Abstract
Background
Electronic health record (EHR) users must regularly review large amounts of data in order to make informed clinical decisions, and such review is time-consuming and often overwhelming. Technologies like automated summarization tools, EHR search engines and natural language processing have been shown to help clinicians manage this information.
Objective
To develop a support vector machine (SVM)-based system for identifying EHR progress notes pertaining to diabetes, and to validate it at two institutions.
Materials and methods
We retrieved 2000 EHR progress notes from patients with diabetes at the Brigham and Women's Hospital (1000 for training and 1000 for testing) and another 1000 notes from the University of Texas Physicians (for validation). We manually annotated all notes and trained a SVM using a bag of words approach. We then used the SVM on the testing and validation sets and evaluated its performance with the area under the curve (AUC) and F statistics.
Results
The model accurately identified diabetes-related notes in both the Brigham and Women's Hospital testing set (AUC=0.956, F=0.934) and the external University of Texas Faculty Physicians validation set (AUC=0.947, F=0.935).
Discussion
Overall, the model we developed was quite accurate. Furthermore, it generalized, without loss of accuracy, to another institution with a different EHR and a distinct patient and provider population.
Conclusions
It is possible to use a SVM-based classifier to identify EHR progress notes pertaining to diabetes, and the model generalizes well.
Keywords: electronic health record, support vector machine, natural language processing, search
Background and introduction
To make good decisions, clinicians are responsible for reviewing, integrating and interpreting a considerable amount of clinical information, often in a short period of time.1 2 Furthermore, these data are often duplicated3 or fragmented4 making accurate clinical decisions more difficult. Clinicians employ a variety of techniques for filtering and summarizing these data. We previously introduced a theoretical model of clinical summarization called AORTIS, which focuses on six steps: aggregation, organization, reduction, transformation, interpretation and synthesis.1
Electronic health records (EHR) generally excel at the first two tasks: aggregation and organization, and can often perform some degree of reduction, although frequently only on coded, structured data,5 for example, by showing only a patient's current medications or the most recent value of a laboratory study. In recent work, we have developed several methods for enhanced summarization of coded, structured data, including methods for problem-oriented knowledge-based reduction of medications and laboratory results using ontologies,6 association rule mining7 and crowd sourcing.8
Consistent with the capabilities of existing EHRs, all of these methods employ structured, coded data. However, unstructured data, mostly in the form of free-text progress notes, are also critical for clinical decision making. In this paper, we turn our attention to such data, with a focus on a common clinical task: identifying all notes pertaining to a particular condition (in our case, diabetes).9
Myriad natural language processing (NLP) techniques exist that are designed to process or classify free-text clinical documents and several recent reviews have described the state of the art in NLP,10–13 and a comprehensive review by Nadkarni et al10 describes the foundations, fundamentals and applications of NLP in a clinical context. This review divides NLP into low-level and high-level tasks. Examples of low-level tasks include tokenization, part-of-speech tagging and chunking, while high-level tasks include named entity recognition, negation identification and information extraction.
Most of the tasks considered by Nadkarni et al10 and, indeed, most work in NLP is ultimately done in support of extraction tasks, such as identifying a patient's smoking status or medication list.14–16 However, classification tasks have also been studied using similar techniques. Sohn and Savova17 developed a classifier that used a three-stage combination of rule-based classification, negation detection and support vector machine (SVM)-based classification to take sentences from clinical notes and identify smoking status with strong results. Solti et al18 similarly developed a classification system to identify cases of acute lung injury in chest x-ray reports using a maximum entropy classifier, and found that their system was more accurate than a keyword-based classifier developed by experts. In this study, we considered a different task: classification of outpatient clinical notes by disease.
For this project, we chose to build a classifier based on a SVM. SVM and their applications in classification and NLP have been described in detail, and the SVM approach has been used in a variety of tasks. For example, Carroll et al19 used a SVM-based approach to identify patients with rheumatoid arthritis using a mix of structured and natural language data. Roberts and Harabagiu20 used SVM as well as conditional random fields to extract concepts from EHR data, as did Minard et al.21 However, all of these systems were focused on extraction of information, rather than classification.
