Abstract
Objectives
To test the feasibility of using text mining to depict meaningfully the experience of pain in patients with metastatic prostate cancer, to identify novel pain phenotypes, and to propose methods for longitudinal visualization of pain status.
Materials and methods
Text from 4409 clinical encounters for 33 men enrolled in a 15-year longitudinal clinical/molecular autopsy study of metastatic prostate cancer (Project to ELIminate lethal CANcer) was subjected to natural language processing (NLP) using Unified Medical Language System-based terms. A four-tiered pain scale was developed, and logistic regression analysis identified factors that correlated with experience of severe pain during each month.
Results
NLP identified 6387 pain and 13 827 drug mentions in the text. Graphical displays revealed the pain ‘landscape’ described in the textual records and confirmed dramatically increasing levels of pain in the last years of life in all but two patients, all of whom died from metastatic cancer. Severe pain was associated with receipt of opioids (OR=6.6, p<0.0001) and palliative radiation (OR=3.4, p=0.0002). Surprisingly, no severe or controlled pain was detected in two of 33 subjects’ clinical records. Additionally, the NLP algorithm proved generalizable in an evaluation using a separate data source (889 Informatics for Integrating Biology and the Bedside (i2b2) discharge summaries).
Discussion
Patterns in the pain experience, undetectable without the use of NLP to mine the longitudinal clinical record, were consistent with clinical expectations, suggesting that meaningful NLP-based pain status monitoring is feasible. Findings in this initial cohort suggest that ‘outlier’ pain phenotypes useful for probing the molecular basis of cancer pain may exist.
Limitations
The results are limited by a small cohort size and use of proprietary NLP software.
Conclusions
We have established the feasibility of tracking longitudinal patterns of pain by text mining of free text clinical records. These methods may be useful for monitoring pain management and identifying novel cancer phenotypes.
Keywords: Natural Language Processing, Pain Management, Metastatic Prostate Cancer, Personalized Medicine, Individualized Medicine, Cancer Pain
Introduction
Pain is a debilitating part of the experience of metastatic cancer. An automated system to categorize and track pain in electronic medical records could provide a powerful means to improve clinical care, and could allow novel ‘high pain’ or ‘low pain’ phenotypes to be defined and studied on a molecular basis. We tested the feasibility of using natural language processing (NLP) of text from clinical encounters to depict meaningfully the experience of pain in patients with metastatic prostate cancer over time.
Background
Worldwide, prostate cancer is the second most commonly diagnosed cancer and the sixth leading cause of cancer death in men.1 In the past decade, significant effort has been made to better understand and reduce the burden of pain on the cancer patient,2–5 the patient's family, caregivers, and society.6 Pain status can predict survival in metastatic prostate cancer,7 and changes in pain status have been examined as a surrogate marker of effectiveness of new therapies.8 9 Several validated pain survey tools have been proposed for routine clinical care.10 11
NLP has been used to quantify associations between diseases, conditions, and symptoms,12–16 for vocabulary discovery,17 and for cohort construction.18–25 NLP applications focusing on pain in clinical records have successfully detected the experience of pain in free text within an electronic medical record.26–28 Some studies suggest that, in some scenarios, NLP of medical record text may perform better than patient-completed surveys in detection of clinically relevant pain.26 28
Although pain has previously been normalized and classified manually for purposes of statistical correlations,29 we used NLP to automatically characterize the experience of pain over thousands of records. To our knowledge, this is the first study to combine NLP, date resolution, and statistical analysis to create a longitudinal study of pain in the clinical record. Our system normalized each mention of pain in longitudinal clinical records by severity classification and number of days before death. We used regression modeling techniques to analyze both the newly structured data and the existing structured data to search for phenotypic correlations with pain in the context of metastatic prostate cancer.
Pain management is fundamental to effective clinical care, and significant pain is a consequence of the disordered biology of many cancers. This study tests the feasibility of automatically tracking patient pain over time using NLP of clinical record text. If NLP-based pain tracking is feasible, further study will be indicated to test the hypothesis that adoption of NLP-based pain tracking within electronic health record systems could provide significant added value in clinical care and in advancing research in disease phenotyping.
Methods
Patient cohort
Thirty-three men from the PELICAN (Project to ELIminate lethal CANcer) integrated clinical/molecular autopsy study of metastatic prostate cancer were the subjects of this study. Subjects joined the institutional review board-approved study between 1995 and 2005. The mean age of the study subjects at the time of diagnosis of prostate cancer was 62 years (range 42–75). The mean interval between diagnosis and death was 6.3 years (range 0.8–15.4). Of the 33 subjects in the study, 27 were Caucasian, five were African-American, and one was of Hispanic background. Six subjects were seen only in community hospital inpatient, clinic and private office settings; the remaining 27 subjects were followed in a combination of oncology center and community hospital clinic settings.
Clinical records obtained
The study obtained and analyzed all available paper, electronic, radiologic, radiation therapy, and pathology medical records for each subject. Subjects provided a list of institutions and physician offices where medical care was received, and copies of medical records from the various institutions and offices were obtained.
Creation of electronic records for each study subject
A total of 23 887 pages of paper records were converted into electronic text using methods described in the online appendix. The electronic record included laboratory values, radiology reports, pathology reports, and records of inpatient and outpatient encounters with providers. The text recorded in 4409 inpatient and outpatient encounters is called the ‘PELICAN corpus’ and is the focus of this study.
Integrated Life Sciences Research (ILSR) database and removal of identifiers
The full curated electronic text of each paper record was placed in the ILSR database, a system created to support the PELICAN Study. The average number of inpatient or outpatient records per year between diagnosis and death was 32 (range 4–212). Subject date of birth, date of death, race/ethnicity, all available serum prostate-specific antigen (PSA) concentrations, body weight measurements, body height measurements, and radiation therapy records were separately tabulated in ILSR by project data curators.
