Abstract
Objective
Natural language processing (NLP) tasks are commonly decomposed into subtasks, chained together to form processing pipelines. The residual error produced in these subtasks propagates, adversely affecting the end objectives. Limited availability of annotated clinical data remains a barrier to reaching state-of-the-art operating characteristics using statistically based NLP tools in the clinical domain. Here we explore the unique linguistic constructions of clinical texts and demonstrate the loss in operating characteristics when out-of-the-box part-of-speech (POS) tagging tools are applied to the clinical domain. We test a domain adaptation approach integrating a novel lexical-generation probability rule used in a transformation-based learner to boost POS performance on clinical narratives.
Methods
Two target corpora from independent healthcare institutions were constructed from high frequency clinical narratives. Four leading POS taggers with their out-of-the-box models trained from general English and biomedical abstracts were evaluated against these clinical corpora. A high performing domain adaptation method, Easy Adapt, was compared to our newly proposed method ClinAdapt.
Results
The evaluated POS taggers drop in accuracy by 8.5–15% when tested on clinical narratives. The highest performing tagger reports an accuracy of 88.6%. Domain adaptation with Easy Adapt reports accuracies of 88.3–91.0% on clinical texts. ClinAdapt reports 93.2–93.9%.
Conclusions
ClinAdapt successfully boosts POS tagging performance through domain adaptation requiring a modest amount of annotated clinical data. Improving the performance of critical NLP subtasks is expected to reduce pipeline error propagation leading to better overall results on complex processing tasks.
Keywords: Natural Language Processing, NLP, POS Tagging, Domain Adaptation, Clinical Narratives
Introduction
Electronic health record systems store a considerable amount of patient healthcare information in the form of unstructured, clinical notes. These narratives contain information about a patients’ sociological and medical history, state of health, medical prognoses, and past and present treatment plans. If properly transformed into a coded form, this information can be leveraged for more advanced medical applications supporting evidence-based medicine, clinical decision support, and research activities.1–6
Clinical natural language processing (NLP) systems have been devised to process unstructured text and transform it into a desired coded form to support these many healthcare-related activities. These systems commonly decompose complex processing tasks into a series of consecutive subtasks in which subsequent stages are dependent on the output from previous stages. These processing models are known as processing pipelines.7 8 Pipelines can be made up of varying subtask components depending on the complexity and nature of the end processing objective. A well-known problem in this processing paradigm is cascading error propagation caused by the residual error that is generated in each subtask.8 This compounding residual error affects the overall performance of NLP systems whose end objective is some higher-level task such as concept extraction or complex inference to mimic human-like language understanding. NLP used to support clinical care demands a higher degree of accuracy as results are incorporated into critical decisions related to patient care. It is therefore important that NLP pipeline systems perform as optimally as possible.
The methods currently used in NLP are strongly influenced by machine learning and statistical techniques. These methods are predicated on the availability of large volumes of annotated training data for supervised learning, model development, and benchmarking. Obtaining large volumes of annotated data in the clinical domain remains a barrier to realizing fully the potential benefits of clinical NLP.9 Stringent healthcare privacy laws continue to impede the sharing of clinical data across institutions. Within institutions, annotated clinical text is sparse and expensive to produce. Unique medical terminology and complex disease processes commonly require knowledgeable medical staff to help in the annotation process. These factors have all played into the limited availability of clinically annotated text to support modern, statistically based, clinical NLP methods.
To overcome these barriers, alternative approaches to traditional training methods are being explored. One such method is domain adaptation. Domain adaptation is an approach by which plentiful, out-of-domain training data are leveraged with a limited set of target domain data such that the underlying probability distribution of the target domain is more effectively represented by the aggregate data than by the limited target data alone.
Most statistically based learning techniques rely on the assumption that training data and test data share a common underlying probability distribution. Through domain adaptation, the large volume of out-of-domain, source-labeled data can be leveraged with a small amount of in-domain, target data, with the goal of optimizing the model for the target domain. This approach allows existing NLP tools with good performance characteristics in general English domains to be adapted to clinical target domains for a fraction of the cost of generating new models requiring large volumes of expensive annotation.
In this study, we evaluate domain adaptation of part-of-speech (POS) tagging. We selected this task because it is a well-studied area for domain adaptation, yet as we will show, suggested methods have not generalized well when applied to clinical narratives. POS tagging is an important syntactic process whose performance can greatly affect subsequent downstream processes such as syntactic parsing and semantic inference. We demonstrate that through domain adaptation, we can reduce the residual error in POS tagging in a cost-effective manner leveraging current out-of-domain algorithms with a modest amount of in-domain (clinical) annotated data. We confirm that clinical narratives have different linguistics characteristics than those of general English and biomedical texts. We show that state-of-the-art POS taggers with accuracies upward of 97% quickly drop to accuracies in the 80%s when applied to clinical narratives using their general English or biomedical source domain models.
Furthermore, we introduce a new error-correction approach to domain adaptation for POS tagging that outperforms other popular methods. This performance enhancement is achieved by introducing a transformation-based learner, reducing the adaptation problem to that of error correction going from the source to target domain. We introduce a novel, hill-climbing lexical generation probability rule into the transformation-based learner that dominates the traditional symbolic rules used in most transformation-based learner applications.
We also apply an unambiguous lexicon derived from the SPECIALIST lexicon10 to help with unknown word identification and unique medical vocabulary. This approach requires moderately easy extraction of unambiguous terms (terms with only one POS) from the SPECIALIST lexicon10 and a modest amount of target, clinical report data to boost POS tagging accuracies back to acceptable levels for clinical NLP.
We believe this approach has broad applicability to several subtasks found in NLP pipelines and provides a fairly generalizable approach to reduce pipeline propagation error in NLP tasks.11–15
Background
Unique characteristics of clinical text
The ability for statistical NLP methods to operate optimally on clinical narratives is determined by how well the sample data used in model training represents the probability distribution of this subdomain. Meystre et al16 define clinical texts as texts written by clinicians in the clinical setting. These texts have been well studied and have been shown to contain structural and linguistic characteristics that differ from general English or biomedical text.16–21 This has led to classifying these texts as a sub-domain language.17 Sub-domain languages are characterized by distinct linguistic features not found in other corpora such as general English. Some of the features unique to clinical text include distinct informational categories (eg, disease, procedure, body location), specific co-occurrence patterns (eg, patient verb symptom in body location), paraphrastic patterns (eg, medication dose frequency route), omissions of contextual information and telegraphic statements (eg, ‘bilateral infiltrates noted’, interpreted to mean that bilateral infiltrates in the lungs are noted), temporal patterns (eg, ‘right perihilar infiltrative change present, please correlate clinically’), and medical acronyms and specialized medical terminology. These unique sub-language features make clinical texts difficult to process accurately using statistically based NLP tools developed from non-medical domains. This means that the underlying statistical models used in these methods need to be retrained from representative clinical training data or adapted to reflect accurately the underlying sub-language probability distribution.
