Abstract
Objective
Online health knowledge resources contain answers to most of the information needs raised by clinicians in the course of care. However, significant barriers limit the use of these resources for decision-making, especially clinicians’ lack of time. In this study we assessed the feasibility of automatically generating knowledge summaries for a particular clinical topic composed of relevant sentences extracted from Medline citations.
Methods
The proposed approach combines information retrieval and semantic information extraction techniques to identify relevant sentences from Medline abstracts. We assessed this approach in two case studies on the treatment alternatives for depression and Alzheimer's disease.
Results
A total of 515 of 564 (91.3%) sentences retrieved in the two case studies were relevant to the topic of interest. About one-third of the relevant sentences described factual knowledge or a study conclusion that can be used for supporting information needs at the point of care.
Conclusions
The high rate of relevant sentences is desirable, given that clinicians’ lack of time is one of the main barriers to using knowledge resources at the point of care. Sentence rank was not significantly associated with relevancy, possibly due to most sentences being highly relevant. Sentences located closer to the end of the abstract and sentences with treatment and comparative predications were likely to be conclusive sentences. Our proposed technical approach to helping clinicians meet their information needs is promising. The approach can be extended for other knowledge resources and information need types.
Keywords: clinical decision support, information needs, knowledge summary, natural language processing
Background
In a 1985 seminal study, Covell et al1 observed that physicians raised two questions for every three patients seen in an outpatient setting. In 70% of the cases these questions were not answered. More recent research2 has produced similar results, with little improvement compared with the findings of Covell et al.1 These information needs are gaps in medical knowledge that providers need to fill in order to make, confirm, or carry out decisions on patient care.3 Knowledge gaps lead to suboptimal decisions, lowering the quality of care,4–6 and represent important missed opportunities for self-directed learning and for changes in practice patterns.7 Although studies have shown that online health knowledge resources provide answers to most of the clinicians’ questions, significant barriers limit the use of these resources to support patient care decision-making.2 8 A critical barrier is that finding relevant information, which may be located in several documents, takes an amount of time and cognitive effort that is incompatible with the busy clinical workflow. A promising approach to reduce these barriers is the automatic summarization of multiple sources that account for the context of a particular information need. In this paper we assess the feasibility of such an approach, in which we extract sentences from Medline citations by integrating existing semantic information retrieval and semantic information extraction techniques. We evaluated the system output in two case studies: depression and Alzheimer's disease.
Observing patient care information needs among primary care clinicians, Ely et al created a taxonomy of 64 information need types.9 The taxonomy follows a Pareto distribution—that is, approximately 20% of the information need types accounted for 80% of the information needs that clinicians raised. Studies have also shown that information needs are influenced by contextual factors related to the patient, clinician, care setting, and the task at hand.3 10 Context-aware information retrieval solutions such as ‘Infobuttons’ help clinicians to meet some of their information needs.11 12 However, present solutions still require clinicians to scan information within multiple documents while relevant content is typically contained in a few short passages. This is especially true when clinicians need to compare multiple approaches to a particular patient care problem. The long-term goal of our research is to summarize automatically the literature available on a set of clinical topics that might be relevant in the care of a particular patient. By automatically summarizing the literature, we expect to reduce clinicians’ time and cognitive effort when seeking information to support patient care decisions. We call the final product of this approach a ‘knowledge summary’. The present study focuses on one of the important components of generating a knowledge summary—that is, extracting sentences from the literature that are relevant to a particular clinical topic. Specifically, we designed and assessed a method that contrasts multiple treatment approaches for a given medical problem by extracting sentences from Medline citations. Questions related to treatment comprised 25% of the information needs raised by physicians in the study by Ely et al.9
Methods
The methods are divided into two parts: (1) description of the system that generates knowledge summaries; and (2) system evaluation.