A related but different problem is EHR search. In this problem, which is a special case of the general document classification task, the goal is not to extract concepts but, instead, to identify relevant documents pertaining to a user's query. A recent paper by Tawfik et al22 described a similar semantic search engine for EHR. Although they did not assess the accuracy of their search algorithms, they were able to show that users with access to a semantic search engine in their EHR were able to complete sample tasks with less than half the number of clicks and nearly twice as fast as users without such a search tool. Other studies of EHR search engines have also been reported, and generally found high user satisfaction, although they have not reported on the accuracy of the search algorithm.23–25
In this study, we describe a SVM-based approach for searching and classifying EHR progress notes. We apply this method to the task of identifying notes about diabetes. The system is designed to answer the question ‘which of this patient's notes pertain to diabetes?’ Such a system could be used as part of a problem-oriented summarization system or EHR search engine, allowing a clinician selectively to filter a patient's progress notes to identify just those notes pertaining to diabetes. We also formally validate the system at two sites and report the accuracy and performance characteristics of our algorithm.
Methods
Creation of data samples
We employed three data samples in our study: a training sample, a test sample and an external validation sample. The training and test samples were both drawn from medical records at the Brigham and Women's Hospital (BWH), Boston, Massachusetts, USA. BWH is a large academic medical center in Boston with a self-developed EHR. We selected a random sample of 1000 patients who had been seen at BWH and had a coded entry of diabetes mellitus (any type) on their problem list. We then retrieved all outpatient notes for this sample of 1000 patients and selected a random sample of 2000 notes describing these patients’ visits. Because we sampled at random, some patients may have had more than one note in our sample, while others might have had none. After annotation, we divided the sample of 2000 notes randomly, assigning 1000 notes to our training sample and 1000 notes to our test sample.
For validation, we retrieved a random sample of 1000 notes for patients with diabetes (using the same coded definition of diabetes) from the University of Texas Faculty Physicians (UTP), a large, multispecialty ambulatory academic practice in Houston, Texas, USA. UTP physicians use a commercially developed outpatient EHR system (Allscripts Enterprise, Chicago, Illinois, USA). This external validation sample was chosen because it represents an entirely distinct patient and provider population.
The study was reviewed and approved by the Partners HealthCare Human Subjects Committee and The University of Texas Health Science Center at Houston Committee for the Protection of Human Subjects.
Sample annotation
Two annotators (SH (fourth year medical student) and AK (physician)) independently reviewed all notes from the test and training samples. Each note was assigned a score of 1, 2 or 3, where 1 indicated that the note had no mention of diabetes, 2 indicated that the note mentioned diabetes only in passing (eg, in a past medical history section), while 3 indicated that the note specifically addressed diabetes (eg, diabetes control, medication changes, endocrinology referral) and/or its long-term complications (eg, podiatry or ophthalmology referrals). To measure inter-annotator agreement, both weighted and unweighted Cohen's κ coefficients were calculated. After calculating inter-annotator agreement, all cases in which disagreement was present were reviewed collaboratively by the annotators and consensus was reached. We designed a procedure in which the first author (AW) would adjudicate any notes in which consensus could not be reached, but this procedure was not needed as consensus was reached by SH and AK on all notes.
In addition, one of the annotators (SH) reviewed the validation sample of 1000 notes from the University of Texas Physicians practice and graded them using the same scale described above.
Feature extraction
Before training the SVM, it was necessary to extract features from the notes that could be used for classification. We used a ‘bag of words’ approach, so each note was converted to a feature vector. The first step of the bag of words process is to create a lexicon of frequently occurring words. We counted the number of times each unique word was used in the training set of notes—if a word was used multiple times in a single note in the training set, all instances were counted. We then included all words that occurred more than a threshold number of times as features in our model. We tested various values for this threshold, ranging from 100 to 1000 (ultimately settling on 250). For each of these lexicons, we then checked each note for the presence of each of the words in the lexicon, and constructed a feature vector that contained a 0 if the lexicon word was not present in the note and a 1 if it was present.
Notably, we also experimented with several potential enhancements to our feature extraction approach, including feature selection, stop word filtering and various approaches for named entity recognition, synonym resolution and negation detection. However, none of these approaches exceeded the performance of our bag of words approach and, as such, we do not detail them here.