General POS taggers
For the most part, optimal POS tagging has been achieved. Several successful, statistically based approaches have reached accuracies upward of 97% on general English grammar.22–27 Most of these accuracies have been recorded using Penn Treebank,28 Wall Street Journal (WSJ) data in which there exists a large volume of labeled data. We describe a representative subset of state-of-the-art taggers that will be included in our evaluation to confirm their operating characteristics on general English as a baseline. We also show how their performance is significantly reduced when their out-of-domain, general English tagging models are applied to the clinical text domain.
The Stanford POS tagger is a high-performing open-source tagger that uses a maximum entropy method to learn a log-linear conditional probability model and reports a tagging accuracy of 97.24% on Penn Treebank WSJ data.23 24 It includes two general English models trained from Penn Treebank WSJ data. The first model, english-left3words-distsim.tagger is based on a standard left-to-right third-order conditional Markov model considering the three left words to the target word. The second model, english-bidirectional-distsim.tagger is based on a bidirectional dependency network considering preceding and following word context to the target word.
The OpenNLP POS tagger is an open source tagger that is also based on maximum entropy.29 30 Although we could not find tagger accuracies reported, our evaluation found it to be on a par with the Stanford tagger tested on Penn Treebank WSJ data using the packaged en-pos-maxent.bin general English model.
The LBJ POS Tagger is an open-source tagger produced by the Cognitive Computation Group at the University of Illinois.31 32 It is based on a two-layer neural network in which the first layer represents POS tagging input features and the second layer represents POS multi-classification nodes. Winnow33 34 is a weight training algorithm used to optimize the neural network model. This tagger reported a tagging accuracy of 96.6% on its packaged general English model trained and tested on Penn Treebank WSJ data.
The LingPipe POS tagger is implemented using a bigram hidden Markov model (HMM).35 It includes two models trained from biomedical domain corpora, GENIA and MedPost. The GENIA corpus is made up of 2000 MEDLINE abstracts tied to MeSH terms: human, blood cells, and transcription factors.36 The MedPost corpus is made up of 5700 sentences from random subsets of MEDLINE biological abstracts.37 Our interest in LingPipe was to evaluate how closely biomedical text linguistic structures paralleled clinical narrative linguistic structures.
Easy Adapt is a POS tagger based on a perceptron model that uses a best candidate search space pruning algorithm that optimizes the weights of the perception using supervised learning.38 Easy Adapt also supports a method of domain adaptation that is discussed in detail below, which produced superior results when evaluated against other domain adaptation approaches.39
POS tagging in the clinical text domain
Results reported in the literature on POS tagging on clinical texts demonstrate limited consistency and reproducibility. We found no studies that addressed the generalizability of results across institutions or that use corpora made up of a broad sample of different clinical narrative types. This may reflect the stringent privacy laws around the use and distribution of private health information.9 Most of the studies that have been conducted on POS tagging in the clinical domain have been done on biomedical abstracts or a single clinical report type. We review a subset of these studies below.
Smith et al37 constructed a HMM POS tagger trained with bigram frequencies from MEDLINE abstracts. Their tagger reported a tagging accuracy of 97%. Although the results of Smith et al37 were impressive, Coden et al21 and Meystre et al16 provide evidence that the linguistic and distributional characteristics of biomedical texts and clinical texts are not the same.
Campbell and Johnson20 applied the transformation-based tagger of Brill40 to a corpus of discharge summaries and reported an accuracy of 96.9%. Their methods make mention of modifications made to the tagset to ensure consistency of the documented guidelines used by human annotators. If tagsets are modified in such a way as to categorize POS at a more course-grained level, this can lead to overstated tagger accuracies. The tagger may not have to differentiate between as many ambiguous case categories. Also, a note type from a single institution does not provide evidence of generalizability across institutions or validity across different report types.
Pakhomov et al41 described a proprietary corpus that was constructed from clinical notes. They reported a tagging accuracy of 94.7% over 100 650 tokens using the HMM-based TnT tagger.42 This same corpus is described as the MED corpus and was used in a study conducted by Savova et al.43 That study evaluated the OpenNLP tagger,29 30 which is integrated into the clinical text analysis and knowledge extraction system (cTAKES). The OpenNLP tagger retrained using the MED corpus reported a tagging accuracy of only 93.6%. This same tagger and model when tested by Fan et al44 on progress notes in a recent study produced accuracies of 85–88%.
Approaches to domain adaptation
Domain adaptation is a method used to adapt learning algorithms trained from a source domain where labeled data are readily available to a target domain where labeled data are limited. The goal of this method is to retain similar cross-domain characteristics learned from the source domain and couple those characteristics with new distinct attributes related to and learned from the limited labeled data available in the target domain. The hope is that this approach will perform better than an approach developed from either the source or target labeled data alone. The realized benefit is that out-of-domain tools can be adapted to new domains only requiring a modest amount of newly annotated target data.
Several approaches to domain adaptation have been researched. The most common approach is simply to combine the source and target labeled data, and train a new model. Coden et al21 evaluated this method using Penn Treebank WSJ, labeled data as source data and the MED clinical notes corpus (as described in Pakhomov et al)41 as the target data. They also considered a method that augments the aggregated source-target labeled data with an unambiguous lexicon derived from the target domain. They reported trigram HMM model tagger accuracies of 92.87% without lexicon, and 92.88% accuracy with lexicon.
Liu et al45 explored an interesting domain adaptation method known as sample selection.46 In that method, heuristics are used to identify training cases from the target domain that are believed most informative and will provide the most benefit in retraining a statistical machine learner. In their study, they compared two different word frequency-based selection heuristics. A maximum entropy tagger is then retrained using a training set constructed from combined Penn Treebank WSJ source-labeled data and heuristic selected target-labeled cases. They reported accuracies of 92.7% and 81.2% on two implemented heuristics. They also reported a tagging accuracy of 93.9% using the same source WSJ data combined with all available target data representing slightly over 21 000 tagged words. The study did not address generalizability across different clinical note types or healthcare institutions.