System architecture
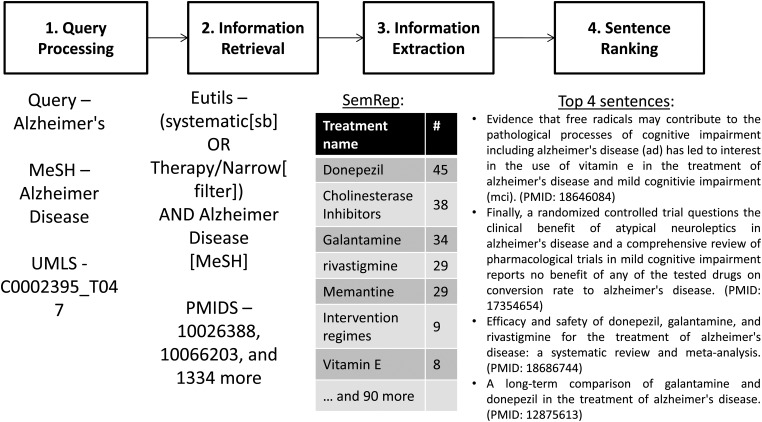
The system to generate knowledge summaries is built as a pipeline that combines the following natural language processing (NLP) tools and resources: Unified Medical Language System (UMLS) Metathesaurus13 for extracting concepts, SemRep14 for extracting semantic predications, and the TextRank algorithm15 for ranking the sentences that contain those semantic predications. Figure 1 depicts the system architecture and flow.
Figure 1.
System architecture with an example.
Step 1: Query processing
As shown in figure 1, the query for a clinical topic (eg, treatment alternatives for Alzheimer's disease) is initially processed with a UIMA (Unstructured Information Management Architecture)-based16 concept extraction pipeline that maps concepts in narrative text to the UMLS Metathesaurus.13 The components in the pipeline are tokenization, lexical normalization, UMLS Metathesaurus look-up, and concept screening and Medical Subject Heading (MeSH) conversion.
First, the clinical Text Analysis and Knowledge Extraction System17 tokenization component, which was adapted from the open NLP suite18 for biomedical text, splits the query into multiple tokens.
Second, lexical normalization (converting words to a canonical form) is performed using an efficient in-memory data structure similar to a hash table.19 The Lexical Variant Generation terminology from the UMLS Metathesaurus is compressed by (1) converting the terms to lower case; (2) removing the terms where the normalized word has more than one token; and (3) removing the terms that have the same base form.
Third, a UMLS Metathesaurus look-up is performed using a well-known efficient algorithm called Aho-Corasick string matching.20 Our implementation of the Aho-Corasick algorithm21 loads the normalized tokens and their substrings as the individual states of the corresponding finite state machine. The transitions between the different states represent the different terms formed by the original tokens.
Fourth, the UMLS concepts in the query that are members of the semantic group treatment (Drug and Therapeutic or Preventive Procedure) or disorder (Abnormality, Dysfunction, Disease or Syndrome, Finding, Injury or Poisoning, Pathologic Function, and Sign or Symptom) are selected.
Finally, the screened concepts are mapped to MeSH headings for information retrieval (Step 2) and the UMLS concepts unique identifiers (CUIs) are used for information extraction (Step 3). For example, the query in figure 1 (Alzheimer's disease) is interpreted as a request for treatment alternatives to that particular disorder. In Step 1, ‘Alzheimer's’ is tagged to contain the corresponding UMLS concept C0002395 of type T047 (Disease or syndrome; grouped under ‘Disorders’).
Step 2: Information retrieval
In this step the system retrieves a set of relevant documents. We developed additional rules over the National Library of Medicine Entrez Programming Utilities (E-utilities)22 to retrieve clinically relevant abstracts that describe treatment alternatives for a given condition. The algorithm attempts to emulate a search cascade that gives preference to stronger evidence (eg, systematic reviews vs individual randomized trials) whenever available.23 It also uses the Haynes Clinical Query filters to retrieve high-quality clinical studies.24
The algorithm consists of: (1) retrieval of abstracts that match the MeSH terms of the condition of interest and are indexed with a publication type of systematic review or are retrieved using the Haynes Clinical Queries therapy filter24 tuned for precision; (2) when the number of abstracts returned is smaller than an arbitrary threshold (100 in this study), the PubMed query is progressively relaxed using the Clinical Queries therapy filter tuned for recall and searching with both the MeSH terms and their corresponding keywords. These steps are described as pseudo code in Algorithm 1. Step 2 in figure 1 illustrates the clinical topic ‘treatment of Alzheimer's disease’ translated into an E-utilities query.
