Abstract
Objective
Biomedical research increasingly relies on the integration of information from multiple heterogeneous data sources. Despite the fact that structural and terminological aspects of interoperability are interdependent and rely on a common set of requirements, current efforts typically address them in isolation. We propose a unified ontology-based knowledge framework to facilitate interoperability between heterogeneous sources, and investigate if using the LexEVS terminology server is a viable implementation method.
Materials and methods
We developed a framework based on an ontology, the general information model (GIM), to unify structural models and terminologies, together with relevant mapping sets. This allowed a uniform access to these resources within LexEVS to facilitate interoperability by various components and data sources from implementing architectures.
Results
Our unified framework has been tested in the context of the EU Framework Program 7 TRANSFoRm project, where it was used to achieve data integration in a retrospective diabetes cohort study. The GIM was successfully instantiated in TRANSFoRm as the clinical data integration model, and necessary mappings were created to support effective information retrieval for software tools in the project.
Conclusions
We present a novel, unifying approach to address interoperability challenges in heterogeneous data sources, by representing structural and semantic models in one framework. Systems using this architecture can rely solely on the GIM that abstracts over both the structure and coding. Information models, terminologies and mappings are all stored in LexEVS and can be accessed in a uniform manner (implementing the HL7 CTS2 service functional model). The system is flexible and should reduce the effort needed from data sources personnel for implementing and managing the integration.
Keywords: Translational Medical Research, Interoperability, Semantics, Terminology, Ontology, LexEVS
Introduction
Biomedical research increasingly relies on the integration of information from multiple data sources, obtained either primarily for the purposes of research, such as trial data and genetic samples, or through secondary use of routinely collected data, for example, electronic health records (EHR). However, the heterogeneity of these data sources represents a major challenge to the research task.1–3 Two levels of heterogeneity can be distinguished: structural and terminological. First, information models are used to represent the organization of data structures in information systems.4–6 Variation in their forms and approaches generates structural heterogeneity of the data models. Second, numerous medical coding systems (terminologies) are used to represent diagnoses, procedures, and treatments in health databases,7 frequently with many-to-many mappings between them, creating semantic heterogeneity, sometimes also referred to as terminological heterogeneity.8
Rector8 mentions that these two types of heterogeneity, structural and semantic, are not independent as there are mutual constraints between the information models and coding systems.9 This interdependence corresponds to what Rector calls the ‘binding’ between an information model and a coding system, and presents a notorious source of ambiguity in clinical systems.4 At the time of coding, implicit knowledge is sometimes used but not formally represented in the information model. Some models function under the closed world assumption, whereby omission implies falsehood, while others support the open world assumption in which omission merely states that the information is not available. Further complexity is caused by differences in granularity, depth, coverage and composition (single term vs expressions) between models.
This article proposes a unified framework for the integration of heterogeneous information models and terminologies to construct a single solution for structural and semantic interoperability. This approach is currently being adopted in TRANSFoRm, a EU FP7 project that aims to support the integration of clinical and translational research data comprehensively in the primary care domain.10 11
Background and significance
Structural and semantic interoperability in biomedical data has been explored in a number of initiatives. Given our interest in translational medicine and data reusability, we focus here on those allowing federated queries from multiple clinical repositories and EHR.
There have been attempts to create generic information models to serve as standards, including the OpenEHR reference model, the informatics for integrating biology and the bedside (i2b2) model, the HL7 reference information model and the clinical data acquisition standards harmonization (CDASH).12–15 An ongoing international collaboration between standards organizations and industry partners, the clinical information modeling initiative, aims at bringing together a variety of approaches to clinical data modeling (HL7 templates, openEHR archetypes, etc) as a series of underlying reference models.16 A similar endeavor is ongoing with the biomedical research integrated domain group in the research area.17 Nevertheless, many existing data sources are not designed according to these initiatives.
Approaches to structural heterogeneity can be grouped in two categories: extract-transform-load (ETL) systems and mediators systems. In the former, the different data sources to be integrated (eg, data warehouses) are all expected to conform to some structural model. This is achieved by carrying out an ETL process on an existing relational database to transfer the data into a single target model. Multiple projects have been built on this approach. The shared health research information network (SHRINE) aims at bringing together various i2b2 clinical data repositories.13 18 19 The i2b2 model is also used by other projects like TRANSMART.20 The Stanford translational research integrated database environment, an initiative from Stanford, uses the HL7 reference information model as a foundation for their model while EU-ADR developed its own common model.21 22 Finally, the electronic primary care research network (ePCRN) project, focusing on the primary care domain, based its structure on the American Society for Testing and Materials continuity of care record information model.23 24
Other systems use a mediator approach to address structural heterogeneity. Some central schema is mapped to the local schemas of individual data sources, which retain their original structure. These central schemas were initially described as ontologies.25 Projects such as advancing clinico-genomic trials in the cancer domain leveraged this approach.26 Other projects implemented mediators in different ways. The biomedical informatics research network (BIRN) and its follow-up initiative the neuroscience information framework are using an XML approach.27–29 The cancer biomedical informatics grid (caBIG) is a long-standing National Cancer Institute (NCI)-driven initiative to federate healthcare data with sources represented as unified modeling language (UML) models.30–32 A similar modeling approach is used by the federated Utah research and translational health e-repository (FURTHeR) and electronic health record for clinical research.33 34 None of these implementations use vocabulary services to support their structural aspects.