Blitzer et al47 investigated adapting a POS tagger from the Penn Treebank WSJ source domain to a target domain of MEDLINE biomedical abstracts using a method introduced as structural correspondence learning. In this approach, pivot features such as <the token to the right of word> are identified as features that occur frequently across both domains of unlabeled data and also behave contextually in the same way. In this example, the word that is left of the mentioned pivot feature may be unambiguously identified in the source labeled data. Using this pivot feature, target domain cases are tagged that are considered in high correspondence with the defined pivot feature. Blitzer et al47 reported an improvement from 87.9% baseline to 88.9%. A critical aspect of this approach is that high quality pivot features can be identified across both domains.
Daumé and Marcu39 demonstrated that the commonly used source-target labeled data aggregation method as well as other high performing approaches to domain adaptation can be outperformed using a feature space augmentation approach. In this method, known as Easy Adapt, the learner's feature space is expanded to contain three versions of the original feature space: a general version, a source-specific version, and a target-specific version. The general version feature space represents features shared across both source and target labeled POS cases. The source-specific version feature space is unique to the source labeled data and the target-specific version is unique to the target labeled data. The three feature space versions are then aggregated to make up the feature set used for training. More formally stated, if the original input space is defined as  =ℝF for F>0 then the augmented feature space is χ=ℝ3F. This approach reported an error rate of 3.61% on POS tagging adaptation from WSJ text to PubMed biomedical abstracts. It is worth noting that this method of domain adaptation generalizes well to most machine learning algorithms and adaptation problems.
=ℝF for F>0 then the augmented feature space is χ=ℝ3F. This approach reported an error rate of 3.61% on POS tagging adaptation from WSJ text to PubMed biomedical abstracts. It is worth noting that this method of domain adaptation generalizes well to most machine learning algorithms and adaptation problems.
Materials and methods
POS annotated datasets
Three corpora were used to carry out the experiments in this study. The source domain corpus was made up of Penn Treebank WSJ text. Two corpora made up of clinical narratives were used as target domains. The distributional characteristics of each corpus are shown in table 1.
Table 1.
Distributional corpus characteristics
| Corpus | No of sentences | No of tokens | No of unique words |
|---|---|---|---|
| WSJ non-clinical corpus | 8887 | 210 413 | 17 196 |
| IHC clinical corpus | 524 | 10 233 | 2442 |
| Pitt clinical corpus | 1036 | 12 227 | 2100 |
| Combined IHC and Pitt clinical corpus | 1560 | 22 460 | 3558 |
IHC, Intermountain Healthcare; Pitt, University of Pittsburgh; WSJ, Wall Street Journal.
The Intermountain Healthcare clinical (IHC) corpus was constructed from a uniform distribution of the 10 most common, electronically recorded, clinical report types in operation at two large community hospitals, Intermountain Medical Center and LDS Hospital in Salt Lake City, Utah. The report types are described in box 1.
Box 1. Ten most common clinical report types.
Progress note
Consultation report
Discharge summary
Operative report
Surgical pathology report
History and physical report
Emergency department report
x-Ray chest two views frontal lateral
Emergency department visit note
x-Ray chest one view portable
Reports were selected from encounters over a 2-year period from January 2008 to December 2009. One hundred and ninety-seven randomly selected sentences (3672 tokens) from this clinical text corpus were annotated for POS using the Penn Treebank tag set28 and guidelines28 48 supplemented with guidelines conforming to the philosophy of the SPECIALIST lexicon.10 49 The SPECIALIST lexicon10 49 is heavily made up of compound nouns. For example, in the SPECIALIST lexicon10 49 ‘fecal occult blood’ is considered a clinical symptom annotated as a compound noun. Inter-annotator agreement among two annotators measured using the Fleiss50 κ test statistic was 0.953 (95% CI 0.941 to 0.965). An additional 327 sentences were then annotated between the two annotators for a total of 524 clinical sentences.
In addition, a second target clinical corpus was obtained from the Biomedical Language Understanding Lab51 at the University of Pittsburgh. This corpus known in this study as the Pitt clinical corpus consists of 11 emergency department reports and 35 radiology reports. The reports were randomly selected from documents deposited into the MARS medical archival system at the University of Pittsburgh Medical Center52 between March and April 2007.
The set of emergency department reports contains 534 sentences (6236 tokens); the set of radiology reports 502 sentences (5991 tokens). All reports in the corpus were manually annotated for POS using the Penn Treebank tag set and guidelines,28 48 supplemented with guidelines specifically addressing the annotation of POS information in clinical reports. These supplemental guidelines expand on complex tagging decisions for cases that arise frequently in clinical text but are less acute in general English, for example, choosing between verbal, nominal, and adjectival tags for words ending in ‘-ing’. The report set was annotated in four rounds. Inter-annotator agreement measured using the Cohen53 κ test statistic ranged from 0.86 to 0.94.
The two individual clinical corpora were also combined to form a larger target clinical corpus as part of the evaluation. We use 10-fold cross-validation in our evaluations.
POS tagger experiments
Four top performing POS taggers were selected (OpenNLP tagger,29 Stanford tagger,23 24 LBJ tagger,31 and LingPipe tagger)35 and evaluated against the three corpora using their out-of-the-box trained models as described in table 2. The focus of these experiments was to confirm the operating characteristics when applied to clinical narratives. Included in these evaluations were models developed on biomedical abstracts and a domain adapted clinical model generated from the proprietary MED corpus.41 Biomedical abstracts intuitively would seem linguistically closer to clinical texts.
Table 2.
Top performing taggers with out-of-the-box models
| POS tagger | Algorithm | Model | Training corpus description |
|---|---|---|---|
| OpenNLP tagger | Maximum entropy | en-pos-maxent.bin | Penn Treebank WSJ |
| OpenNLP tagger | Maximum entropy | postagger.model.bin.gz | Mayo Clinical Model—cTAKES |
| Stanford tagger | Maximum entropy | english-bidirectional-distsim.tagger | Penn Treebank WSJ |
| Stanford tagger | Maximum entropy | english-left3words-distsim.tagger | Penn Treebank WSJ |
| LBJ tagger | Winnow neural network | N/A | English Penn Treebank WSJ |
| LingPipe tagger | HMM | pos-en-bio-genia.HiddenMarkovModel | GENIA (MEDLINE abstracts w/MeSH terms: human, blood cells, and transcription factors) |
| LingPipe tagger | HMM | pos-en-bio-medpost.HiddenMarkovModel | Medpost (MEDLINE biological abstracts) |
cTAKES, clinical text analysis and knowledge extraction system; HMM, hidden Markov model; WSJ, Wall Street Journal.