Algorithm 1. Information retrieval strategy to retrieve abstracts relevant to a particular treatment topic.
pmidSET ← []
if topic not treatment type then
goto END
end if
MIN_PMIDS=100
join=AND
BEGIN:
query=''
for eachconcept in the topic search do
ifconcept is disorder or treatment then
query=query+join+mesh-form(concept) [MeSH]
end if
end for
BEGIN1:
pmidSET ← eutils(systematic[sb] AND query)
pmidSET ← eutils(Therapy/Narrow[filter] AND query)
ifpmidSET.size()<MIN_PMIDS then
pmidSET ← eutils(Therapy/Broad[filter] AND query)
end if
if pmidSET.size()<MIN_PMIDS && join=AND then
join=OR
goto BEGIN
end if
ifpmidSET.size()<MIN_PMIDSthen
query=topic search
goto BEGIN1
end if
END:
return pmidSET
Step 3: Information extraction
The system uses SemRep14 to extract relevant semantic predications from the documents retrieved in Step 2. SemRep extracts semantic predications from text using UMLS concepts and associations (eg, aromatase inhibitors treat breast carcinoma). A total of 56 million predications of 26 types (eg, TREATS, CAUSES, INHIBITS) from more than 21 million Medline citations are currently available in a relational database.
Algorithm 2. Information extraction method to retrieve semantic predications relevant to a particular treatment topic.
if topic not treatment type then
goto END
end if
subjects ← []
objects ← []
for eachconcept in the topic search do
ifconcept is disorder then
objects ← objects+concept
else ifconcept is treatment then
subjects ← subjects+concept
end if
end for
ifsubjects.size()==0 && objects.size()> 0 then
return predications whose object's CUI is one of the objects' CUIs
else ifsubjects.size()> 0 && objects.size()> 0 then
answers ← predications whose object's CUI is one of the objects' CUIs AND subject's CUI is one of the subjects' CUIs
ifanswers.size()>MIN_ANSWERS then
returnanswers
end if
returnanswers+predications whose object's CUI is one of the objects' CUIs OR subject's CUI is one of the subjects' CUIs
else ifsubjects.size()> 0 then
return predications whose subject's CUI is one of the subjects' CUIs
else then
return predications whose object's name is one of the objects' UMLS preferred terms
This predication database is queried to retrieve the semantic predications of type ‘TREATS’ that are relevant to the clinical topic of interest. The output is then summarized by eliminating uninformative predications.25 26 Algorithm 2 details how the queries are created to retrieve the relevant semantic predications.
For example, in Step 2 of figure 1 the PubMed query retrieved 1336 relevant abstracts. In Step 3 the predication database is queried for the predications of the type TREATS from these abstracts where the concept C0002395 is an object. The predications are then pruned according to the relative granularity of the concepts. This removes generic uninteresting treatment concepts such as ‘Pharmaceutical preparations’ and ‘Therapeutic procedure’ from the list of treatments. The most frequent of the 97 treatment options for Alzheimer's disease are shown in Step 3 in figure 1.
Step 4: Sentence ranking
We adapted the TextRank algorithm15 to rank the sentences retrieved in the previous step after the pruning process. A similar algorithm (PageRank27) has been used by Google in its search engine. This approach allows us to take into account the similarity between the query and the sentence and also that among the individual sentences. The semantic predications extracted from the above step correspond to a sentence in a Medline abstract. Since there could be several sentences for each treatment alternative, it is important to select the most important ones to be included in a knowledge summary.