The terminological needs of various projects are handled internally. The SHRINE project uses a pivot terminology and BIRN stores term mappings in a relational database.35 36 The smart open services for European patients (epSOS) project is developing an ontology to address the multilingual and mapping needs of its community.37 38 Nevertheless, terminology servers are often involved like Apelon DTS in FURTHeR and Bioportal in ONCO-I2B2.39 40
The LexEVS terminology server, having originally been developed in the context of the caBIG initiative, is being used by several projects (eg, ePCRN, NCI thesaurus browser).24 41 42 The web-based server bioportal also uses it as part of its infrastructure.43 LexEVS permits unification of all loaded terminologies under the LexGrid format (including ontologies expressed as ontology web language).44 It allows a range of deployment options, from a local installation to a grid service, and is available under an open source license. V.6 of LexEVS implements the HL7 common terminology services 2 (CTS 2) service functional model (SFM), although it does not conform to the HL7 CTS 2 OMG specification because the specification was finalized after V.6 was released.45 46 Prior to our efforts, LexEVS implementations have mostly been used to support terminological information.
Binding between information models and terminologies presents a challenge in its own right. A number of projects mentioned above have developed their own solutions; nevertheless, standards for metadata registries have been created to address this question (eg, ISO 11179).47 Projects such as eMERGE and caBIG use the cancer data standard repository (caDSR).48 It stores data elements described by a definition of what is represented as well as the list of valid values. caBIG binds its UML models with the terminologies through use of these data elements. eMERGE also uses the caDSR to harmonize local genotype and phenotype data elements. The binding of structure and terminology has also been addressed in the context of HL7 with the TermInfo initiative currently focusing on the use of SNOMED CT in HL7 V3.49
All of these projects consider structural and semantic aspects of interoperability to be distinct, leading them to be managed separately, although the separation between structure and terminology is drawn differently in different projects. Recognizing their dependencies and that terminological and structural operations share a common set of requirements (through binding and mappings), we hypothesized that a unified ontology-based knowledge framework can facilitate interoperability between heterogeneous sources, without having to create a separation and different tools for management. Based on our analysis of terminological solutions, we investigated whether LexEVS was a functional tool to implement this approach.
In the next section, we present the framework and describe the generic approach for each of its components. We then test this method on a clinical study example from the TRANSFoRm project, focusing on integrating two primary care data repositories, the NIVEL primary care database (NPCD)50 of the Netherlands Institute for Health Services Research (NIVEL)51 and the general practice research database (GPRD)52 of the UK's Medicines and Healthcare Products Regulatory Agency.53
Materials and methods
The main aims of our work are to simplify the handling of heterogeneous data sources for the users and to minimize the interoperability implementation workload for the data sources. We believe the mediation paradigm best meets these goals.25 Instead of using ETL to enforce a uniform information model, our framework uses mappings to relate local models to a general information model (GIM). This also facilitates user operations as they only need to interact with the general model and do not need to be familiar with each data source's information model.
The mediation framework has been constructed according to the local-as-view principle.54 In this approach, each source schema is defined as a set of views on the global schema, as opposed to the global-as-view principle in which the global schema is defined in terms of the sources. So the GIM does not have to be derived directly from any source. Rather, it should be built to construct a sound and logical view of the domain of interest in order to make sure all required concepts are present. This ensures scalability, as adding a new source does not necessitate a modification of the GIM. It also presents a more stable model to the user.
In our framework, GIM is represented as an ontology, allowing it to be stored in the LexEVS terminology server together with the data source models (DSM) and the terminologies. Mappings between GIM and data sources can then be uniformly created, stored and leveraged as described below. In parallel, similar methods can be used to handle terminological operations.
Architecture overview
The modeling infrastructure resides entirely within a terminology server, enabling unification of structural and semantic modeling and operations within this server. Several types of models are present:
The GIM
Models describing each data source (DSM)
Mapping sets between the sources and the GIM—one set per source
Terminologies used to code the data elements (eg, International Classification of Diseases (ICD)-10 codes…)
Mappings between terminologies.
An overview of how the different models interact together is presented in figure 1, which shows a user query being sent to mulitple data sources. Security and other administrative issues have been intentionally left out of this list in order to focus on the relevant steps for this demonstration.
The query is expressed using GIM concepts
The mediation engine generates a specific query for each data source
The data sources fulfill the requests
- The returned dataset has its structure aligned with the GIM
- DSM to extract which terminology was used to code a given concept in the source
- If possible and desired, the system can semantically align resulting coded values based on the terminologies used by one of the sources or a separate terminology. This operation uses:
- Terminologies and mappings between terminologies to transcode the values.
Figure 1.

Architecture supporting model interactions based on LexEVS for query mediation-based query resolution.
General information model
The GIM is used to represent a unified view of the domain concepts and their relationships. For example, date of birth, diagnosis and patient are all relevant concepts in a clinical care context. Each concept also has intrinsic properties. Given the data integration function of the ontology and its role as a mediation schema, we chose a realist approach using basic formal ontology (BFO) 1.1 as the foundation of the model.55 56 The implementation of BFO as a formal, description logics ontology allows easier interaction with projects using semantic web technologies (like epSOS), or other parts of projects implementing the framework. For example, the provenance service and the decision support service from TRANSFoRm both rely on ontologies and will need to interact closely with the unified integration framework.
Figure 2 illustrates how ‘gender’ and its relevant attributes represented in GIM are rendered once loaded in LexEVS.
Figure 2.

General information model—partial representation of ‘gender’ attributes in LexEVS.
The ‘codedWith’ properties of the concept support binding between the information model and the relevant terminology (or value set) and contribute to its semantics representation. In this case, it indicates that values for this concept are to be represented with the terminology named ‘gim_gender’ stored in LexEVS. Multilingual capabilities are handled natively within LexEVS by combining property values with a language descriptor. When a translation is provided, this allows the model also to propose a multilingual solution without resorting to another system.