Domain adaptation experiments
We evaluated Easy Adapt against a new method of domain adaptation called ClinAdapt. This newly suggested method of domain adaptation turns POS tagging into a form of error correction. This is a three-step process. The first step is to base tag each target domain sentence using a tagger model trained from the source domain. We selected the OpenNLP tagger from the four evaluated taggers based on its performance and speed at which it tags. Base tagging was done using the out-of-the-box en-pos-maxent.bin model trained on general English WSJ text as defined in table 2. The second step in the process was supported by the construction of an unambiguous lexicon of terms derived from the SPECIALIST lexicon.10 Terms contained within the SPECIALIST lexicon that are defined as having only one POS were extracted and made up the unambiguous lexicon. This step of the process retags the initially tagged words that exist in the constructed unambiguous lexicon. The intuition is that if a word exists in a clinically oriented lexicon with one POS then that POS must be correct. In the final step, a transformation-based learner makes final tag corrections to the words in the sentence by applying rules that were generated from the available target domain (clinical) data, thereby adapting that task of POS tagging to the target domain.
Transformation-based learners traditionally use symbolic rule templates representing linguistic features related to the task at hand, such as POS tagging. For example, proper nouns typically start with a capital letter. So a template may exist that changes an erroneous POS tag to a proper noun if the first letter of the word is capitalized. These templates are instantiated with the linguistic features surrounding a candidate tagging error in an attempt to identify the template instantiation across all templates that corrects the most errors in a training iteration.54 Box 2 defines the rule templates that are instantiated and compete against one another to correct the most errors in each iteration of training. The rule template that corrects the most net-errors (number of errors corrected by the proposed rule minus number of errors introduced by the proposed rule) over the entire corpus is selected as the best rule for that iteration. The winning rule instantiation is then applied to the training corpus correcting those errors associated with the rule and then the process repeats itself. This training process continues generating a sequence of rule transforms that corrects errors as each rule is applied to the corpus. For example, the rule template given by equation (1) is interpreted as:
| 1 |
Box 2. Transformation-based learner rule templates.
Lexical generation probability rule
if {p(tp|tci−1) p(wi|tp) p(tci+1|tp)>p(tci|tci−1) p(wi|tci) p(tci+1|tci)} then change tci to tp
Symbolic linguistic rules
if isAcronym(wi) then change wi tag tci to tp
if isSymbol(wi) then change wi tag tci to tp
if containsDigit(wi) then change wi tag tci to tp
if startsWithCapitalLetter(wi) then change wi tag tci to tp
if hasPrefix(wi, prefixa) then change wi tag tci to tp
if hasSuffix(wi, suffixa) then change wi tag tci to tp
if isPlural(wi) then change wi tag tci to tp
if isTag(tci−1, taga) then change wi tag tci to tp
if isTag(tci−2, taga) then change wi tag tci to tp
if isTag(tci−2, taga) & isTag(tci−1, tagb) then change wi tag tci to tp
if isTag(tci−3, taga) & isTag(tci−2, tagb) & isTag(tci−1, tagc) then change wi tag tci to tp
if isTag(tci+1, taga) then change wi tag tci to tp
if isTag(tci+2, taga) then change wi tag tci to tp
if isTag(tci+1, taga) & isTag(tci+2, tagb) then change wi tag tci to tp
if isTag(tci+1, taga) & isTag(tci+2, tagb) & isTag(tci+3, tagc) then change wi tag tci to tp
if isTag(tci−1, taga) & isTag(tci+1, tagb) then change wi tag tci to tp
if isWord(wi−1, worda) then change wi tag tci to tp
if isWord(wi−2, worda) & isWord(wi−1, wordb) then change wi tag tci to tp
if isWord(wi+1, worda) then change wi tag tci to tp
if isWord(wi+1, worda) & isWord(wi+2, wordb) then change wi tag tci to tp
if isWord(wi−1, worda) & isWord(wi+1, wordb) then change wi tag tci to tp
If the POS tag (tci−1) of the preceding word is assigned POS tag (taga) then change the tag of the current word (wi) from POS tag (tci) to the new POS tag (tp). An instantiation of this template during a training iteration may be ‘if the POS tag of the preceding word is an adjective then change the tag of the current word from a verb to a noun’. This rule would be applied across the entire corpus correcting errors, and in some cases introducing errors when the verb was the correct POS tag and should not have been changed to a noun. The rule correcting the most net-errors would then be selected as the best rule transform for that training iteration. For each rule template, all combinations of tags are considered as linguistic features instantiated in the template producing a correction score for each instantiation. The rule with the best score wins that round of training and is applied to the corpus. Training halts when all the rule template instantiations correct less than three errors in a given training iteration.
The transformation-based tagger of Brill47 only considered symbolic linguistic rules not taking advantage of the many benefits that statistical machine learning approaches provide.55 To extend this algorithm, we introduced a novel statistically based, hill-climbing rule predicated on lexical generation probability given by equation (2).
 |
2 |
This method introduces a Markov model-based mathematical optimization technique into the transformation-based learner model.
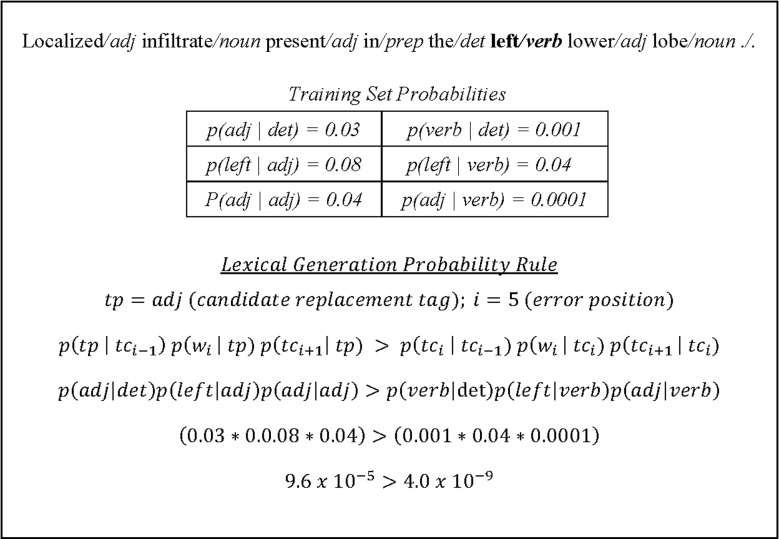
To facilitate discussion we introduce some notation. TP is defined as the set of POS tags defined by the Penn Treebank tag set.28 A POS tagged sentence is given by a sequence of words {w1 … wn} that make up the sentence, and a sequence of currently assigned POS tags given by {tc1 … tcn}. Furthermore, we define a newly proposed POS tag drawn from the set TP as tp. Then the rule template is applied as follows. Given a sentence {w1 … wn}, a current tag sequence {tc1 … tcn}, and a newly proposed replacement tag  for an incorrectly tagged word wi, tp replaces tci if the former probability with the proposed replacement tag tp is greater than the probability with the current incorrect tag tci. This rule attempts to find the optimal tag for the word in the sentence using a Markov model. In figure 1, we work through an example for clarity.