Pain status categorization
A multidisciplinary team consisting of NLP software developers, medical subject matter experts (SMEs), and statisticians developed a pain categorization model based on a conservative four-tiered pain scale: no pain (category 0); some pain (category 1); controlled pain (category 2); severe pain (category 3).
Natural language processing
We used ClinREAD, a proprietary healthcare-domain-oriented, rule-based NLP system (Lockheed Martin, Bethesda, Maryland, USA) built on AeroText (Rocket Software, Newton, Minnesota, USA) and previously successfully used by members of the study team in the Informatics for Integrating Biology and the Bedside (i2b2) obesity challenge.22 30 ClinREAD was chosen because of its availability to the project team and team familiarity with its use. Other valid approaches, including machine learning, were not used because of lack of available resources for the current project. The first stage of the current project involved iterative development and evaluation of NLP-based pain extraction and qualification (severity, anatomy, and date) in the 4409-record PELICAN corpus, for the purposes of discovery over a closed dataset. During this stage, we made iterative modifications to our entire system, data model, normalization rules, and vocabulary (details in online appendix). We tested the generalizability of the NLP methods on 889 unannotated, deidentified discharge summaries provided courtesy of i2b2.31
The system rated each mention of experienced or explicitly denied pain on the basis of the context in which it was found (table 1). We developed 42 pain severity contextual rules, such as (complete list in online appendix table 2):
[pain severity modifier] [body location] [pain term]
[pain term] [to be] [adv] [PainSeverityComplement]
[pain term] … [pain severity complement] out of [10|ten]
Table 1.
Example pain severity indicators
| No pain (category 0) | Some pain (category 1) | Controlled pain (category 2) | Severe pain (category 3) | |
|---|---|---|---|---|
| Example modifiers (occurring before pain mention) | No | Some | Controlled | Severe |
| Without | Mild | Controlling | Significant | |
| No complaint | Intermittent | Treatment for | Crushing | |
| Denied any | Occasional | Essentially controlled | Excruciating | |
| Absence of | Negligible | To control this | Exquisite | |
| Example complements (occurring after pain mention) | Relieved | Dull | Controlled | Intractable |
| Resolved | Not too bad | Managed | [8–10] | |
| 0 | [1–7] | Well managed | [Eight–ten] | |
| Zero | [One–seven] | Unbearable | ||
| Persistent | Uncontrollable |
Vocabulary from the Unified Medical Language System (UMLS; version 2010AB)29 was imported via the Metathesaurus from 35 level 0 source vocabularies (see online appendix table 3). We selected 16 semantic types based on the domain of the data as shown in online appendix table 4. Lookup tables were created from each set of synonymous terms in order to associate each phrase with a preferred term and a UMLS concept unique identifier (CUI). A filtering process similar to that of Roberts et al32 was used to remove irrelevant terms. After filtering, a total of 675 000 terms and phrases were contained in the study vocabulary.
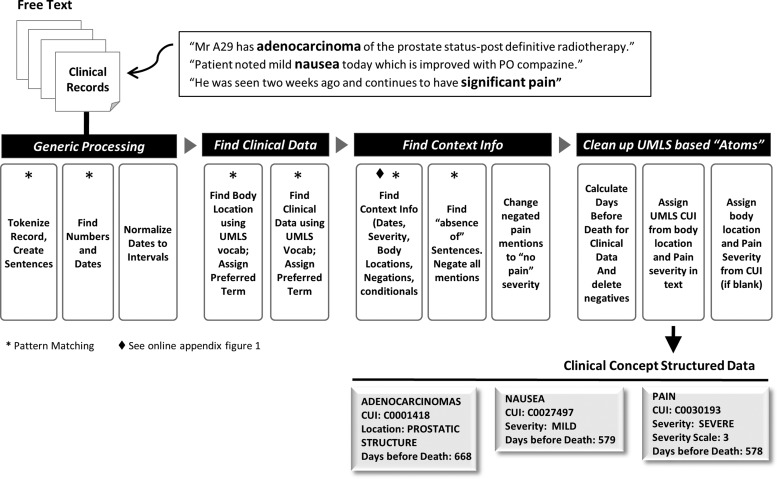
We combined the vocabulary terms with context patterns in order to recognize internal dates, negatives, conditionals, and pain severity. These context patterns were developed manually. ClinREAD, like MedLEE,33 is rule-based. Each clinical concept (‘sign or symptom’, ‘finding’, ‘injury or poisoning’, ‘disease or syndrome’, or ‘neoplastic process’) is associated with a date and a body location; see online appendix for further detail. The system resolved incomplete dates (eg, ‘in July’) based on the date of the encounter, and resolved relative dates (eg, ‘four days prior to admission’) based on the previous date mention. Each resolved date is represented as a range (startdate, enddate). This date resolution component was based on the development team's previous work34–37 and is described in the online appendix. When dates were missing, the date of the clinical encounter was used as the default. Date associations were used to normalize the clinical concept to the number of days before death, for each individual study subject. This calculation is enabled through the conversion of the midpoint of absolute date ranges to the modified Julian format.38 Each mention of pain was associated with a severity level from the four-tiered pain scale. A subset of 637 strings from semantic type ‘sign or symptom’ were identified as indicating pain, listed in online appendix table 5. The NLP algorithm used for the study is summarized in figure 1 and as follows.
- Generic processing
- Text tokenization and sentence detection.
- Find mentions of dates and numbers; identify the date of the encounter to be used in date resolution.
- Resolve dates in document order, and calculate Julian format of the midpoint of each date range.