Our sentence ranking approach includes two steps. First, for structured abstracts we exclude sections (ie, objectives, selection criteria, and methods) that typically do not contain background statements or study conclusions. Second, we represent each unique sentence as a vertex in a graph. Each pair of sentences is connected with an edge whose weight is determined by the cosine word similarity between the sentences. Using a formula similar to Google's PageRank, the probability with which a random reader of the graph reaches a sentence is determined. Sentences sharing more words with the sentences already read will be read next as per this model. Step 4 in figure 1 shows the five top sentences selected for the treatment of Alzheimer's disease.
Case study evaluation
For the evaluation study we used a case study approach by assessing the output of the knowledge summary system on the treatment of two conditions: depression and Alzheimer's disease. These topics were selected because the treatment of these conditions is complex: there is little definite evidence regarding the best treatment approach, multiple treatment alternatives are available, and the optimal treatment option depends on a series of contextual constraints such as patient age and comorbidities.
The case study topics were selected after the system was developed and therefore were not used to guide system development.
Sentences retrieved by the system on each topic were rated independently by two physicians in our team (GDF, RM) according to four attributes that are desirable in a ‘strong’ sentence for clinical decision-making: (1) topic-relevant; (2) conclusive; (3) comparative; and (4) contextually-constrained.
Topic-relevant sentences describe one or more treatment alternatives for the condition of interest. Specifically, sentences about the condition of interest but focused on aspects other than treatment such as prevention and diagnosis were considered not to be relevant, as illustrated in sentence (1) in box 1. In addition to sentences, abstracts were also rated according to topic relevancy. Since non-relevant sentences are not useful, regardless of the other three criteria, only relevant sentences were rated on all four criteria.
Box 1. Example sentences.
Not topic-relevant: ‘There is insufficient randomized evidence to support the routine use of antidepressants for the prevention of depression or to improve recovery from stroke’. (Pubmed ID: 15802637)
Conclusive and contextually-constrained: ‘There is marginal evidence to support the use of tricyclic antidepressants in the treatment of depression in adolescents, although the magnitude of effect is likely to be moderate at best’. (Pubmed ID: 10908557)
Comparative: ‘Escitalopram versus other antidepressive agents for depression’. (Pubmed ID: 19370639)
Conclusive and comparative: ‘We found no strong evidence that fluvoxamine was either superior or inferior to any other antidepressants in terms of efficacy and tolerability in the acute phase treatment of depression’. (Pubmed ID: 20238342)
Not topic-relevant: ‘Observational studies suggest that some preventive approaches, such as healthy lifestyle, ongoing education, regular physical activity, and cholesterol control, play a role in prevention of AD’. (Pubmed ID: 16529393)
Conclusive sentences comprise a statement about one or more treatment alternatives for the condition of interest, either as background information (eg, current state of knowledge) or study conclusion, as in sentence (2) in box 1. Conclusive sentences are more useful for clinical decision-making than those that describe the study objectives or methods, as in sentence (3) in box 1.
Comparative sentences contrast two or more treatment approaches for the condition of interest. For example, sentence (4) in box 1 compares fluvoxamine versus other antidepressants in the treatment of depression. Sentences that compare a treatment option with a placebo were not labeled as comparative sentences. When considering treatment alternatives for a given condition, sentences that compare multiple alternatives are typically more useful than sentences with no comparison or comparison with a placebo.
Contextually-constrained sentences include specific clinical situations in which a treatment alternative is applicable such as care setting, comorbidity, and age group, as in sentence (2) in box 1. Contextual constraints are important because a relevant sentence may become completely irrelevant if the patient at hand does not meet these constraints. For example, sentence (2) in box 1 is only relevant if the patient is an adolescent.
Finally, we tested the following three null hypotheses: (1) sentence relevancy is not associated with its TextRank probability; (2) sentence conclusiveness is not associated its TextRank probability; and (3) sentence conclusiveness is not associated with the relative position of the sentence in the abstract. Sentence position was operationalized as the sentence absolute position divided by the total number of sentences in the abstract. We used the Student t statistics to test this hypothesis, with a two-tailed significance level of 0.05. The Holm's step-down procedure was used to adjust for multiple comparisons.26
Results
The depression case retrieved 228 sentences from 122 PubMed abstracts and the Alzheimer's disease case retrieved 336 sentences from 194 abstracts. Overall, the two raters agreed on 80.3% of the ratings (κ=0.78).