SVM training
Next, we trained SVM using LIBSVM 3.126 with a Gaussian radial basis function kernel using a grid search to find optimal values of the kernel parameter, γ, and the cost parameter, C, which controls the trade-off between false positives and false negatives. The parameters were chosen that optimized the accuracy of the SVM on the training set under cross-validation. After the optimal SVM was found, we applied it to the test set to yield probability estimates for each note in the test set. We then compared these estimates to our previous manual annotations of the test set and formulated a receiver operating characteristic (ROC) curve and computed the area under the curve (AUC). For each point on the curve, we also calculated precision, recall, and the F-measure. We used the ROCR module of the R statistical package to make these calculations.27
Although it is possible to train a multi-class SVM, most work on SVM and most standard evaluations are designed for a binary classification problem. In order to facilitate the evaluation, we converted our three-level annotation scheme into a binary scheme, separating notes that mentioned or discussed diabetes from those that did not mention diabetes at all: a grade of 1 represented a negative case for the SVM and a grade of 2 or 3 represented a positive case.
Results
Inter-annotator agreement
Figure 1 shows the flow of codes through the annotation and consensus process. The overall agreement between the two annotators for the BWH set was 97.7%, and the κ was 0.960 for the three-level agreement. As our grades are ordinal, we also computed a linearly weighted κ, which takes into account the fact that a difference of one grade is smaller than a difference between two grades. The calculated linearly weighed κ was 0.970. These κ represent very good agreement. Annotation of notes took each reviewer approximately 20 h.
Figure 1.
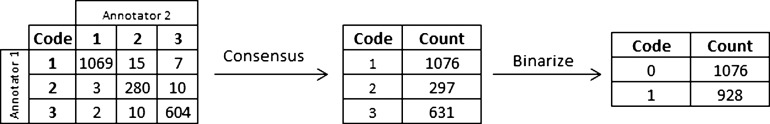
Flow of codes through annotation and consensus process.
SVM model training
As described in the Methods section, we trained the SVM models on the training set, and optimized the three parameters (two kernel function parameters for the SVM and the lexicon threshold parameter for the feature selection) using a grid search and cross-validation. The best model used cost parameter c=211=2048, γ=2−13=0.0001220703125 and a lexicon threshold of at least 250 occurrences per word. The resulting model had a total of 275 support vectors (163 in the positive class and 112 in the negative class), and used 126 features (frequent words). Model training took approximately 2 h to complete the entire grid search. Although it is difficult to assess the influence of individual features in a kernel-transformed SVM, the five features with the highest f scores were ‘diabetes’, ‘a1c’, ‘metformin’, ‘dm’ and ‘insulin’ (all features are converted to lower case). It is important to note that these are not necessarily the most influential features in the actual SVM model but, instead, are features that are present in the final model and, independently, are strong predictors of whether a note pertains to diabetes.
Model evaluation and performance on the test set
After the model was trained, we applied it to the test set, which consisted of an additional 1000 annotated notes. Overall, model performance was excellent. The ROC curve from the test dataset analysis is shown in figure 2. The AUC was 0.956 and the best F statistic was 0.934, with corresponding precision of 0.952 and recall of 0.916. This represents a substantial improvement over a free-text search for the word ‘diabetes’, which has a F statistic of 0.800 (shown on figure 2 as a blue x).
Figure 2.
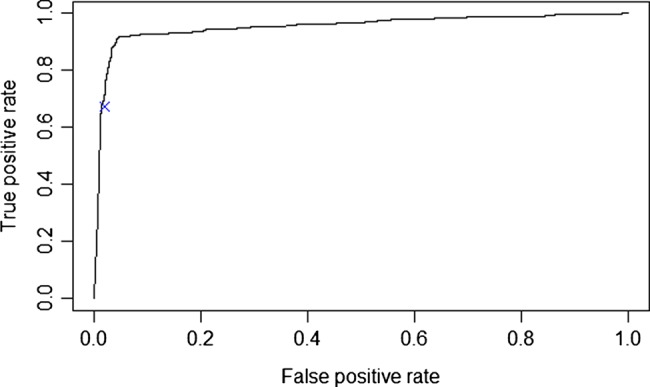
Receiver operating characteristic curve showing performance of support vector machine classifier on test set with comparison to text-search (blue x).