- Find clinical data (concept extraction)
- Find mentions of body locations using UMLS vocabulary; look up preferred term.
- Find mentions of clinical concepts using UMLS vocabulary; look up preferred term.
- Disambiguate overlapping UMLS vocabulary based on confidence scores associated with each contextual rule. Semantic type disambiguation of overloaded terms was rudimentary; we used context in some cases but relied heavily on default assumptions for terms commonly used in the context of prostate cancer (especially for abbreviations). For the purposes of discovery over a closed corpus, the current disambiguation was effective, as shown by our evaluated performance.
- Find context information
- Find context of UMLS vocabulary to identify negations, conditionals, and hypotheticals, and to associate pain severity level, body locations and explicit dates. Context rules were manually built. The negation context rules used much of the same vocabulary as the NegEx algorithm.39 Additional rules to distinguish family history, conditionals, and hypotheticals were manually developed from examination of the dataset.
- Find negated list sentences (‘The patient denies nausea, headache, fevers, or chills.’) and negate all concept mentions within them. These predictable sentences were common in the dataset.
- Convert negated instances of pain concepts to severity scale 0.
- Clean clinical concept data
- Associate dates and body locations with UMLS clinical concepts. Use the date of the encounter when an explicit date for the event cannot be determined.
- Assign CUIs based on vocabulary, severity, semantic type, and body location.
- Update structured data concepts that lack severity or body locations where the CUI implies them (‘severe headache’, C0239889).
- Delete non-pain negatives, conditionals, and hypotheticals from the structured dataset in order to create a set of actual, experienced clinical concepts.
- Calculate ‘days before death’ for each clinical concept, using the associated Julian formatted date and a lookup table listing the Julian formatted date of death for each subject in the PELICAN cohort.
Figure 1.
Natural language processing algorithm. CUI, concept unique identifier; UMLS, Unified Medical Language System.
Although our system is proprietary, it could be replicated using other tools. One might start with any system that extracts concepts and identifies assertions as defined for the 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text.38 One could then integrate a temporal reference extraction and normalization tool such as HeidelTime,40 GATE with the Tagger_DateNormalizer plugin,41 or DANTE,42 filter out the pain-related concepts, as listed in appendix table 5, and identify the level of pain using rules defined in appendix table 2 and the lookup table defined in appendix table 11.
Study database
NLP processing of the records database produced structured data on each pain mention in each clinical record for all 33 study subjects. These data were combined with demographic and other separately curated data about each subject into a single study database suitable for statistical analysis.
Identification of correlates of severe pain
We undertook a univariate logistic regression analysis to identify correlates of severe pain for use in a multivariate model; factors investigated included receipt of various drugs (eg, opioids, chemotherapy, steroids), body mass index (BMI), receipt of palliative radiation, and frequency of utilization of health services—that is, we correlated severe pain, as derived by NLP processing, with clinical and demographic factors from the structured (ie, non-NLP-based, pre-existing) portion of the study database. For this analysis, ‘severe pain’ was any reading of controlled or severe pain—that is, any reading of 2 or 3 during a month of observation versus any other reading (−1=no data, 0=explicit report of no pain, 1=reported pain not described as controlled or severe); see online appendix for further details. We then constructed a multiple regression model to assess the strength of associations between the occurrence of severe pain and all defined variables for which p was less than 0.1 in the univariate analysis. Inclusion of a dichotomous variable indicating ‘last year before death’ controlled for time effects.43 All statistical analyses were conducted with SAS V.9.2 using the Proc Logistic procedure.
Visualization of patients’ experience of pain
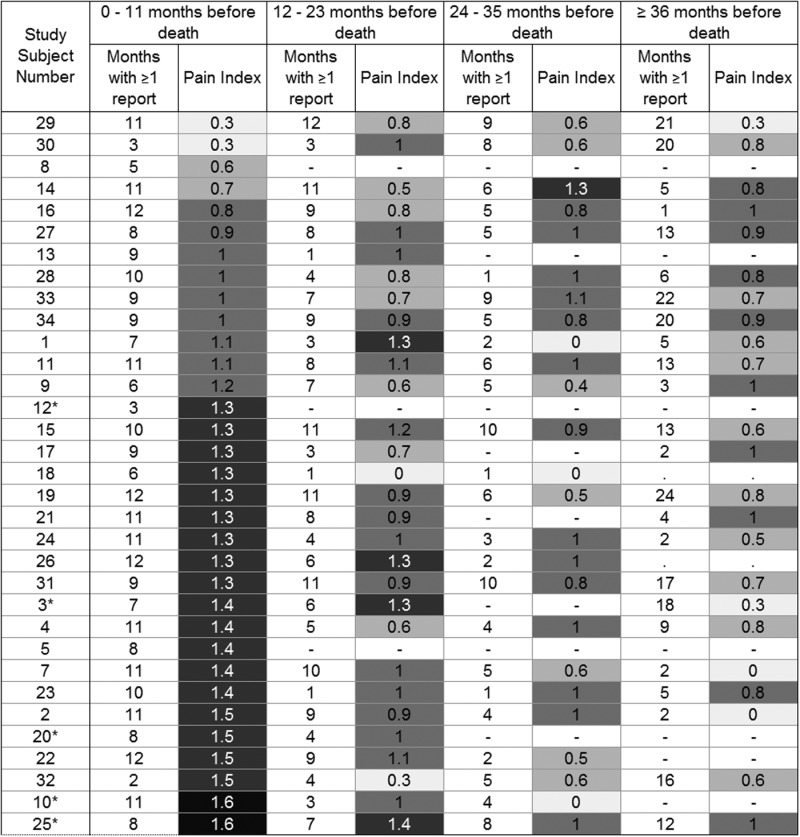
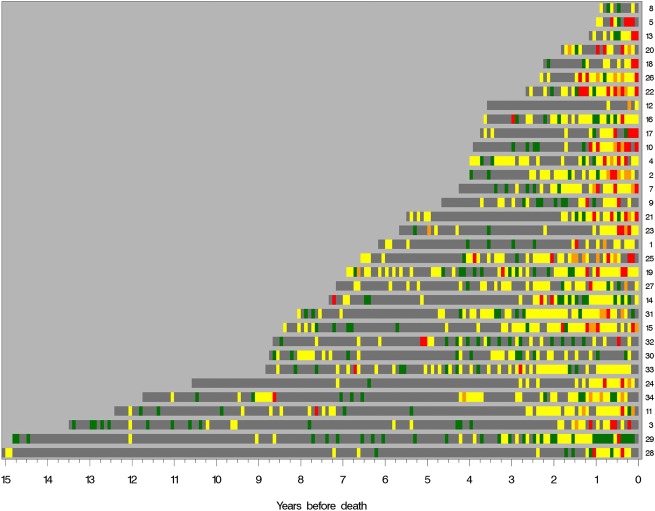
We determined a pain index value for each subject during four intervals before death, with pain index defined as the mean monthly maximum pain value (max_pain) for months in which a pain report was available; the monthly max_pain values used were no pain=0, some pain=1, and controlled pain or severe pain=2 (see figure 3). We then obtained longitudinal views of pain status in each subject by plotting color-coded monthly max_pain values from diagnosis until death (figure 2). When no pain status report was available for a given month, we used the most recent pain status as the imputed value for a given subject.
