inform Decis Mak 2010;10:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Department of Health and Human Services Consolitated Health Informatics—Standards Adoption Recommendation, Allergy. 2006. [cited 7 Oct 2011]. http://www.hhs.gov/healthit/chiinitiative.html (accessed 2 Oct 2011)
- 50.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med 1998;37:394–403 [PMC free article] [PubMed] [Google Scholar]
- 51.Elkin PL, Brown SH, Carter J, et al. Guideline and quality indicators for development, purchase and use of controlled health vocabularies. Int J Med Inf 2002;68:175–86 [DOI] [PubMed] [Google Scholar]
- 52.First DataBank MedKnowledge [cited 13 Dec 2011]. http://www.fdbhealth.com/fdb-medknowledge/
- 53.U.S. Food and Drug Administration Food Allergen Labeling and Consumer Protection Act of 2004 (Public Law 108–282, Title II). [cited 4 Apr 2012]. http://www.fda.gov/food/labelingnutrition/FoodAllergensLabeling/GuidanceComplianceRegulatoryInformation (accessed 5 Sept 2011)
- 54.National Library of Medicine Unified Medical Language System (UMLS), Convergent Medical Terminology. 2011. [cited 26 Dec 2011]. http://www.nlm.nih.gov/research/umls/Snomed/cmt.html (accessed 5 Sept 2011)
- 55.Ceusters W, Elkin P, Smith B. Negative findings in electronic health records and biomedical ontologies: a realist approach. Int J Med Inf 2008;76(Supplement 3):s326–s33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flouris G, Huang Z, Pan JZ. Inconsistencies, negations and changes in ontologies. In: Proc of AAAI 2006;21:1295–300 [Google Scholar]