Data source models
A new DSM is defined for every data source to be supported. The goal of this stage is to provide enough information to the system in order to translate a query based on the GIM into the local language used to query the source. The exact nature of the properties and relations will be related to the underlying type of source to be modeled.
For example, a SQL data source ‘SA’ would have hierarchical relations such as hasTable and hasField with other relations representing the relations between the tables (oneToMany, OneToOne…) with the keys on each side. Another data source ‘SB’ could be an XML document, with XPath as its query language. A model fulfilling the same goal can be created describing nodes, elements and attributes.
A DSM fragment is illustrated in figure 3, representing a field. In terms of concept properties, we have some similarities with the GIM but also specific properties for a SQL source concept.
Figure 3.

Data sources models—partial representation in LexEVS of a field named ‘GESLACHT’ from a source SQL database.
The objectType property gives the nature of the concept (field) while the name of the object is in the description. Multiple textual presentations (here Dutch and English) can be created to provide translations in order to facilitate the use of the information in multiple contexts. As with the GIM, ‘codedWith’ properties hold the name and versions of the terminology (or local value set) used to code data for this concept (a field in this example). Note that this does not need to be the same terminology in all DSM and GIM. This allows a DSM to register the specific terminology (or value set) used to code the information locally, irrespective of what is registered with GIM.
Mappings between a source and the GIM
A mapping set does not need to duplicate the concepts from the model but simply reference them via their code and coding scheme name. A relation is then created for each correspondence between a GIM concept and a DSM concept.
We developed a generic mapping model defining data transformation operations to align source data values with the GIM, supporting not only one-to-one mappings but also more complex cases. One-to-one operations include simple mappings such as a date corresponding to a date/time value, while a more complex case would consist of two distinct but related fields. For example, a symptom (a code from a terminology) can possibly denote multiple entity types (in GIM). For example, ‘abdominal pain’ can be used to code a ‘presenting complaint’, a ‘symptom’ or even sometimes a ‘final diagnosis’ if no clear diagnosis emerges during the consultation. Some data sources, instead of having three fields representing the three possible entity types, will have two fields: one storing the actual symptom code and one for the entity type. For example, field A would store the value ‘abdominal pain’, while field B would store the entity type ‘presenting complaint’ in the same record, to distinguish it from someone with a diagnosis of abdominal pain as part of their medical history.
In this case, instead of linking directly from the source to the GIM, an intermediate concept is created in the mapping set. This intermediate concept will hold the condition for this relation to be true. So, if our example maps to some concept AP154 in GIM, the mapping would proceed as Field A→Condition 1 (Field B=‘Value 1’)→GIM AP154, that is, Field A represents GIM concept AP154 only if Field B=‘Value 1’. Intermediate concepts can also be chained in order to combine different operations.
The model also supports the creation of a virtual element to capture implicit knowledge. For example, it could represent a laboratory unit that might not be physically present in the data source because it is always the same in the context of that source. Similarly, the mapping model can support yes/no fields (eg, a column denoting the presence or absence of diabetes), which combines both the structural and terminological elements.
Terminologies
The UMLS presents a unified view of a large number of relevant biomedical terminologies.57 It includes over two million concepts from various vocabularies and millions of relationships. By using concept unique identifiers—used to relate codes in different terminologies but with a similar meaning—and semantic groups, it facilitates terminology alignment. The UMLS can be loaded directly in LexEVS 6, which supports all its features.
Additional LexEVS loaders are easily created to load terminologies that are not yet supported. This was exemplified by the creation of a loader for the anatomical therapeutic chemical classification system (ATC 2011) in collaboration with the LexEVS developers.
Mappings between terminologies
Once terminologies are loaded in LexEVS, mappings between them can be created in a similar way as for the data models. For some of them, relationships are readily available and can be simply loaded into LexEVS. This is typically the case for terminologies integrated in the UMLS.
For others, local mappings have to be created. For example, if a hospital uses a local coding set to identify its laboratory tests, it could be loaded into LexEVS. Subsequently, mappings between this local set and logical observation identifiers names and codes could be created. This would allow translations from the local site to a more standard terminology, thereby facilitating interoperability with other groups without having to recode data locally or create a duplicate data warehouse.
When more than two terminologies are used, mapping sets can be created between each of them or only to some selected central (pivot) terminology, which then acts as a hub for translating concepts. A pivot terminology is optional in the GIM framework and left for the users to decide on. In the absence of a designated terminology, the user can choose one of the terminologies supported in the selected sources to which the others will attempt to map.
Results
The first implementation of GIM was realized as part of the EU FP7 TRANSFoRm project, which aims at supporting patient safety through integration of clinical and research settings, workflows and data.11 The technology developed can facilitate the interactions with individual EHR systems for trial recruitment and follow-up, as well as diagnostic support. The TRANSFoRm project also relies on a workbench to explore clinical and research data repositories. To achieve this, significant challenges need to be overcome in the areas of interoperability and methods for data integration.
Clinical data integration model: GIM instantiation in TRANSFoRm
The clinical data integration model (CDIM) is the GIM instantiation in TRANSFoRm, and covers concepts relevant to data integration in primary care research such as medication, diagnosis, and laboratory tests. It is implemented as an ontology web language ontology based on the BFO 1.1.56 It imports the general medical science,58 the vital sign ontology59 and the information artifact ontology.60 The ontology also integrates concepts from existing ontologies such as the ontology for biomedical investigations,61 the gene ontology62 and the translational medicine ontology63 when possible.