Figure 3.
Pain index by subject in last months of life. Five African-American subjects have asterisks added to their study subject numbers.
Figure 2.
Study subject pain ribbons ‘algograph’ display. Maximum pain reported by each subject from prostate cancer diagnosis until time of death from prostate cancer. Pain data are reported for each month. Grey, no report; green, no pain; yellow, pain; orange, controlled pain; red, severe pain.
To test the possibility of visualizing a summary of pain records from a group of subjects, we displayed the fraction of study subjects in each pain severity up to the time of death (see online appendix figure 2A), as well as the worst pain severity detected for each subject for each month up to death (see online appendix figure 2b).
Results
The purpose of the project was discovery over a closed dataset and a study of feasibility. To evaluate and improve the performance of the NLP algorithm on the dataset, we completed multiple rounds of SME evaluation (GSB and RJT). Across all patient encounters, the NLP algorithm identified 6387 pain mentions (mean 1.45 pain mentions per record) and 13 827 drug mentions.
Evaluation of NLP method within PELICAN clinical text records
After development, we evaluated performance on the closed PELICAN corpus using the AeroText Answer Key Editor. The SMEs separately corrected 32 automatically annotated full text clinical encounter records randomly selected from the entire study set to create ‘answer keys’. These 32 records contained 207 mentions of pain. The NLP developers had no influence on the correction of the annotations. Inter-annotator agreement on pain mention (exact token match in the text and normalized concept name), pain start and end date (exact match), body location of pain (exact match), and pain severity integer are shown in table 2A, B. We assessed inter-annotator agreement by scoring one annotation set against the other. The entire team then met to discuss and adjudicate the two sets of corrections. Pain mentions on which there were disagreements were resolved to form the gold standard answer key; see online appendix table 6 for examples. We then assessed system performance compared against the gold standard answer key, requiring correct answers (region of text, normalized concept name, body location, and pain severity integer) to be exact matches. Recall is the percentage of pain mentions in the record that were correctly identified by the NLP system. Precision is the percentage of pain mentions identified by the NLP system that are correct. F-measure is the harmonic mean of precision and recall, and provides a measure of overall accuracy. F-measure for pain mention detection was 0.95, and for overall average pain severity assignment was 0.81 (see also table 3).
Table 2.
Inter-annotator agreement on (A) pain mention, pain start and end date, and body location on the PELICAN corpus, (B) pain severity on the PELICAN corpus, (C) pain mention, pain start and end date, and body location on the i2b2 corpus, and (D) pain severity on the i2b2 corpus
| A—PELICAN corpus | B—PELICAN corpus | C—i2b2 corpus | D—i2b2 corpus | ||||
|---|---|---|---|---|---|---|---|
| Agreement | Agreement | Agreement | Agreement | ||||
| Pain mention | 0.97 | No pain | 0.93 | Pain mention | 0.88 | No pain | 0.81 |
| Start date of pain | 0.93 | Some pain | 0.91 | Start date of pain | 0.79 | Some pain | 0.86 |
| End date of pain | 0.91 | Controlled pain | 0.79 | End date of pain | 0.74 | Controlled pain | 1.00 |
| Body location of pain | 0.76 | Severe pain | 0.85 | Body location of pain | 0.71 | Severe pain | 0.67 |
| Severity of pain overall average | 0.88 | Severity of pain overall average | 0.85 | ||||
i2b2, Informatics for Integrating Biology and the Bedside; PELICAN, Project to ELIminate lethal CANcer.
Table 3.