Validation of SVM model on UTP notes
For validation, we then applied the same model (trained on BWH data) to the external validation set, which contains notes from UTP. The AUC for the validation data set was 0.947 and the best F statistic was 0.935, with corresponding precision of 0.951 and recall of 0.920. Figure 3 shows a ROC curve comparing the performance of the model on the BWH test set and the UTP external validation set.
Figure 3.
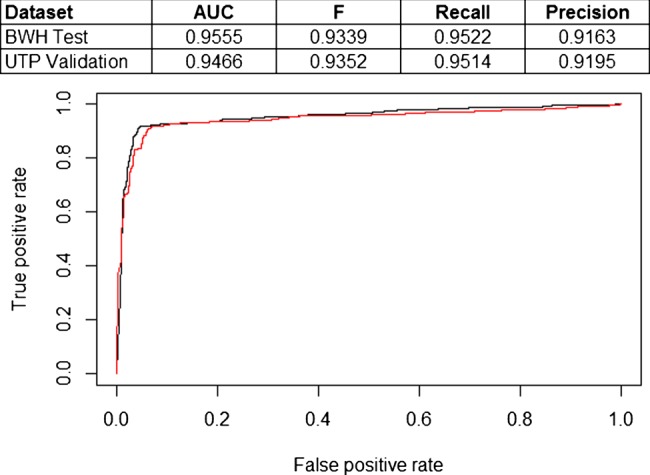
Comparison of receiver operating characteristic curves for support vector machine model trained on Brigham and Women's Hospital (BWH) training set and tested on BWH test set (black line) and University of Texas Faculty Physicians (UTP) external validation set (red line). AUC, area under the curve.
Discussion
We successfully developed a simple method to identify diabetes-specific progress notes using a SVM note classifier with high performance. Like all classifiers, the SVM model requires a trade-off between precision and recall, so institutions utilizing a SVM must decide whether it is more important to retrieve a large number of notes at the cost of increasing the number of notes that are not about diabetes (ie, false positives); or whether it is more important to retrieve only notes that are about diabetes at the cost of increasing the number of false negative notes (ie, notes about diabetes that were not retrieved). This trade-off actually represents a distinct advantage over keyword search methods—any given keyword search has a fixed precision and recall—in the case of the keyword search we compared our results against, the precision was quite high (most notes returned were really about diabetes) but the recall was low (many notes about diabetes that did not contain the specific keyword were not found).
Another strength of our approach is the external validation we conducted. In addition to showing how the same data mining techniques could be used with different EHRs, we found that the performance of our model, trained on BWH data, was almost equivalent to that obtained using UTP data, suggesting that our model is generalizable across settings and not fixed to our particular local medical or colloquial dialect. This external validation (as well as the BWH test set and our cross-validation process) also helped guard against overfitting, which is a common concern in studies such as ours. Because our model fitting process had several parameters (the SVM cost parameter C, the kernel parameter γ and the threshold for lexicon inclusion), we could have selected parameters that overfit our test data, which would have made our model less generalizable. We guarded against this in several ways. First, we used cross-validation as we selected our parameters, so overfit models would be penalized during the parameter search process. Second, we assessed the performance of our model on both an independent test set as well as an independent, external validation set. Performance remained high on both the test and external validation sets, suggesting that overfitting from our approach was minimal.
The performance of our system was quite strong and, in fact, considerably stronger than has been found by other investigators using similar techniques. We believe that this is due to two main factors. First, our classification task was straightforward—identifying notes that are about diabetes appears to be relatively unambiguous, as evidenced by the high inter-annotator agreement for our human annotators. Second, our approach suggests that there may be sublanguage differences between notes pertaining to diabetes and other clinical notes—NLP classification systems that exploit sublanguage differences are often particularly effective.28 29
Some limitations of our study warrant discussion. First, our study focused on only one classification method—SVM. It is possible that other advanced data mining methods could have worked as well or better than the current model. A second limitation is our focus on diabetes—we did not test our methods on other conditions and, although we believe the method is likely to generalize to other conditions, this is not yet proved.
Conclusion
In conclusion, we designed an effective SVM-based method to filter notes about patients with diabetes and showed that our model generalized from an institution with a locally developed EHR to another institution with a distinctly different patient and provider population and a commercially developed EHR and retained excellent performance. Future study should focus on whether our method can be applied to other diseases.