The resulting ontology has 457 classes (102 unique to CDIM) and 73 object properties (1 sub-property unique to CDIM). Twenty-one novel CDIM classes had to be introduced to represent and manage temporal aspects necessary in TRANSFoRm. All required concepts, as defined by use cases, could be modeled in CDIM. Figure 4 presents a subset of CDIM adapted to illustrate a subset of queries related to the diagnosis of diabetes.
Figure 4.
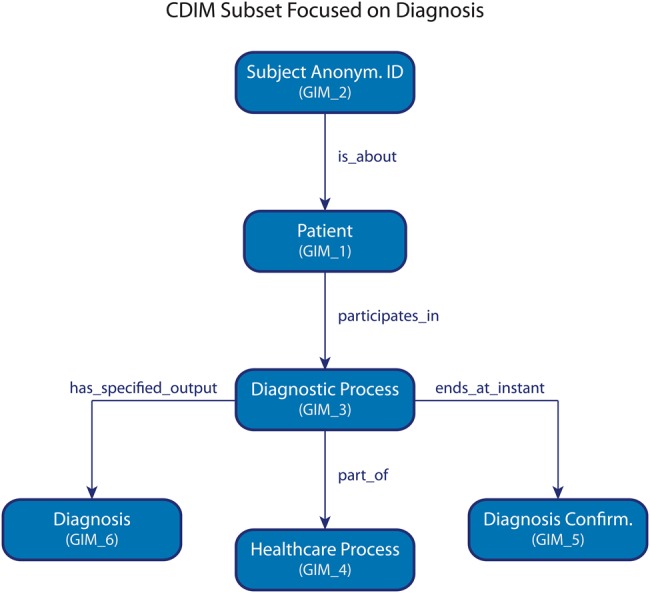
Clinical data integration model subset focused on diagnosis. Identifiers are in parentheses.
Instantiation of structural models, terminologies and mappings
Two clinical data repositories were used to evaluate the suitability of the framework for the project: NPCD from the Netherlands and GPRD from the UK. Both their structures and the terminologies used to code information are different. For example, medication is coded with the British national formulary (BNF) codes in GPRD but the ATC classification is used in NPCD, with diagnoses coded with read codes V.2 in GPRD and ICPC V.1 in NPCD.
Structural models in XML were created for both sources using a semi-automated tool and then loaded into LexEVS. The NPCD database extract we used contained 60 521 anonymized patient records, whereas the GPRD extract made available for the project contained 5000 patient entries. Eight tables (181 fields) in NPCD and 10 tables (107 fields) in GPRD were considered in the structural models.
CDIM was mapped with 44 elements in NPCD and 47 in GPRD. High level classes such as ‘processual entity’ are part of CDIM and are essential to knowledge modeling but are not expected to be used as mapping targets as they are too generic. Twenty-nine mappings (32%) were one-to-one direct relations between CDIM concepts and a data source structural element. The other mappings included concatenation operations and conditional mappings (including related tables). No virtual elements were necessary for the current data source mappings. Figure 5 illustrates an example of a conditional mapping. Precise and comprehensive knowledge of each data source and its real-life usage was essential to achieve satisfactory mappings and query results. Not all fields of the data sources are targets for mappings, nor are all concepts in CDIM mapped to each data source; their coverage typically differs from CDIM. Nevertheless, all the relevant entities for the use cases were successfully mapped. Figure 5 presents those mappings necessary to illustrate the examples in figure 6.
Figure 5.

Mapping examples between general information model and data sources models (general practice research database and NIVEL primary care database). Identifiers are in parentheses.
Figure 6.

Examples of query resolution as applied to TRANSFoRm using clinical data integration model (figure 4), its mappings to the data sources models (figure 5) and terminologies. Highlighted segments represent each level-specific addition based on information from the models served by LexEVS.
Based on our use case and available data sources, we focused on ICD-9 and ICD-10 codes, International Classification of Primary Care (ICPC) V.1 codes, read codes V.2 for diagnoses, the ATC, the BNF for drugs, as well as on logical observation identifiers names and codes for laboratory tests.
Evaluation
We evaluated the applicability of the GIM approach to TRANSFoRm's clinical trial use cases. We focused on the retrospective diabetes cohort study.64 This use case aims at identifying eventual associations between single nucleotide polymorphism and diabetes complications or responses to oral antidiabetic drugs. Twenty-six relevant queries were identified and were all successfully implemented, in conjunction with appropriate terminological values. For example:
Patients ≥35 years old
AND
((with a diagnosis of diabetes accompanying a prescription or an episode of care)
OR (taking metformin OR a sulfonylurea medication in last 5 years)
OR (having a laboratory test of glycosylated hemoglobin >6.5%
OR a random glucose >11.0 mmol/l
OR a fasting glucose >7.0 mmol/l))
Figure 6 demonstrates different features of the LexEVS implementation of the framework. The first example illustrates how to create the local source query based on information contained within CDIM and the DSM. The latter would contain field and table relations required to derive the SQL statement. By utilizing the mappings shown in figure 5, the query is translated in the local source query format.
Similar principles can be applied for multiple sources, but as shown in the first example of figure 6, the resulting dataset structure is based on the local source. In the example, it is not clear that ‘DIAGNOSE’ and ‘medcode’ carry a similar meaning, especially as this equivalence is only true if a condition on the field ‘constype’ is applied. By adjusting the local query to maintain a reference to CDIM, the resulting datasets from two data sources (NIVEL and GPRD) can be assembled in a coherent structure as in example 2.