Accuracy of natural language processing algorithm extraction of pain mentions regarding (A) pain mention, pain start and end date, and body location on the PELICAN corpus, (B) pain severity on the PELICAN corpus, (C) post-development pain mention, pain start and end date, and body location on the i2b2 corpus, (D) post-development pain severity on the i2b2 corpus
| A—PELICAN corpus | B—PELICAN corpus | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | Inc | FN | FP | Recall | Precision | F-Measure | TP | Inc | FN | FP | Recall | Precision | F-Measure | ||
| Pain mention | 153 | 0 | 7 | 9 | 0.96 | 0.94 | 0.95 | Explicitly no pain | 19 | 3 | 2 | 0 | 0.79 | 0.86 | 0.83 |
| Start date of pain | 145 | 8 | 7 | 9 | 0.91 | 0.90 | 0.90 | Some pain | 75 | 18 | 1 | 8 | 0.80 | 0.74 | 0.77 |
| End date of pain | 145 | 8 | 7 | 9 | 0.91 | 0.90 | 0.90 | Controlled pain | 17 | 1 | 2 | 0 | 0.85 | 0.94 | 0.90 |
| Body location of pain | 51 | 1 | 23 | 3 | 0.68 | 0.93 | 0.80 | Severe pain | 20 | 0 | 2 | 1 | 0.91 | 0.95 | 0.93 |
| Severity of pain overall average | 131 | 22 | 7 | 9 | 0.82 | 0.81 | 0.81 | ||||||||
|
C—i2b2 corpus |
D—i2b2 corpus |
||||||||||||||
| TP | Inc | FN | FP | Recall | Precision | F-Measure | TP | Inc | FN | FP | Recall | Precision | F-Measure | ||
| Pain mention | 105 | 0 | 6 | 17 | 0.95 | 0.86 | 0.90 | Explicitly no pain | 18 | 1 | 0 | 3 | 0.95 | 0.82 | 0.88 |
| Start date of pain | 74 | 31 | 6 | 17 | 0.67 | 0.61 | 0.64 | Some pain | 67 | 10 | 2 | 14 | 0.85 | 0.74 | 0.79 |
| End date of pain | 73 | 32 | 6 | 17 | 0.66 | 0.60 | 0.63 | Controlled pain | 5 | 0 | 3 | 0 | 0.63 | 1.00 | 0.81 |
| Body location of pain | 64 | 0 | 42 | 10 | 0.60 | 0.86 | 0.73 | Severe pain | 4 | 0 | 1 | 0 | 0.80 | 1.00 | 0.90 |
| Severity of pain overall average | 94 | 11 | 6 | 17 | 0.85 | 0.77 | 0.81 | ||||||||
FN, false negative; FP, false positive; i2b2, Informatics for Integrating Biology and the Bedside; Inc, incorrect; PELICAN, Project to ELIminate lethal CANcer; TP, true positive.
Evaluation of NLP methods within i2b2 discharge summaries
We further evaluated the generalizability of our NLP methods using a blind test set from 889 unannotated, deidentified discharge summaries from i2b2.31 Detailed methods are provided in the online appendix. A test set of 30 discharge summaries (containing 111 pain mentions) was chosen and kept unknown to the NLP developers at all times during the evaluation process. The remaining i2b2 records were designated the ‘development set.’ Ground truth was created using the same process as the PELICAN evaluation, with the added control that the annotation process was supervised by a developer not involved in the project. Inter-annotator agreement on the i2b2 corpus is shown in table 2C, D. The SMEs adjudicated each disagreement to obtain an approved gold standard. Several differences were noted by the SMEs between the i2b2 and the PELICAN clinical record corpora, including an increased frequency of ambiguously dated pain mentions in the i2b2 corpus, as shown by the low inter-annotator agreement for start and end dates in table 2C. Further discussion can be found in the online appendix, as can the adjudicated annotations for our 30-report i2b2 test set.
The NLP system was run, as built, on the blind i2b2 test set and scored against the approved gold standard using the AeroText scoring tool. The initial extraction F-measure for pain mentions in the new test set was 0.87; see appendix for complete scores. A 10-hour development process was then conducted to adjust for stylistic differences in the new corpus. The system was scored again, and the system F-measures on pain mentions and pain severity increased to 0.90 and 0.81, respectively.
Date association accuracy was significantly lower than for the PELICAN corpus, falling for start date from 0.90 for PELICAN to only 0.64 for i2b2. We believe that this was the result of a larger number of ambiguous date references in the i2b2 corpus and differences in the annotation guides used by the SMEs to annotate the two corpora; see online appendix for further discussion.
Post-development measures of the NLP extraction over the i2b2 corpus are given in table 3; the final NLP extraction of pain in the i2b2 test set is given in the online appendix. Developers remained blind to the test set throughout the development process. The blind evaluation on an independent dataset showed that, with 10 h of development time to adjust for corpus stylistic differences, the NLP system developed for this project is generalizable beyond the PELICAN corpus.
Pain phenotype exploration
Overall, pain increased markedly during the last 2 years of life (figure 2). Metastatic prostate cancer was the listed cause of death in all study cases, and none of the subjects was found to have significant additional contributing causes of death. In the final year of life, subject pain index varied widely, from 0.3 to 1.6, with a roughly equal distribution of subjects across this spectrum. The five African-American study subjects clustered at the high end of the pain index spectrum (range 1.3–1.6) (table 4).
Table 4.
Multivariate regression analysis of natural language processing-detected pain and clinical variables detects clinically expected associations between pain status and administration of palliative radiation and opioid drugs, as well as months before death, and number of inpatient and outpatient visits
| Variable | OR point estimate | 95% CI | p Value |
|---|---|---|---|
| Received palliative radiation | 3.61 | 1.90 to 6.84 | <0.0001 |
| Log PSA concentration | 1.09 | 0.86 to 1.38 | 0.492 |
| Age at diagnosis | 1.00 | 1.00 to 1.00 | 0.655 |
| African-American | 1.56 | 0.80 to 3.05 | 0.190 |
| Subject BMI <90% of maximum | 1.02 | 0.62 to 1.67 | 0.935 |
| Chemotherapy administered | 1.75 | 1.01 to 3.04 | 0.046 |
| NSAID administered | 1.34 | 0.78 to 2.29 | 0.284 |
| Opioid drug administered | 6.91 | 4.07 to 11.74 | <0.0001 |
| Steroid drug administered | 1.21 | 0.70 to 2.06 | 0.497 |
| In last year of life | 2.52 | 1.46 to 4.36 | 0.001 |
| Number outpatient visits | 1.29 | 1.12 to 1.48 | 0.001 |
| Number inpatient visits | 1.30 | 0.75 to 2.26 | 0.350 |
BMI, body mass index; NSAID, non-steroidal anti-inflammatory drug; PSA, prostate-specific antigen.