Table 1 provides descriptive statistics for the case study ratings. Overall, 515 out of 564 (91.3%) sentences retrieved in both case studies were rated as relevant to the topic of interest. About one-third of the relevant sentences were conclusive and a smaller percentage compared two or more treatment alternatives. While 44% of the depression sentences included contextual constraints, only 6.4% of the Alzheimer's disease sentences had constraints.
Table 1.
Case study ratings
| Depression n/N | Alzheimer's disease n/N | |
|---|---|---|
| Relevant abstract | 113 (92.6%)/122 | 164 (84.5%)/194 |
| Relevant sentence | 218 (95.6%)/228 | 297 (88.4%)/336 |
| Conclusive | 68 (31.2%)/218 | 104 (35.0%)/297 |
| Comparative | 39 (17.9%)/218 | 13 (4.4%)/297 |
| Contextually-constrained | 96 (44.0%)/218 | 19 (6.4%)/297 |
In both case studies most of the non-relevant sentences were related to the condition of interest, but the focus was on the diagnosis or prevention of the condition instead of treatment alternatives, as illustrated in sentences (1) and (5) in box 1.
In both case studies the TextRank probability was not significantly different in relevant versus non-relevant sentences (depression: 0.51 vs 0.48; p=0.45; Alzheimer's disease: 0.53 vs 0.49; p=0.18). In the depression case study, the TextRank probability was not significantly different in conclusive versus non-conclusive sentences (0.48 vs 0.52; p=0.07) while, in the Alzheimer's disease case study, the TextRank probability was significantly higher in non-conclusive than conclusive sentences (0.55 vs 0.50; p=0.01).
Overall, conclusive sentences were located closer to the end of the abstract than non-conclusive sentences (0.51 vs 0.26; p<0.00001).
Discussion
In this study we assessed the feasibility of automatically generating clinical knowledge summaries using semantic information retrieval and extraction techniques. The strength of the sentences retrieved by the system was rated based on four attributes: relevant, conclusive, comparative, and contextually-constrained.
Overall, the system retrieved a high rate of relevant sentences (96% for depression and 88% for Alzheimer's disease). This is highly desirable, given that clinicians’ lack of time is one of the main barriers to using knowledge resources at the point of care.2 In a previous study by Fiszman et al, the average precision of SemRep for predications on the treatment of four diseases was 73%.28 The higher relevancy found in our study may be attributed to the information retrieval strategy (Step 2) which focuses on retrieving high-quality clinical studies. In addition, Fiszman et al had a stricter definition of relevancy since only predications that represented drugs in the gold standard were considered relevant.
Sentence rank was not significantly associated with relevancy. This finding is possibly due to the overall high relevancy found in our study, which leaves little room for improvement. Nevertheless, relevancy could be further enhanced by improving the precision of SemRep. Another potential approach is to explore domain-specific summarization methods as suggested by Reeve et al.29
Only about one-third of the sentences retrieved included a conclusive statement. Retrieving conclusive sentences is challenging but could be approached through a combination of methods such as sentence position, comparative predications,30 and linguistic cues such as hedges. In our study, conclusive sentences were located much closer to the end of the abstract than non-conclusive sentences. In addition, structured abstracts include a Conclusion section that is typically composed of conclusive sentences. Although only a small number of Medline citations contain a structured abstract, the percentage of structured abstracts in Medline increased from 2.4% in 1992 to 20.3% in 2005.31 Finally, sentences with treatment and comparative predications (eg, treatment A HIGHER_THAN treatment B) may be more likely to be conclusive sentences.