Although both result sets now share an identical structure, the terminologies used to code the information are different. In some situations, alignment might not even be possible, at least not in a completely automated fashion as with ATC and BNF for medication types. In this diabetes example, we consider the ‘coded with’ properties in the local DSM, as previously described. For GPRD, ‘Non-insulin dependent diabetes mellitus’ in read codes V.2 can be related to an ICD-10 code (E11) by following mappings in LexEVS. The same can be done for NIVEL with ICPC-1 code T90.02 to ICD-10 code E11. The final unified dataset is homogenous and consistent semantically as in example 3.
Discussion
Achieving interoperability between health data sources such as EHR and registries is a challenging but crucial endeavor for both designers and users of health information technology. The structural and terminological aspects of data source interoperability, while intrinsically linked, have traditionally been handled separately.65 66 From a structural perspective, a number of projects have adopted a common model to which each source is expected to comply, whether when inputting data (eg, CDASH in the clinical research domain)15 or when data are being extracted (eg, EU-ADR focusing on adverse event analysis).22
Other projects have opted for a mediation approach, with a centralized knowledge model, often represented as an ontology. XML and UML designs are also possibilities, as utilized in the BIRN and FURTHeR projects, respectively. Our framework is built around GIM as the central knowledge model, expressed as an ontology with a realist approach based on BFO 1.1.
The semantic challenges are addressed either through dedicated project-specific tools or through terminological servers, such as the one used in the ePCRN project. The GIM framework is novel in that it uses a terminological server not only for handling semantic interoperability, but for structural aspects as well.
Binding both terminological and structural aspects, when they are managed separately, is a challenge that has previously been handled through the use of metadata registries such as caDSR, as used in the caBIG and eMERGE projects.30 67 The registries allow data elements to be created in which a definition and a list of permissible values is attached. Our framework avoids this situation by handling the binding in the mediation structure, in which both sets of models are located already. It allows data elements present in existing data sources to be described and integrated readily in the context of GIM and allows the use of local code value sets easily as they are stored in the framework.
Our approach represents a step beyond the traditional interoperability paradigm involving a different set of tools for dealing with structural, terminological and binding challenges, in that we present a unified framework that provides an integration solution for these facets inside a single structure. Our LexEVS implementation of GIM, as demonstrated in the TRANSFoRm project, allows a query to be expressed using clinical concepts from a single generic model that is represented as an ontology, and allows its translation into source-specific queries, which then return the results from each source, simplifying and standardizing the interoperability task.
Strengths and limitations
One of the biggest barriers to the usage of federated data sources is the resource and effort expected from the data sources to participate in a collaborative structure.3 In order to mend heterogeneity between two data sources, related elements must be mapped to each other. Whether structural models, such as database schemas, or terminologies are to be aligned, the processes share a common subset of requirements.68 Multiple approaches have been developed to address the issue.57 69 Our infrastructure does not necessitate a priori substantial changes to the structure of the data source. If desired, ETL may be used to transform the initial data schema into a derived schema closer to GIM, and this could facilitate the use of direct mappings. If an organization already has a data warehouse, it might be used as is, thereby reducing integration effort and avoiding data duplication.
The architecture presented decouples the interoperability modeling aspects from the application itself. For some data sources, especially EHR, exposing the structure of their databases might not be possible or desirable. In this case, an instance of LexEVS can be installed on a local server, allowing query translation to happen at the local level.
From the maintenance perspective, the addition of a new piece of information to a source will necessitate mappings to the relevant GIM terms before becoming usable.9 Note that our approach can leverage the GIM semantic richness to make this mapping step easier.70 This occurred with the CDIM implementation of GIM in the TRANSFoRm project, in which we use ‘codedWith’ properties to suggest concepts that might share similar semantics. Similarly, distance between concepts in the graph can be used to suggest related concepts. Mappings within TRANSFoRm are currently created manually but should it be expanded, mapping tools will be required in order to support its development. Our LexEVS implementation supports most attributes necessary to allow such work.70 This has recently been identified as a core challenge to the field by Shvaiko and Euzenat,68 and we believe that our approach can contribute to an alignment infrastructure, fostering collaboration.
There are a number of advantages to using LexEVS as the implementation technology. The GIM ontology is stored in the LexEVS terminology server, allowing us to leverage its two optimization axioms: ‘fully restrict then query’ and ‘lazy loads’. The former minimizes resource requirements by allowing the system to restrict any query fully, including operations on sets (eg, intersections, unions or differences) before running it against the data source. The latter technique preferentially loads only certain types of information in the first pass while retaining a pointer to load more information dynamically should this be needed. Together, these facilitate efficient query mediation on heterogeneous data sources.
Our approach also benefits directly from LexEVS capabilities for handling versioning. Multiple versions of the models, terminologies and mappings can coexist in the system, and be maintained independently from our framework, removing the need for a separate implementation of versioning. Similarly, multilingual capabilities supported by LexEVS can be used for many operations without resorting to an ancillary tool.
Once loaded and functional, the framework can leverage intrinsic capabilities of LexEVS to create value sets (ie, subsets of related concepts), which can then be used to handle terminological needs (eg, codes used to represent drugs to treat diabetes) and manage GIM concept groups. For example, relevant concepts related to laboratory tests can be grouped in order to facilitate searching and browsing. This is different from other efforts in which structural models are stored in project-specific structures. Using LexEVS to manage GIM and DSM automatically provides the methods that implement the HL7 CTS 2 SFM, and ultimately HL7 CTS 2 OMG, ensuring that the implementation remains maintainable and reusable.71
The level of automation for query translation and results aggregation depends on the possibility of creating meaningful mappings between relevant terms.72 73 We showed in our example that mappings between different terminologies can be utilized to automate the process fully for some situations. Nevertheless, some terminology pairs do not lend themselves to such an exercise. These include the ATC and BNF terminologies for therapeutic substances.74 75 Their approach to classification varies in granularity, depth and coverage, leading for some terms to one-to-many mappings or absence of related concept. In such a scenario, the infrastructure can readily support a user interface in which similar, but not necessarily equivalent, terms in different terminologies used by different sources could be suggested, edited and finally approved by the user instead of being automatically chosen.