The system detected no severe or controlled pain in two subjects (8 and 30). The number of clinical encounter records available per year between diagnosis and death for these two subjects was 32 and 19, indicating that the lack of severe pain reports in these two subjects was not due to a lack of clinical encounters. We found no evidence that these subjects died earlier in the course of their disease from non-cancer causes. Since bone pain is the major source of pain in men with metastatic prostate cancer, we reviewed bone scan findings in these two subjects, and both demonstrated widespread bone changes consistent with metastatic prostate cancer, similar to scan results from all other study patients.
Correlates of severe pain
In the initial univariate analysis, all considered variables except for receipt of definitive radiation and maximum recorded BMI correlated significantly with severe pain. African-American ethnicity was borderline associated with severe pain (OR 1.5, p=0.09). Receipt of opiates (OR 25.6, p<0.001), palliative radiation (OR 13.8, p<0.0001), and being in the last year of life (OR 9.9, p<0.001) were strongly associated with severe pain. See online appendix for detailed univariate analysis results.
In the multivariate analysis, only five of the 12 remaining factors were significantly associated with severe pain (p<0.1): receipt of palliative radiation, opioids, or chemotherapy; being in the last year of life; and the number of outpatient visits (table 4). Receipt of non-steroidal anti-inflammatory drugs (NSAIDs), corticosteroids or sex-steroid-manipulating drugs were not significantly associated. These findings are consistent with current clinical practice, where palliative radiation44 45 and opioids46 are treatments typically reserved for severe pain, and NSAIDs, corticosteroids, and sex-steroid drugs are used more generally across the pain spectrum.46 Similarly, the last year of life is clinically known to be when severe pain is most common46 and when clinical encounters are most frequent. The multivariate model found no significant association with increasing serum PSA concentration, age at diagnosis, or decline in BMI to <90% maximum after controlling for the effects of time. There was a non-significant trend associating African-American ethnicity with more severe pain.
The model and findings were robust, explaining 83% of the variability in the data. When we excluded six patients who were seen only in a community setting and who had fewer recorded clinical encounters, the patterns of association remained unchanged. Moreover, when we removed from the model all variables that were not significant in the univariate analysis, the strengths of the associations (adjusted ORs) of the remaining variables and p values changed only marginally.
Discussion
In multivariate regression analysis, pain status detected by NLP correlated statistically with parameters clinically known to be associated with increased pain. Conversely, pain status detected by NLP was not associated with parameters not expected to be clinically associated with pain status, such as administration of definitive radiation with curative intent. These results suggest that meaningful NLP-based pain status monitoring is feasible. While this project used a rule-based NLP system, machine-learning-based NLP tools should be tested in future work.
Text in longitudinal data is valuable for the study of symptoms such as pain, where the clinical unstructured description may be more complete than it is in structured data.28 NLP techniques convert such unstructured data into structured data, which is typically more amenable to rigorous analysis and display.
Relief of pain is essential in the management of many acute and chronic diseases, and convenient automated monitoring of patient pain status could provide a valuable new tool for improving quality of life and care. Real-time, easy-to-interpret views of the pain status history of an individual patient or a group of patients, as shown in figure 2 and online appendix figure 2, could allow busy clinicians to identify patients most in need of increased pain management intensity, and allow researchers to perform visual and quantitative comparison of groups of subjects participating in clinical trials of novel therapies or novel clinical interventions.
NLP-based determination of pain status may help to identify clinically significant molecular differences between prostate cancers. For example, a study of the molecular differences in the cancers of the two men who apparently experienced no severe pain could provide important clues to the biological determinants of severe pain in metastatic prostate cancer. Similarly, the trend toward increased pain experienced by the five African-Americans compared with the other men in the study is consistent with an oncology clinical trial which found that African-American men were more likely than white men to have extensive disease and bone pain.47
Limitations and future directions
Our study has several limitations. First, the dataset was relatively small, covering just 33 patients. Second, it was difficult to distinguish pain mentions that were not related to the subjects’ metastatic prostate cancer. Although SME review of the records revealed only rare examples of pain not related to prostate cancer in the current study, future studies should implement formal methods to identify and link pain to a relevant disease source. Third, it was difficult to distinguish pain control status from the patient's current experience of pain. This study defaulted to ‘controlled’ pain as one of the pain categories because there were multiple records where the patient was noted to be taking opioids for pain, but no current pain level was provided. Fourth, we may have slightly biased our annotation of the PELICAN corpus by using system outputs to initialize annotations. This technique has been shown to improve consistency, reduce annotation time,48 and improve inter-annotator agreement.49 50 We minimized possible bias by having the annotators work independently and by submitting the results to team scrutiny and collaborative discussion. In essence, the answer key was generated by compiling the answers of four (overlapping) SMEs: two humans, the system itself, and the team as a whole. The similar evaluation results obtained on the separate i2b2 corpus, which used isolated test and development sets, suggest that any bias was minimal. Finally, the use of proprietary ClinREAD and AeroText NLP software may limit reproducibility. However, this limitation is at least partially mitigated by our provision of detailed rules, as well as results from our analysis of the i2b2 corpus. Investigators interested in further analysis of the study dataset using other methods and under appropriate confidentiality protection are invited to contact the senior author.