Interestingly, conclusive sentences in the Alzheimer's case study had a significantly lower TextRank probability than non-conclusive sentences. Conclusive sentences are often shorter and limited to a subject and object. This may lead to a lower similarity with the query terms and other sentences, which are factors used by the cosine similarity and TextRank methods. Despite the significant association, sentence rank may not be useful for identifying conclusive sentences given the small difference in the rank means.
A small percentage of the sentences retrieved by the system compared treatment alternatives. Direct treatment comparisons are useful, since clinicians are often faced with deciding between two or more treatment alternatives for a particular patient. The main reason for the small percentage is likely to be the lack of comparative studies in the biomedical literature; most studies compare an intervention with a placebo. Nevertheless, it is still possible to improve the retrieval of comparative sentences from Medline citations. A potential approach is to retrieve drug comparative predications such as treatment A COMPARED_WITH treatment B; treatment A HIGHER_THAN treatment B; and treatment A SAME_AS treatment B.30 Another approach is to retrieve abstracts that have a publication type field of ‘Comparative Study’.
Almost half of the sentences in the depression case study contained contextual constraints but a much smaller proportion of such sentences were retrieved for Alzheimer's disease. This difference may be due to the fact that depression is a much more well-researched condition with more treatment options than Alzheimer's disease. In addition, depression occurs in almost all age groups and its presentation can be altered by several other conditions, while Alzheimer's disease affects only elderly subjects. The identification of contextual constraints can be improved by retrieving predication types such as PROCESS_OF and COEXISTS_WITH, both within treatment sentences and other sentences in the abstract.
Study limitations
This study has five main limitations. First, the system evaluation consisted of two case studies, limiting the generalizability of our findings. However, these preliminary results provide useful insights regarding the feasibility of the proposed approach and potential areas for improvement.
Second, the threshold (ie, 100 documents) used in our information retrieval algorithm is arbitrary and not empirically established as an optimal threshold. Further studies are needed to identify a threshold that achieves optimal recall without overloading clinicians with too much information.
Third, our method was focused on treatment. It is not known whether a similar performance would be obtained in other similarly prevalent information needs such as disease diagnosis. From a technical standpoint, the system pipeline could be extended to other types of information needs by adapting the information retrieval strategy (Step 2) and exploiting other semantic relationships (Step 3). Kilicoglu et al32 provide an exhaustive list of the types of semantic relationships produced by SemRep, which may be used for such an extension.
Fourth, an important limitation of our study was that we did not assess the recall of the system. Hence, no conclusions can be made regarding the comprehensiveness of the knowledge summary for the case study topics. Nevertheless, a previous assessment of SemRep on the treatment of four diseases yielded an average recall of 98%.28 Future studies need to assess the overall recall of the knowledge summary system, tackling other factors besides SemRep that affect the knowledge summary recall: (1) recall of the information retrieval step (see second limitation); and (2) recall after aggregation of summary from sentences (see section on future studies below).
Last, we used an inclusive definition of topic relevancy that considers any sentence that describes one or more treatment alternatives for the condition of interest to be relevant. On the other hand, real patient care scenarios impose additional constraints to information needs, restricting the definition of relevancy. The relevancy of the system output in a real patient care environment is therefore likely to be lower than that reported in our study. However, high topic relevancy is a first necessary step towards achieving a useful solution for patient care.
Future studies
The present study has raised several areas that warrant further investigation. First, this study focused on sentence extraction, which is only one of the necessary steps for the long-term goal of generating a clinical knowledge summary. Other steps also need to be investigated to compose a usable solution, such as methods to aggregate and visualize the retrieved sentences into an understandable summary. Previous research in sentence simplification33 and natural language generation34 would be useful in this effort.
Second, our approach will need to be adapted to extract sentences for different types of resources and information needs. For example, the performance of SemRep using full-text resources is unknown.
Third, the present study raised several promising directions for improving the identification of strong sentences. We are currently investigating some of these directions, such as using sentence position and comparative predications to identify conclusive sentences, and using Medline metadata fields and comparative predications to identify comparative sentences.