Applicability
The infrastructure is currently being deployed in the pan-European TRANSFoRm project, with a view to deploying it in other EU and US translational research projects in academia and industry. Specific TRANSFoRm activities that require combined semantic and structural integration include:
Support for dynamic and persistent linkage between data sources for widely scalable epidemiological studies.
Support for clinical decision support embedded in the EHR, enabling capture and recording of clinical diagnostic cues in a controlled form.
Support for real time linkage to a variety of different EHR systems for extraction of clinical data elements into an electronic case report form and write-back of controlled data elements to the EHR to serve as an eSource for regulated clinical trials.
Deploying CDIM as a unified framework in this setting allows the project tools to have full control over the content and structure of queries sent to data sources, and demonstrated its applicability to multiple deployment scenarios, including distributed installations. This study showed that this unified framework, supported by LexEVS, is a suitable platform in which to achieve these tasks in the context of two exemplar databases. The tool chosen in TRANSFoRm was LexEVS. Nevertheless, in a different context, other tools such as Bioportal might also have the potential to support the framework.
Conclusion
In this paper, we presented a novel, unifying approach to address interoperability challenges in heterogeneous data sources, by representing structural and semantic models in a single framework. This represents a significant departure from the previous strategies for addressing interoperability in translational research, and it has been successfully demonstrated within the context of the clinical research studies of the EU TRANSFoRm project.
The advantage of this approach is that the systems using the architecture can rely solely on GIM concepts, abstracting over both the structure and coding specificities of the data sources. Information models, terminologies and mappings are all stored in LexEVS and can be accessed using the same methods (implementing the HL7 CTS 2 SFM). The system is flexible, and should reduce the integration effort required from the data sources, thereby lowering the cost of entry of this type of research for smaller institutions, and removing the need for larger institutions to invest in additional data warehousing.
Acknowledgments
The authors would like to thank their colleagues from the TRANSFoRm project for their support and insightful discussions regarding this endeavor. In particular, Peter Leysen, Hilde Bastiaens, and Anna Nixon Andreasson developed the use cases. Nasra Khan provided data extracts and processing at NIVEL, and Nick Wilson at CPRD. Jean-Karl Soler assisted in terminology mappings. Lei Zhao developed terminology services in TRANSFoRm. The authors also thank the members of the LexEVS development team for their invaluable help.
Footnotes
Contributors: JFE created, designed and tested the generic framework, created the ontology and drafted the manuscript. AB and OD participated in the design of the generic framework and worked on the ontology development. AB, OD, VC and BCD helped to draft the manuscript. AT, TNA and BCD contributed to the conception and design of the TRANSFoRm project and its architecture. JFE, VC, AB, MMM, TNA and AT participated in the implementation of the framework in TRANSFoRm. JFE, AB and TNA contributed to the TRANSFoRm vocabulary services. MMM, JFE and RAV worked on the TRANSFoRm data sources model structures. All authors critically reviewed the manuscript and approved the final version.
Funding: This work was supported in part by the European Commission—DG INFSO (FP7 247787).
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Cimino J J. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med 1998;37:394–403 [PMC free article] [PubMed] [Google Scholar]
- 2.Cimino JJ. In defense of the Desiderata. J Biomed Inform 2006;39:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sujansky W. Heterogeneous database integration in biomedicine. J Biomed Inform 2001;34:285–98 [DOI] [PubMed] [Google Scholar]
- 4.Rector AL, Qamar R, Marley T. Binding ontologies and coding systems to electronic health records and messages. Appl Ontol 2009;4:51–69 [Google Scholar]
- 5.Eichelberg M, Aden T, Riesmeier J, et al. A survey and analysis of electronic healthcare record standards. ACM Comput Surv 2005;37:277–315 [Google Scholar]
- 6. Haux R, Kulikowski C, editors. IMIA Yearbook of Medical Informatics 2006. Methods Inf Med 2006;45(Suppl 1):S136–44. [Google Scholar]
- 7. Geissbuhler A, Kulikowski C, editors. IMIA Yearbook of Medical Informatics 2008. Methods Inf Med 2008;47(Suppl 1):67–79. [Google Scholar]
- 8.Rector AL. Clinical terminology: why is it so hard? Methods Inf Med 1999;38:239–52 [PubMed] [Google Scholar]
- 9.Qamar R, Kola JS, Rector AL. Unambiguous data modeling to ensure higher accuracy term binding to clinical terminologies. AMIA Annu Symp Proc 2007;2007:608–13 [PMC free article] [PubMed] [Google Scholar]
- 10.Delaney B. TRANSFoRm: translational medicine and patient safety in Europe. In: Grossman C, Powers B, McGinnis JM, eds. Digital infrastructure for the learning health system: the foundation for continuous improvement in health and health care: workshop series summary. Washington, DC: National Academies Press, 2011: 198–202 [PubMed] [Google Scholar]
- 11.TRANSFoRm Project http://www.transformproject.eu (accessed 11 Apr 2012).