Conclusions
Electronic health records have greatly facilitated detection and understanding of disease phenotypes and their relationship with genetic and non-genetic factors.51–55 The study reported here, which we believe to be the first to use NLP to obtain longitudinal pain status information in a cohort of patients, shows that NLP-based monitoring of patient pain status is feasible and generalizable to new datasets, and provides a number of phenotype-oriented observations useful for guiding future research. Future studies should focus on comparison of pain-status tracking by NLP versus other validated pain survey tools, and on practical integration of the two methods in settings where electronic health records are in routine use.
Supplementary Material
Acknowledgments
We thank the men and their families who participated in the PELICAN integrated clinical–molecular autopsy study of prostate cancer. We also thank Mario A Eisenberger, Michael A Carducci, V Sinibaldi, T B Smyth, and G J Mamo for oncologic and urologic clinical support. M Rohrer and M Padmanaban provided database support. W B Isaacs facilitated initial development of the PELICAN study by GSB. Deidentified i2b2 clinical records used in this research were provided by the i2b2 National Center for Biomedical Computing funded by U54LM008748 and were originally prepared for the Shared Tasks for Challenges in NLP for Clinical Data organized by Dr Ozlem Uzuner, i2b2 and SUNY. We also thank the JAMIA reviewers for helpful suggestions about article content.
Footnotes
Contributors: NHH, RJT, LS, JAH, LCC, and GSB collaboratively directed and designed the study (LCC retired in 2010). NHH conducted the NLP analyses, with assistance from DA. LS and RL conducted the statistical analyses. RL managed data integration and developed the graphic representations. NHH, RJT, LS, JAH and GSB wrote the manuscript. GSB proposed the current study and graphic representations, and founded, designed and directs the PELICAN (Project to ELIminate lethal prostate CANcer) project.
Funding: Autopsy study of prostate cancer support 1994–1998 from CaPCURE. Support for natural language processing project from Lockheed Martin Information Systems and Global Solutions.
Competing interests: None.
Ethics approval: This study was conducted with the approval of Johns Hopkins Medicine Institutional Review Board.
Provenance and peer review: Not commissioned, externally peer reviewed.
Open Access: This is an Open Access article distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 3.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/3.0/
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69–90 [DOI] [PubMed] [Google Scholar]
- 2.Sandblom G, Carlsson P, Sennfalt K, et al. A population-based study of pain and quality of life during the year before death in men with prostate cancer. Br J Cancer 2004;90:1163–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson JC, Wood J, Feuer D. Prostate cancer: palliative care and pain relief. Br Med Bull 2007;83:341–54 [DOI] [PubMed] [Google Scholar]
- 4.Kaya E, Feuer D. Prostate cancer: palliative care and pain relief. Prostate Cancer Prostatic Dis 2004;7:311–15 [DOI] [PubMed] [Google Scholar]
- 5.Pinski J, Dorff TB. Prostate cancer metastases to bone: pathophysiology, pain management, and the promise of targeted therapy. Eur J Cancer 2005;41:932–40 [DOI] [PubMed] [Google Scholar]
- 6.Sennfalt K, Carlsson P, Sandblom G, et al. The estimated economic value of the welfare loss due to prostate cancer pain in a defined population. Acta Oncol 2004;43:290–6 [DOI] [PubMed] [Google Scholar]
- 7.Halabi S, Vogelzang NJ, Kornblith AB, et al. Pain predicts overall survival in men with metastatic castration-refractory prostate cancer. J Clin Oncol 2008;26:2544–9 [DOI] [PubMed] [Google Scholar]
- 8.Armstrong AJ, Garrett-Mayer E, Ou Yang YC, et al. Prostate-specific antigen and pain surrogacy analysis in metastatic hormone-refractory prostate cancer. J Clin Oncol 2007;25:3965–70 [DOI] [PubMed] [Google Scholar]
- 9.Berry DL, Moinpour CM, Jiang CS, et al. Quality of life and pain in advanced stage prostate cancer: results of a Southwest Oncology Group randomized trial comparing docetaxel and estramustine to mitoxantrone and prednisone. J Clin Oncol 2006;24:2828–35 [DOI] [PubMed] [Google Scholar]
- 10.Pincus T, Yazici Y, Bergman MJ. RAPID3, an index to assess and monitor patients with rheumatoid arthritis, without formal joint counts: similar results to DAS28 and CDAI in clinical trials and clinical care. Rheum Dis Clin North Am 2009;35:773–8, viii [DOI] [PubMed] [Google Scholar]
- 11.Gater A, Abetz-Webb L, Battersby C, et al. Pain in castration-resistant prostate cancer with bone metastases: a qualitative study. Health Qual Life Outcomes 2011;9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. IMIA Yearbook 2008: Access to Health Information 2008:128–44 [PubMed] [Google Scholar]
- 13.Wang X, Chused A, Elhadad N, et al. Automated knowledge acquisition from clinical narrative reports. AMIA Annu Symp Proc 2008;2008:783–7 [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C-H, Wu C-H, Yang H-C.Text mining of clinical records for cancer diagnosis innovative computing, information, and control. 2007.