Finally, tagging treatment sentences with contextual constraints enables semantic integration between electronic health record systems and knowledge resources. Such an integration could be achieved by indexing sentences according to the context dimensions and terminologies defined in the Health Level Seven (HL7) Context-Aware Knowledge Retrieval Standard.35
Conclusion
For the two case studies, the system retrieved a high percentage of topic-relevant sentences. However, a smaller percentage of sentences were conclusive, comparative, or contextually-constrained. Future studies are needed to develop and test heuristics that help identify these sentences. Overall, this seems to be a feasible approach to constructing context-specific semantic knowledge summaries to support clinicians’ decision-making in patient care.
Footnotes
Contributors: SRJ: conception, design, development, interpretation of results, drafting of the manuscript, final approval. GDF: conception, analysis of data, interpretation of results, drafting of the manuscript, final approval. RM: conception, analysis of data, interpretation of results and critical revision of manuscript, final approval. CW: conception, analysis of data and critical revision of manuscript, final approval. MF: conception, development and critical revision of manuscript, final approval. JM: conception, critical revision of manuscript, final approval. HL: conception, design, development and critical revision of manuscript, final approval.
Funding: JM and RM have been partially funded for this research through NSF Award 1117286 and the NIH CTSA Award UL1TR000083. GDF was supported by grant number K01HS018352 from the Agency for Healthcare Research and Quality.
Competing interests: None.
Provenance and peer review: Not commissioned; internally peer reviewed.
Data sharing statement: The system or a web service of it will be available to non-profit researchers with no competing interest upon contacting the corresponding author. At a later time we will disseminate it as an open source software which will be available to everyone.
References
- 1.Covell DG, Uman GC, Manning PR. Information needs in office practice: are they being met? Ann Intern Med 1985;103:596–9 [DOI] [PubMed] [Google Scholar]
- 2.Ely JW, Osheroff JA, Chambliss ML, et al. Answering physicians' clinical questions: obstacles and potential solutions. J Am Med Inform Assoc 2005;12:217–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Currie LM, Graham M, Allen M, et al. Clinical information needs in context: an observational study of clinicians while using a clinical information system. AMIA Annu Symp Proc 2003;11:190–4 [PMC free article] [PubMed] [Google Scholar]
- 4.Leape LL, Bates DW, Cullen DJ, et al. Systems analysis of adverse drug events. ADE Prevention Study Group. JAMA 1995;274:35–43 [PubMed] [Google Scholar]
- 5.Solomon DH, Hashimoto H, Daltroy L, et al. Techniques to improve physicians' use of diagnostic tests: a new conceptual framework. JAMA 1998;280:2020–7 [DOI] [PubMed] [Google Scholar]
- 6.van Walraven C, Naylor CD. Do we know what inappropriate laboratory utilization is? A systematic review of laboratory clinical audits. JAMA 1998;280:550–8 [DOI] [PubMed] [Google Scholar]
- 7.Green ML, Ciampi MA, Ellis PJ. Residents' medical information needs in clinic: are they being met? Am J Med 2000;109:218–23 [DOI] [PubMed] [Google Scholar]
- 8.Ely JW, Osheroff JA, Maviglia SM, et al. Patient-care questions that physicians are unable to answer. J Am Med Inform Assoc 2007;14:407–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ely JW, Osheroff JA, Gorman PN, et al. A taxonomy of generic clinical questions: classification study. BMJ 2000;321:429–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Fiol G, Haug PJ. Classification models for the prediction of clinicians' information needs. J Biomed Inform 2009;42:82–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maviglia SM, Yoon CS, Bates DW, et al. KnowledgeLink: impact of context-sensitive information retrieval on clinicians' information needs. J Am Med Inform Assoc 2006;13:67–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Fiol G, Haug PJ. Infobuttons and classification models: a method for the automatic selection of on-line information resources to fulfill clinicians' information needs. J Biomed Inform 2008;41:655–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32(Database issue):D267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilicoglu H, Shin D, Fiszman M, et al. A PubMed-Scale Repository of Biomedical Semantic Predications. Bioinformatics 2012;Epub ahead of print [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mihalcea R, Tarau P. TextRank: Bringing order into texts. In Lin D, Wu D (Eds). Proceedings of EMNLP. Barcelona, Spain: Association for Computational Linguistics, 2004:404.– [Google Scholar]
- 16.Apache Unstructured Information Management Architecture. 2012. http://uima.apache.org (accessed 17 September 2012). [DOI] [PMC free article] [PubMed]
- 17.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Apache OpenNLP. 2012. http://opennlp.apache.org (accessed 17 September 2012).