for an incorrectly tagged word wi, tp replaces tci if the former probability with the proposed replacement tag tp is greater than the probability with the current incorrect tag tci. This rule attempts to find the optimal tag for the word in the sentence using a Markov model. In figure 1, we work through an example for clarity.
Figure 1.
Lexical generation probability rule execution example.
In this example, the word ‘left’ is an adjective incorrectly marked as a verb. Estimating the probabilities from the training set and applying the lexical generation rule template where tp is an adjective, we would increment the score of this template instantiation by one for correctly modifying the tag to an adjective because it results in a greater probability over being marked as a verb. This same process would take place for every possible replacement tag in the set TP, and the highest scoring template among all the template instantiations would be selected. This also includes competing against the symbolic rule instantiations.
The lexical generation probability rule template relies on bigram tag probabilities and word-tag conditional probabilities that are estimated from the training data. These probability estimates were generated using Kneser–Ney smoothing.56 Smoothing is a technique used to reallocate a portion of the available probability mass to low frequency words, unknown words, and unseen tag sequences that were not encountered in the training corpus but may be seen in the test corpus. Kneser–Ney smoothing56 considers the number of different contexts a word or a tag has appeared in when assigning probability estimates. This method of smoothing more accurately quantifies realistic POS tag sequences and word-tag combinations when calculating the product of the conditional components in the lexical generation probability rule.
We evaluated Easy Adapt and ClinAdapt on the two clinical corpora and the combined clinical corpus using 10-fold cross-validation.
Results
POS tagger evaluations
Our results in table 3 report the tagger accuracies on each of the three corpora using the out-of-the-box models available with each tagger. Each tagger trained and tested on general English WSJ text reported accuracies of 96.9–97.3% as would be expected. When tested on clinical narratives these taggers dropped in accuracy by 8.5–15.5%. The cTAKES tagger, trained on clinical data performed slightly better than the standard general WSJ trained models in all but one case. The LingPipe models trained on biomedical abstracts performed poorly on all three corpora.
Table 3.
Out-of-the-box POS tagger performance
| WSJ corpus (%) | IHC clinical corpus (%) | Pitt clinical corpus (%) | Combined IHC and Pitt clinical corpus (%) | |
|---|---|---|---|---|
| OpenNLP tagger (maximum entropy—WSJ) | 97.1 | 87.6 | 82.5 | 84.9 |
| OpenNLP tagger (maximum entropy—Mayo clinical model—cTAKES) | 96.9 | 87.9 | 88.4 | 88.1 |
| Stanford tagger (maximum entropy—bi-directional WSJ) | 97.1 | 85.7 | 86.8 | 86.2 |
| Stanford tagger (maximum entropy—left 3 words WSJ) | 97.1 | 85.7 | 88.6 | 87.2 |
| LBJ tagger (winnow neural network—WSJ) | 97.3 | 87.3 | 81.8 | 84.3 |
| LingPipe tagger (HMM—GENIA) | 78.5 | 81.9 | 81.4 | 81.6 |
| LingPipe tagger (HMM—Medpost) | 74.4 | 83.1 | 82.9 | 83.0 |
cTAKES, clinical text analysis and knowledge extraction system; HMM, hidden Markov model; WSJ, Wall Street Journal.
Domain adaptation evaluations
Easy Adapt reported results of 88.3% on the IHC clinical corpus, 91.0% on the Pitt clinical corpus, and 89.3% on the combined clinical corpus as shown in table 4. Baseline training of Easy Adapt on source only and target only datasets was confirmed to produce lower accuracies as would be expected. If this was not the case, there would be no benefit from domain adaptation as simply retaining learners on target data would be sufficient.
Table 4.
Domain adaptation results
| IHC clinical corpus | Pitt clinical corpus | Combine IHC and Pitt clinical corpus | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Known word (%) | Unknown words (%) | Total (%) | Known word (%) | Unknown words (%) | Total (%) | Known word (%) | Unknown word (%) | Total (%) | |
| Easy Adapt (source only) | 89.7 | 65.6 | 84.5 | 91.3 | 51.3 | 78.3 | 90.5 | 56.4 | 81.1 |
| Easy Adapt (target only) | 87.8 | 70.7 | 85.1 | 91.2 | 74.3 | 89.0 | 89.6 | 74.4 | 87.9 |
| Easy Adapt (source+target) | 89.7 | 74.0 | 88.3 | 92.1 | 80.1 | 91.0 | 90.4 | 75.5 | 89.3 |
| ClinAdapt—base tagger (target only) | 94.7 | 89.6 | 89.8 | 97.4 | 91.4 | 92.1 | 95.9 | 90.6 | 91.1 |
| ClinAdapt (w/lexicon) | |||||||||
| Step 1: base tagging (source only) | 89.1 | 80.1 | 87.6 | 85.6 | 62.0 | 82.5 | 87.0 | 68.4 | 84.9 |
| Step 2: lexicon | 90.5 | 82.3 | 89.2 | 85.3 | 71.8 | 83.6 | 87.4 | 76.1 | 86.1 |
| Step 3: transformation-based learner (target only) | 95.9 | 82.8 | 93.8 | 97.1 | 72.8 | 93.9 | 94.9 | 76.1 | 93.2 |
| ClinAdapt (wo/lexicon) | |||||||||
| Step 1: base tagging (source only) | 89.1 | 80.1 | 87.6 | 85.5 | 62.7 | 82.5 | 87.0 | 67.9 | 84.9 |
| Step 2: transformation-based learner (target only) | 95.6 | 80.2 | 93.2 | 97.0 | 63.2 | 92.6 | 94.8 | 68.0 | 91.8 |
IHC, Intermountain Healthcare; Pitt, University of Pittsburgh.