- 15.Bassøe C-F. Automated diagnoses from clinical narratives: a medical system based on computerized medical records, natural language processing, and neural network technology. Neural Networks 1995;8:313–19 [Google Scholar]
- 16.Roque FS, Jensen PB, Schmock H, et al. Using electronic patient records to discover disease correlations and stratify patient cohorts. PLoS Comput Biol 2011;7:e1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kushima M, Araki K, Suzuki M, et al. Text data mining of the electronic medical record of the chronic hepatitis patient. Hong Kong: IMECS, 2012 [Google Scholar]
- 18.Xu H, Fu Z, Shah A, et al. Extracting and integrating data from entire electronic health records for detecting colorectal cancer cases. AMIA Annual Symposium proceedings 2011;2011:1564–72 [PMC free article] [PubMed] [Google Scholar]
- 19.Uzuner O, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008;15:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savova GK, Ogren PV, Duffy PH, et al. Mayo clinic NLP system for patient smoking status identification. J Am Med Inform Assoc 2008;15:25–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman WW, Christensen LM, Wagner MM, et al. Classifying free-text triage chief complaints into syndromic categories with natural language processing. Artif Intell Med 2005;33:31–40 [DOI] [PubMed] [Google Scholar]
- 22.Childs LC, Enelow R, Simonsen L, et al. Description of a rule-based system for the i2b2 challenge in natural language processing for clinical data. J Am Med Inform Assoc 2009;16:571–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel C, Cimino J, Dolby J, et al. Matching Patient Records to Clinical Trials Using Ontologies. IBM Research Report. May 23, 2007. (RC24265).
- 24.Chute CG. The horizontal and vertical nature of patient phenotype retrieval: new directions for clinical text processing. Proc AMIA Symp 2002;2002:165–9 [PMC free article] [PubMed] [Google Scholar]
- 25.Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62:1120–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pakhomov SV, Jacobsen SJ, Chute CG, et al. Agreement between patient-reported symptoms and their documentation in the medical record. Am J Manag Care 2008;14:530–9 [PMC free article] [PubMed] [Google Scholar]
- 27.Hyun S, Johnson SB, Bakken S. Exploring the ability of natural language processing to extract data from nursing narratives. Comput Inform Nurs 2009;27:215–23; quiz 24–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pakhomov SS, Hemingway H, Weston SA, et al. Epidemiology of angina pectoris: role of natural language processing of the medical record. Am Heart J 2007;153:666–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system. Methods Inf Med 1993;32:281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uzuner O. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc 2009;16:561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uzuner O, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007;14:550–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts A, Gaizauskas R, Hepple M, et al. Combining terminology resources and statistical methods for entity recognition: an evaluation. Proceedings of the Sixth International Conference on Language Resources and Evaluation, LREC 2008, Marrakech, Morocco 2008 [Google Scholar]
- 33.Friedman C, Hripcsak G, DuMouchel W, et al. Natural language processing in an operational clinical information system. Nat Lang Eng 1995;1:83–108 [Google Scholar]
- 34.Childs LC, Cassel D. Extracting and normalizing temporal expresssions. TIPSTER ‘98. Baltimore, MD: Association for Computing Machinery, 1998 [Google Scholar]
- 35.Cassel DM, Taylor SM, Katz GJ, et al. Automated capture and representation of date/time to support intelligence analysis. Intelligence Tools Workshop; 2006; 2006
- 36.NIST 2005 Automatic content extraction evaluation official results (ACE05). 2005
- 37. Negri M, Marseglia L. Recognition and normalization of time expressions: ITC-irst at TERN 2004. Technical Report WP3.7, Information Society Technologies, February 2005.
- 38.Mayer P. Julian Day Numbers. Cited; http://www.hermetic.ch/cal_stud/jdn.htm
- 39.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 40.Strötgen J, Gertz M. Multilingual and cross-domain temporal tagging. Springer: Lang Resources Eval 2012. http://link.springer.com/article/10.1007%2Fs10579-012-9179-y
- 41. Cunningham H. GATE, a general architecture for text engineering. Computers and the Humanities 36.2, 2002:223–54.
- 42. Mazur P, Dale R. The DANTE Temporal Expression Tagger. In: Proceedings of the 3rd Language & Technology Conference (LTC), Poznan, Poland, 5–7 October 2007.
- 43.Murphy TV, Gargiullo PM, Massoudi MS, et al. Intussusception among infants given an oral rotavirus vaccine. New Engl J Med 2001;344:564–72 [DOI] [PubMed] [Google Scholar]
- 44.Prostate Cancer Diagnosis and Treatment NICE Clinical Guidelines, No 58. Cardiff(UK): National Collaborating Centre for Cancer (UK), 2008 [PubMed] [Google Scholar]
- 45.Catton CN, Gospodarowicz MK. Palliative radiotherapy in prostate cancer. Semin Urol Oncol 1997;15:65–72 [PubMed] [Google Scholar]
- 46.Carver AC, Foley KM. Management of cancer pain. In: Kufe DW, Pollock RE, Weichselbaum RR, et al., eds. Holland-Frei cancer medicine. 6th edn Hamilton, ON: BC Decker, 2003 [Google Scholar]
- 47.Thompson I, Tangen C, Tolcher A, et al. Association of African-American ethnic background with survival in men with metastatic prostate cancer. J Nat Can Inst 2001;93:219–25 [DOI] [PubMed] [Google Scholar]
- 48.Hoste V, De Pauw G. KNACK-2002: a richly annotated corpus of Dutch written text. 2006
- 49.Ogren PV, Savova G, Buntrock JD, et al. Building and evaluating annotated corpora for medical NLP systems. AMIA Annu Symp Proc. 2006;2006:1050 [PMC free article] [PubMed] [Google Scholar]
- 50.Uzuner O, Solti I, Xia F, et al. Community annotation experiment for ground truth generation for the i2b2 medication challenge. J Am Med Inform Assoc 2010;17:519–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction. Using electronic medical records as a tool for genome science. Circulation 2010;122:2016–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kullo IJ, Fan J, Pathak J, et al. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc 2010;17:568–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kullo IJ, Ding K, Jouni H, et al. A genome-wide association study of red blood cell traits using the electronic medical record. PLoS ONE 2010;5:e13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet 2010;86:560–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Washington NL, Haendel MA, Mungall CJ, et al. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS biology 2009;7:e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.