- 12.Beale T, Heard S, Kalra D, et al. The openEHR reference model—EHR information model—Release 1.0.2. http://www.openehr.org/releases/1.0.2 (accessed 29 Jun 2012)
- 13.Murphy SN, Mendis M, Hackett K, et al. Architecture of the open-source clinical research chart from informatics for integrating biology and the bedside. AMIA Annu Symp Proc 2007;2007:548–52 [PMC free article] [PubMed] [Google Scholar]
- 14.Schadow G, Mead CN, Walker DM. The HL7 reference information model under scrutiny. Stud Health Technol Inform 2006;124:151–6 [PubMed] [Google Scholar]
- 15.CDASH—Basic Recommended Data Collection Fields for Medical Research http://www.cdisc.org/cdash (accessed 8 Dec 2012).
- 16.Clinical Information Modelling Initiative http://www.openehr.org/326-OE.html?branch=1&language=1 (accessed 8 Dec 2012).
- 17.Fridsma DB, Evans J, Hastak S, et al. The BRIDG Project: a technical report. J Am Med Inform Assoc 2008;15:130–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc 2009;16:624–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010;17:124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szalma S, Koka V, Khasanova T, et al. Effective knowledge management in translational medicine. J Transl Med 2010;8:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowe HJ, Ferris TA, Hernandez PM, et al. STRIDE—an integrated standards-based translational research informatics platform. AMIA Annu Symp Proc 2009;2009:391–5 [PMC free article] [PubMed] [Google Scholar]
- 22.Avillach P, Dufour J-C, Diallo G, et al. Design and validation of an automated method to detect known adverse drug reactions in MEDLINE: a contribution from the EU-ADR project. J Am Med Inform Assoc 2013;20:446–52. [Google Scholar]
- 23.Delaney BC, Peterson KA, Speedie S, et al. Envisioning a learning health care system: the electronic primary care research network, a case study. Ann Fam Med 2012;10:54–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson KA, Fontaine P, Speedie S. The electronic primary care Research Network (ePCRN): a new era in practice-based research. J Am Board Fam Med 2006;19:93–7 [DOI] [PubMed] [Google Scholar]
- 25.Wiederhold G. Mediators in the architecture of future information systems. Comput J 1992;25:38–49 [Google Scholar]
- 26.Martin L, Anguita A, Graf N, et al. ACGT: advancing clinico-genomic trials on cancer—four years of experience. Stud Health Technol Inform 2011;169:734–8 [PubMed] [Google Scholar]
- 27.Gupta A, Ludascher B, Martone ME. Knowledge-based integration of neuroscience data sources. In: Scientific and Statistical Database Management, 2000. Proceedings. 12th International Conference, 2000: 39–52
- 28.Astakhov V, Gupta A, Grethe JS, et al. Semantically based data integration environment for biomedical research. In: Proc of the 19th IEEE Symp Comput Based Med Syst; 22–23 June 2006, Washington, DC: IEEE Computer Society, 2006:171–6 [Google Scholar]
- 29.Ashish N, Ambite JL, Muslea M, et al. Neuroscience data integration through mediation: an (F)BIRN case study. Front Neuroinform 2010;4:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanford J, Mikula R. A model for online collaborative cancer research: report of the NCI caBIG project. Int J Healthc Technol Manag 2008;9:231–46 [Google Scholar]
- 31.González-Beltrán A, Tagger B, Finkelstein A. Federated ontology-based queries over cancer data. BMC Bioinformatics 2011;13(Suppl. 1):S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saltz J, Oster S, Hastings S, et al. caGrid: design and implementation of the core architecture of the cancer biomedical informatics grid. Bioinformatics 2006;22:1910–16 [DOI] [PubMed] [Google Scholar]
- 33.Livne O, Schultz N, Narus S. Federated querying architecture with clinical & translational health IT application. J Med Syst 2011;35:1211–24 [DOI] [PubMed] [Google Scholar]
- 34.Ouagne D, Hussain S, Sadou E, et al. The Electronic Healthcare Record for Clinical Research (EHR4CR) information model and terminology. Stud Health Technol Inform 2012;180:534–8 [PubMed] [Google Scholar]
- 35.Core Ontology—SHRINE https://open.med.harvard.edu/display/SHRINE/Core+Ontology (accessed 18 Apr 2012).
- 36.Bug W, Ascoli G, Grethe J, et al. The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics 2008;6:175–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D3.5.2_Appendix_E_Ontology_Specifications_01.pdf http://www.epsos.eu/uploads/tx_epsosfileshare/D3.5.2_Appendix_E_Ontology_Specifications_01.pdf (accessed 8 Dec 2012).
- 38.epSOS: About epSOS http://www.epsos.eu/home/about-epsos.html (accessed 11 Apr 2012).
- 39.Matney S, Bradshaw R, Livne O, et al. Developing a semantic framework for clinical and translational research. In: AMIA Summit on Translational Bioinformatics; 7–9 March 2011, Bethesda, MD: AMIA, 2011:24 [Google Scholar]
- 40.Segagni D, Tibollo V, Dagliati A, et al. An ICT infrastructure to integrate clinical and molecular data in oncology research. BMC Bioinformatics 2012;13(Suppl. 4):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.NCI Thesaurus Browser. https://cabig-stage.nci.nih.gov/community/tools/NCI_Thesaurus (accessed 8 Dec 2012). [Google Scholar]
- 42.LexEVS 6.0 Architecture https://cabig-kc.nci.nih.gov/Vocab/KC/index.php/LexEVS_6.0_Architecture (accessed 30 May 2011).