Two experiments were conducted using ClinAdapt, one including the SPECIALIST lexicon as a second step in the tagging process and one removing the second step completely (no lexicon). The reason for this was to try and better understand the contribution the integration of a target domain-specific lexicon made. In addition to these two experiments, the ClinAdapt base tagger, OpenNLP, was retrained using clinical target data only as a baseline to confirm base tagger retraining was not sufficient to produce optimal results. Results demonstrated that there is benefit in using a domain adaptation algorithm. ClinAdapt with integrated lexicon reported accuracies of 93.8% on the IHC clinical corpus, 93.9% on the Pitt clinical corpus, and 93.2% on the combined clinical corpora. On each corpus, simply correcting errors by applying the clinical lexicon boosted accuracy by 1.1–1.2% over the base tagger. The additional gains attributed to the transformation-based learner were 4.6–10.3%. Overall, domain adaptation using ClinAdapt (with lexicon) accounted for an increase in accuracy of 6.2–11.4%. Adding the clinical lexicon to ClinAdapt resulted in an overall increase in performance. ClinAdapt with and without the lexicon outperformed Easy Adapt.
The frequency with which the transformation-based learner selected rules when evaluating the combined clinical target corpus is shown in figure 2. The lexical generation probability rule dominated the more traditional symbolic rules being selected as the optimal rule 77.2% of the time. Thirteen symbolic rules shown in the figure shared the remaining 22.8%.
Figure 2.
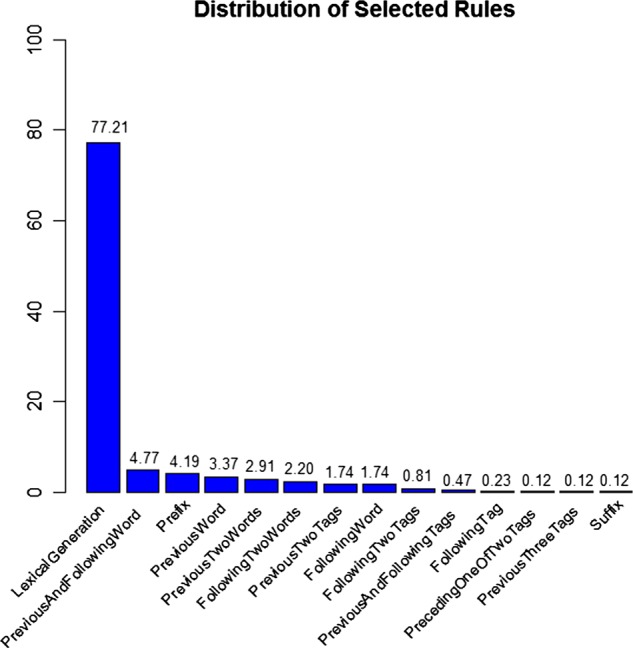
Transformation-based learner rule selection frequency.
To explore the relationship of target training set size to accuracy, the natural logarithm of the performance of Easy Adapt and ClinAdapt were fitted using linear regression as shown by the graph in figure 3. The analysis confirms that added accuracy could be achieved by growing the target training set.
Figure 3.
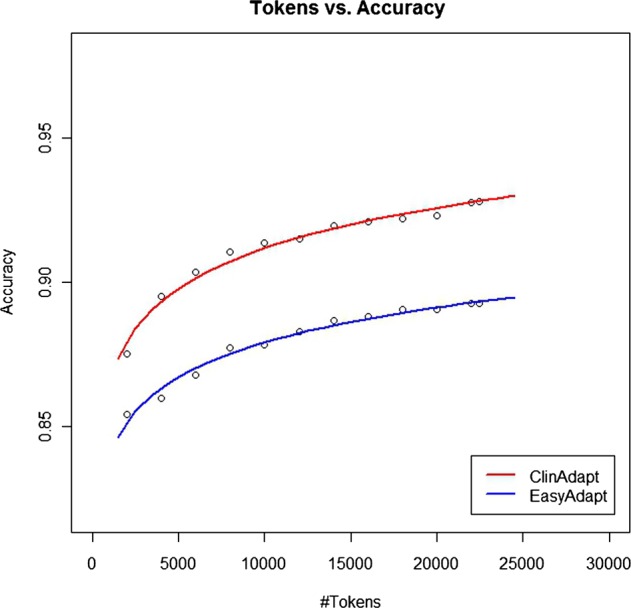
Accuracy as a function of training set size.
Discussion
In this paper, we demonstrated that the linguistic constructions and terminology found in clinical narratives differ from that of general English texts such as newswire and biomedical abstracts. We confirmed this hypothesis by evaluating several state-of-the-art POS taggers with their out-of-the-box models trained on either WSJ text or biomedical abstracts. In all cases, the performance of these taggers dropped significantly when applied to clinical text.
A surprising find was that the cTAKES POS tagger trained on the MED corpus43 performs no better than models trained on general English WSJ text when applied to the clinical corpora from two additional institutions. The fact that these comparisons are possible highlights an important point. Studies in clinical NLP can be more informative if multiple datasets across institutions were used to demonstrate generalizable solutions.9 In this study, we used multiple clinical corpora from independent institutions.
The sparse availability of clinically annotated data remains a barrier to reaching state-of-the-art operating characteristics on statistically based NLP tools when applied to the clinical domain.9 Domain adaptation is a viable method to improve performance and leverage the existing high performance algorithms already available. We introduced a new method of domain adaptation that outperformed leading adaptation methods. It requires modest amounts of clinically annotated data to obtain reasonable operating results. This suggested method changes the adaptation problem to that of error correction thereby increasing efficiency.
Intuitively, clinical narratives are full of unique terminology that may be sparsely encountered depending on the clinical note type. These unique and rarely seen words contribute to performance reductions in statistically based methods that are grounded in learning methods relying solely on textual context for inference.57 We addressed this problem by integrating an unambiguous, domain-specific lexicon, which improves overall POS tagging performance.