- 19.Cormen TH, Leiserson CE, Rivest RL, et al. Introduction to algorithms. Boston: MIT Press, 2001 [Google Scholar]
- 20.Aho AV, Corasick MJ. Efficient string matching: an aid to bibliographic search. Commun ACM 1975;18:333–40 [Google Scholar]
- 21.Torii M, Wagholikar K, Liu H. Using machine learning for concept extraction on clinical documents from multiple data sources. J Am Med Inform Assoc 2011;18:580–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.NLM Entrez Programming Utilities. 2012. http://www.ncbi.nlm.nih.gov (accessed 17 September 2012).
- 23.Swinglehurst DA, Pierce M, Fuller JC. A clinical informaticist to support primary care decision making. Qual Health Care 2001;10:245–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haynes RB, McKibbon KA, Wilczynski NL, et al. Optimal search strategies for retrieving scientifically strong studies of treatment from Medline: analytical survey. BMJ 2005;330:1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, Fiszman M, Shin D, et al. Degree centrality for semantic abstraction summarization of therapeutic studies. J Biomed Inform 2011;44:830–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiszman M, Rindflesch TC, Kilicoglu H. Abstraction summarization for managing the biomedical research literature. In: Proceedings of the HLT-NAACL Workshop on Computational Lexical Semantics. Boston, Massachusetts: Association for Computational Linguistics, 2004, 76–83 [Google Scholar]
- 27.Page L, Brin S, Motwani R, et al. The PageRank Citation Ranking: Bringing Order to the Web. Technical Report: Stanford InfoLab, 1999. Report No.: SIDL-WP-1999-0120. [Google Scholar]
- 28.Fiszman M, Demner-Fushman D, Kilicoglu H, et al. Automatic summarization of MEDLINE citations for evidence-based medical treatment: a topic-oriented evaluation. J Biomed Inform 2009;42:801–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeve LH, Han H, Brooks AD. The use of domain-specific concepts in biomedical text summarization. Inf Process Manage 2007;43:1765–76 [Google Scholar]
- 30.Fiszman M, Demner-Fushman D, Lang FM, et al. Interpreting comparative constructions in biomedical text. In: Proceedings of the Workshop on BioNLP 2007: Biological, Translational, and Clinical Language Processing. Prague, Czech Republic: Association for Computational Linguistics, 2007, 137–44 [Google Scholar]
- 31.Ripple AM, Mork JG, Knecht LS, et al. A retrospective cohort study of structured abstracts in MEDLINE, 1992–2006. J Med Libr Assoc 2011;99:160–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilicoglu H, Rosemblat G, Fiszman M, et al. Constructing a semantic predication gold standard from the biomedical literature. BMC Bioinformatics 2011;12:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonnalagadda S, Gonzalez G. BioSimplify: an open source sentence simplification engine to improve recall in automatic biomedical information extraction. AMIA Annual Symposium Proceedings/AMIA Symposium AMIA Symposium 2010;351–5 [PMC free article] [PubMed] [Google Scholar]
- 34.Cawsey AJ, Webber BL, Jones RB. Natural language generation in health care. J Am Med Inform Assoc 1997;4:473–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Fiol G. Context-aware Knowledge Retrieval ('Infobutton'), Knowledge Request Standard. 2010.. http://www.hl7.org/v3ballot2010may/html/domains/uvds/uvds_Context-awareKnowledgeRetrieval%28Infobutton%29.htm (accessed 2012) [Google Scholar]