- 43.Noy NF, Shah NH, Whetzel PL, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Res 2009;37(web server issue):W170–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pathak J, Solbrig HR, Buntrock JD, et al. LexGrid: a framework for representing, storing, and querying biomedical terminologies from simple to sublime. J Am Med Inform Assoc 2009;16:305–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.CTS2 http://informatics.mayo.edu/cts2/index.php/Main_Page (accessed 14 Jun 2011).
- 46.LexEVS 6.0 CTS2 Guide—EVS—LexEVS—National Cancer Institute—Confluence Wiki https://wiki.nci.nih.gov/display/LexEVS/LexEVS+6.0+CTS2+Guide (accessed 2 Jul 2012).
- 47.caDSR and ISO 11179—caDSR—National Cancer Institute—Confluence Wiki. https://wiki.nci.nih.gov/display/caDSR/caDSR+and+ISO+11179 (accessed 8 Dec 2012).
- 48.Warzel DB, Andonyadis C, McCurry B, et al. Common Data Element (CDE) management and deployment in clinical trials. AMIA Annu Symp Proc 2003;2003:1048. [PMC free article] [PubMed] [Google Scholar]
- 49.Terminfo Project—Overview http://www.hl7.org/Special/committees/terminfo/overview.cfm (accessed 9 Dec 2012).
- 50.NIVEL|LINH http://www.nivel.nl/en/netherlands-information-network-general-practice-jlinh (accessed 28 Jul 2012).
- 51.NIVEL|Netherlands institute for health services research http://www.nivel.nl/en (accessed 11 Apr 2012).
- 52.Clinical Practice Research Datalink—CPRD http://www.cprd.com/intro.asp (accessed 28 Jul 2012).
- 53.Medicines and Healthcare products Regulatory Agency http://www.mhra.gov.uk/#page=DynamicListMedicines (accessed 28 Jul 2012).
- 54.Lenzerini M. Data integration: a theoretical perspective. In: Proceedings of the twenty-first ACM SIGMOD-SIGACT-SIGART Symposium on Principles of Database Systems; 3–6 June 2002, New York, NY: ACM, 2002:233–46 [Google Scholar]
- 55.Smith B, Ceusters W. Ontological realism: a methodology for coordinated evolution of scientific ontologies. Appl Ontol 2010;5:139–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grenon P, Smith B. SNAP and SPAN: Towards dynamic spatial ontology. Spat Cogn Comput 2004;4:69–104 [Google Scholar]
- 57.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32(database issue):D267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheuermann RH, Ceusters W, Smith B. Toward an ontological treatment of disease and diagnosis. AMIA Summit on Translational Bioinformatics 2009;2009:116–20 [PMC free article] [PubMed] [Google Scholar]
- 59.Goldfain A, Smith B, Arabandi S, et al. Vital sign ontology. In: Proceedings of the Workshop on Bio-Ontologies, Vienna, ISMB, 2011:71–4 [Google Scholar]
- 60.information-artifact-ontology —The Information Artifact Ontology (IAO) is an ontology of information entities based on the BFO—Google Project Hosting. http://code.google.com/p/information-artifact-ontology/ (accessed 9 Dec 2012). [Google Scholar]
- 61.Brinkman RR, Courtot M, Derom D, et al. Modeling biomedical experimental processes with OBI. J Biomed Semantics 2010;1(Suppl. 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luciano JS, Andersson B, Batchelor C, et al. The translational medicine ontology and knowledge base: driving personalized medicine by bridging the gap between bench and bedside. J Biomed Semantics 2011;2(Suppl. 2):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leysen P, Bastiaens H, Van Royen P. TRANSFoRm : development of use cases. http://transformproject.eu/Deliverable_List_files/D1.1%20Detailed%20Use%20Cases_V2.1-2.pdf (accessed 28 Feb 2013).
- 65.Qamar R, Rector A. Semantic issues in integrating data from different models to achieve data interoperability. Stud Health Technol Inform 2007;129:674–8 [PubMed] [Google Scholar]
- 66.Park J, Ram S. Information systems interoperability: what lies beneath? ACM Transactions on Information Systems 2004;22:595–632 [Google Scholar]
- 67.Pathak J, Wang J, Kashyap S, et al. Mapping clinical phenotype data elements to standardized metadata repositories and controlled sterminologies: the eMERGE network experience. J Am Med Inform Assoc 2011;18:376–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shvaiko P, Euzenat J. Ontology matching: state of the art and future challenges. IEEE Trans Knowl Data Eng 2013;25:158–76 [Google Scholar]
- 69.Choi N, Song I-Y, Han H. A survey on ontology mapping. ACM SIGMOD Rec 2006;35:34–41 [Google Scholar]
- 70.Shvaiko P, Euzenat J. A survey of schema-based matching approaches. In: Spaccapietra S. (ed) Journal on data semantics IV. Berlin/Heidelberg: Springer, 2005: 146–71 [Google Scholar]
- 71.CTS2—HL7Wiki http://wiki.hl7.org/index.php?title=CTS2 (accessed 28 Jul 2012). [Google Scholar]
- 72.Cimino JJ, Clayton PD, Hripcsak G, et al. Knowledge-based approaches to the maintenance of a large controlled medical terminology. J Am Med Inform Assoc 1994;1:35–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cimino JJ. Terminology tools: state of the art and practical lessons. Methods Inf Med 2001;40:298–306 [PubMed] [Google Scholar]
- 74.Miller GC, Britt H. A new drug classification for computer systems: the ATC extension code. Int J Biomed Comput 1995;40:121–4 [DOI] [PubMed] [Google Scholar]
- 75.BNF.org. http://www.bnf.org/bnf/index.htm (accessed 10 Dec 2012). [Google Scholar]