In this study, we successfully introduced a novel transformation-based learner rule predicated on lexical generation probability. This rule outperformed the more traditional symbolic rules typically used in transformation-based learners. Practically, we have shown how statistically based NLP methods can be integrated with symbolic methods for improved performance. This method should generalize well to other applications of transformation-based learners and domain adaptation.11–15
A limitation of our method is the time it takes to train transformation-based learners. As training sets grow in size, training time increases significantly. This is a well-known problem with transformation-based learners.58 59 In addition, we were unable to obtain a general sense of the number of tags required to reach tagging accuracies achieved in other non-clinical domains.22 Unfortunately, there were not enough data points to extrapolate safely from the natural logarithm fits. It seems reasonable that accuracy would be a logarithmic function of training set size as there are always unknown words being generated in language. Future work will further consider methods to improve the tagging accuracy of unknown words.
Conclusion
NLP tasks are commonly decomposed into subtasks that are chained together in a processing pipeline. The residual error in these subtasks may propagate to unreasonable levels adversely affecting subsequent downstream higher level processing tasks. By improving the performance of the individual subtasks, we expect to reduce the residual propagating error. We have suggested an alternative method of domain adaptation for clinical NLP. This method addresses the issue of limited annotated data to improve performance in the important subtask of POS tagging.
Footnotes
Contributors: JPF was the principle investigator, primary author, conceived the research design, prepared experimental datasets, developed ClinAdapt, and analyzed and interpreted the results. HD assisted in the research design, provided non-clinical datasets, developed and provided Easy Adapt, and edited the manuscript. SLD assisted in research design, analyzed and interpreted results, and edited the manuscript. WWC assisted in the research design, provided University of Pittsburgh clinical datasets, and edited the manuscript. HH provided University of Pittsburgh clinical datasets, authored part of the Methods section, and edited the manuscript. PJH helped shape the research design, provided important intellectual clinical content, and edited the manuscript.
Competing interests: None.
Ethics approval: Ethics approval for this study was obtained from Intermountain Healthcare.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Haug PJ, Christensen L, Gundersen M, et al. A natural language parsing system for encoding admitting diagnoses. Proceedings of AMIA Annual Fall Symposium; 1997:814–18 [PMC free article] [PubMed] [Google Scholar]
- 2.Haug PJ, Koehler SB, Christensen LM, et al. Inventors; Probabilistic method for natural language processing and for encoding free-text data into a medical database by utilizing a Bayesian network to perform spell checking of words. US patent 6292771. Alexandria, VA: United States Patent and Trademark Office, 2001. Sept 18
- 3.Jain NL, Friedman C. Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proceedings of AMIA Annual Fall Symposium; 1997:829–33 [PMC free article] [PubMed] [Google Scholar]
- 4.Jain NL, Knirsch CA, Friedman C, et al. Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. Proceedings of AMIA Annual Fall Symposium; 1996:542–6 [PMC free article] [PubMed] [Google Scholar]
- 5.Jones B, Ferraro JP, Haug P, et al. Performance of a real-time electronic screening tool for pneumonia [abstract]. Am J Respir Crit Care Med 2012;185:A5136 [Google Scholar]
- 6.Vines C, Collingsridge D, Jones B, et al. Emergency department physician experience with a real time, electronic pneumonia decision support tool [abstract]. Acad Emerg Med 2012;19(Suppl. 1):S49 [Google Scholar]
- 7.Chang MW, Do Q, Roth D. Multilingual dependency parsing: a pipeline approach. In: Nicolov N, Bontcheva K, Angelova G, et al. Recent advances in natural language processing IV. Amsterdam: John Benjamins Publishing Co., 2007:55–78 [Google Scholar]
- 8.Finkel JR, Manning CD, Ng AY. Solving the problem of cascading errors: approximate Bayesian inference for linguistic annotation pipelines. Proceedings of EMNLP; 2006:618–26 [Google Scholar]
- 9.Chapman WW, Nadkarni PM, Hirschman L, et al. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc 2011;18:540–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. UMLS Reference Manual. Specialist lexicon and lexical tools. Bethesda, MD: National Library of Medicine (US); http://www.ncbi.nlm.nih.gov/books/NBK9680/ (accessed 7 Jul 2012)
- 11.Campbell DA, Johnson SB. A transformational-based learner for dependency grammars in discharge summaries. Proceedings of ACL; 2002;3:37–44 [Google Scholar]
- 12.Brill E, Resnik P. A rule-based approach to prepositional phrase attachment disambiguation. Proceedings of the 15th Conference on Computational Linguistics; 1994;2:1198–204 [Google Scholar]
- 13.Florian R, Ngai G. Multidimensional transformation-based learning. Proceedings of the 2001 Workshop on Computational Natural Language Learning; 2001;7: 1–8 [Google Scholar]
- 14.Kim J, Schwarm SE, Ostendorf M. Detecting structural metadata with decision trees and transformation-based learning. Proceedings of HLT/NAACL; 2004:137–44 [Google Scholar]
- 15.Jurcicek F, Gašic M, Keizer S, et al. Transformation-based learning for semantic parsing. Proceedings of Interspeech; 2009:2719–22 [Google Scholar]
- 16.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008;35:128–44 [PubMed] [Google Scholar]
- 17.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. J Biomed Inform 2002;35:222–35 [DOI] [PubMed] [Google Scholar]
- 18.Friedman C. Semantic text parsing for patient records. In: Chen H, Fuller S, Friedman C. Medical informatics: knowledge management and data mining in biomedicine. 1st edn New York: Springer-Verlag, 2005:423–48 [Google Scholar]
- 19.Ceusters W, Buekens F, De Moor G, et al. The distinction between linguistic and conceptual semantics in medical terminology and its implication for NLP-based knowledge acquisition. Methods Inf Med 1998;37:327–33 [PubMed] [Google Scholar]
- 20.Campbell DA, Johnson SB. Comparing syntactic complexity in medical and non-medical corpora. Proceedings of AMIA Symposium; 2001:90–4 [PMC free article] [PubMed] [Google Scholar]
- 21.Coden AR, Pakhomov SV, Ando RK, et al. Domain-specific language models and lexicons for tagging. J Biomed Inform 2005;38:422–30 [DOI] [PubMed] [Google Scholar]
- 22.Manning C. Part-of-speech tagging from 97% to 100%: is it time for some linguistics? Proceedings of the 12th International Conference on Intelligent Text Processing and Computational Linguistics; 2011:171–89 [Google Scholar]
- 23.Toutanova K, Manning CD. Enriching the knowledge sources used in a maximum entropy part-of-speech tagger. Joint Sigdat Conference on EMNLP/VLC ; 2000:63–70 [Google Scholar]
- 24.Toutanova K, Klein D, Manning CD, et al. Feature-rich part-of-speech tagging with a cyclic dependency network. Proceedings of of HLT-NAACL; 2003:252–9 [Google Scholar]
- 25.Shen L, Satta G, Joshi A. Guided learning for bidirectional sequence classification. Proceedings of ACL; 2007;45:760–7 [Google Scholar]
- 26.Søgaard A. Simple semi-supervised training of part-of-speech taggers. Proceedings of ACL; 2010:205–8 [Google Scholar]
- 27. Wikipedia: POS tagging (state of the art). http://aclweb.org/aclwiki/index.php?title=POS_Tagging_(State_of_the_art) (accessed 2 Apr 2012)
- 28.Marcus MP, Marcinkiewicz MA, Santorini B. Building a large annotated corpus of English: the Penn Treebank. Comput Linguist 1993;19:313–30 [Google Scholar]
- 29.Baldridge J, Morton T, Bierner G. OpenNLP part-of-speech tagger. http://opennlp.apache.org/ (accessed 2 Apr 2012)
- 30.Ratnaparkhi A. A maximum entropy model for part-of-speech tagging. Proceedings of EMNLP; 1996;1:133–42 [Google Scholar]
- 31.Rizzolo N, Roth D. Learning based java for rapid development of nlp systems. Proceedings of LREC; 2010:958–64 [Google Scholar]
- 32.Roth D, Zelenko D. Part of speech tagging using a network of linear separators. The 17th International Conference on Computational Linguistics; 1998;2:1136–42 [Google Scholar]
- 33.Littlestone N. Learning quickly when irrelevant attributes abound: a new linear-threshold algorithm. Machine Learning 1988;2:285–318 [Google Scholar]
- 34.Littlestone N. Mistake bounds and logarithmic linear-threshold learning algorithms [PhD thesis]. Santa Cruz: University of California, 1990. Technical Report UCSC-CRL-89-11. [Google Scholar]
- 35. LingPipe 4.1.0. http://alias-i.com/lingpipe (accessed 2 Apr 2012)
- 36.Kim JD, Ohta T, Tateisi Y, et al. GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics 2003;19(Suppl. 1):i180–2 [DOI] [PubMed] [Google Scholar]
- 37.Smith L, Rindflesch T, Wilbur WJ. MedPost: a part-of-speech tagger for bioMedical text. Bioinformatics 2004;20:2320–1 [DOI] [PubMed] [Google Scholar]
- 38.Daumé H, III, Marcu D. Learning as search optimization: approximate large margin methods for structured prediction. Proceedings of the 22nd international conference on Machine Learning; 2005:169–76 [Google Scholar]
- 39.Daumé H. Frustratingly easy domain adaptation. Proceedings of 45th Ann Meeting of the Assoc Computational Linguistics; 2007;45:256–63 [Google Scholar]
- 40.Brill E. Transformation-based error-driven learning and natural language processing: a case study in part-of-speech tagging. Comput Linguist; 1995;21:543–65 [Google Scholar]
- 41.Pakhomov SV, Coden A, Chute CG. Developing a corpus of clinical notes manually annotated for part-of-speech. Int J Med Inform 2006;75:418–29 [DOI] [PubMed] [Google Scholar]
- 42.Brants T. TnT: a statistical part-of-speech tagger. Proceedings of ANLP; 2000:224–31 [Google Scholar]
- 43.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan J, Prasad R, Yabut RM, et al. Part-of-speech tagging for clinical text: wall or bridge between institutions? AMIA Annual Symposium Proceedings 2011:382–91 [PMC free article] [PubMed] [Google Scholar]
- 45.Liu K, Chapman W, Hwa R, et al. Heuristic sample selection to minimize reference standard training set for a part-of-speech tagger. J Am Med Inform Assoc 2007;14:641–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwa R. Sample selection for statistical parsing. Comput Linguis 2004;30:253–76 [Google Scholar]
- 47.Blitzer J, McDonald R, Pereira F. Domain adaptation with structural correspondence learning. Proceedings of EMNLP; 2006:120–8 [Google Scholar]
- 48.Santorini B. Part-of-speech tagging guidelines for the Penn Treebank project (3rd revision), Technical Report MS-CIS-90-47. University of Pennsylvania; 1990 [Google Scholar]
- 49.Browne AC, McCray AT, Srinivasan S. The specialist lexicon. Bethesda, MD: Lister Hill National Center for Biomedical Communications, National Library of Medicine; 2000:18–21, NLM Technical Report NLM-LHC-93-01 [Google Scholar]
- 50.Fleiss JL. Measuring nominal scale agreement among many raters. Psychol Bull 1971;76:378–82 [Google Scholar]
- 51. Biomedical Language Understanding (BLU) Lab—University of Pittsburgh. Pittsburgh, PA; http://www.dbmi.pitt.edu/blulab/index.html (accessed 7 Jul 2012)
- 52.Yount RJ, Vries JK, Councill CD. The Medical Archival System: an information retrieval system based on distributed parallel processing. Inf Process Manage 1991;27:379–89 [Google Scholar]
- 53.Wood JM. Understanding and computing Cohen's kappa: a tutorial. WebPsychEmpiricist http://wpeinfo/vault/wood07/Wood07pdf (accessed 2 Apr 2012)
- 54. World of computing. Articles on natural language processing: transformation based learning. http://language.worldofcomputing.net/pos-tagging/transformation-based-learning.html# (accessed 7 Jul 2012)
- 55.Brill E. Some advances in transformation-based part of speech tagging. Proceedings of the Twelfth National Conference on Artificial Intelligence (AAAI-94); 1994:722–7 [Google Scholar]
- 56.Kneser R, Ney H. Improved backing-off for m-gram language modeling. IEEE International Conference on Acoustics, Speech, and Signal Processing; 1995;1:181–4 [Google Scholar]
- 57.Kay M. A life of language. Computat Linguist 2005;31:425–38 [Google Scholar]
- 58.Ngai G, Florian R. Transformation-based learning in the fast lane. Proceedings of NAACL; 2001:40–7 [Google Scholar]
- 59.Carberry S, Vijay-Shanker K, Wilson A, et al. Randomized rule selection in transformation-based learning: a comparative study. Nat Lang Eng 2001;7:99–116 [Google Scholar]