Footnotes
Contributors: All authors meet the ICMJE criteria for authorship. Their contributions were: AW: conception and design, acquisition of data, analysis and interpretation of data, drafting the article and final approval of the version to be published. ABM: conception and design, acquisition of data, analysis and interpretation of data, critical revision of the manuscript for important intellectual content and final approval of the version to be published. SH: acquisition of data, critical revision of the manuscript for important intellectual content and final approval of the version to be published. AK: acquisition of data, critical revision of the manuscript for important intellectual content and final approval of the version to be published. DFS: conception and design, analysis and interpretation of data, critical revision of the manuscript for important intellectual content and final approval of the version to be published.
Funding: This work was supported by Office of the National Coordinator contract #10510592. The funder was not involved in the design, conduct or evaluation of the research.
Competing interests: None.
Ethics approval: The study was reviewed and approved by the Partners HealthCare Human Subjects Committee and The University of Texas Health Science Center at Houston Committee for the Protection of Human Subjects.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Feblowitz JC, Wright A, Singh H, et al. Summarization of clinical information: a conceptual model. J Biomed Inform 2011;44:688–99 [DOI] [PubMed] [Google Scholar]
- 2.Hall A, Walton G. Information overload within the health care system: a literature review. Health Info Libr J 2004;21:102–8 [DOI] [PubMed] [Google Scholar]
- 3.Wrenn JOSD, Bakken S, Stetson PD. Quantifying clinical narrative redundancy in an electronic health record. J Am Med Inform Assoc 2010;17:49–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgeois FCOK, Mandl KD. Patients treated at multiple acute health care facilities. Arch Intern Med 2010;170:1989–95 [DOI] [PubMed] [Google Scholar]
- 5.Laxmisan A, McCoy AB, Wright A, et al. Clinical summarization capabilities of commercially-available and internally-developed electronic health records. Appl Clin Inform 2012;3:80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCoy AB, Wright A, Laxmisan A, et al. A prototype knowledge base and SMART app to facilitate organization of patient medications by clinical problems. AMIA Annu Symp Proc 2011;43:888–94 [PMC free article] [PubMed] [Google Scholar]
- 7.Wright A, Chen ES, Maloney FL. An automated technique for identifying associations between medications, laboratory results and problems. J Biomed Inform 2010;43:891–901 [DOI] [PubMed] [Google Scholar]
- 8.McCoy AB, Wright A, Laxmisan A, et al. Development and evaluation of a crowdsourcing methodology for knowledge base construction: identifying relationships between clinical problems and medications. J Am Med Inform Assoc 2012;19:713–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright A, Maloney FL, Feblowitz JC. Clinician attitudes toward and use of electronic problem lists: a thematic analysis. BMC Med Inform Decis Mak 2011;11:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc 2011;18:544–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohno-Machado L. Realizing the full potential of electronic health records: the role of natural language processing. J Am Med Inform Assoc 2011;18:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chapman WW, Nadkarni PM, Hirschman L, et al. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc 2011;18:540–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008:128–44 [PubMed] [Google Scholar]
- 14.Uzuner O, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008;15:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savova GK, Ogren PV, Duffy PH, et al. Mayo Clinic NLP system for patient smoking status identification. J Am Med Inform Assoc 2008;15:25–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng QT, Goryachev S, Weiss S, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak 2006;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohn S, Savova GK. Mayo Clinic smoking status classification system: extensions and improvements. AMIA Annu Symp Proc 2009;2009:619–23 [PMC free article] [PubMed] [Google Scholar]
- 18.Solti I, Cooke CR, Xia F, et al. Automated classification of radiology reports for acute lung injury: comparison of keyword and machine learning based natural language processing approaches. Proc IEEE Int Conf Bioinform Biomed 2009;2009:314–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll RJ, Eyler AE, Denny JC. Naive electronic health record phenotype identification for rheumatoid arthritis. AMIA Annu Symp Proc 2011;2011:189–96 [PMC free article] [PubMed] [Google Scholar]
- 20.Roberts K, Harabagiu SM. A flexible framework for deriving assertions from electronic medical records. J Am Med Inform Assoc 2011;18:568–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minard AL, Ligozat AL, Ben Abacha A, et al. Hybrid methods for improving information access in clinical documents: concept, assertion, and relation identification. J Am Med Inform Assoc 2011;18:588–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tawfik AA, Kochendorfer KM, Saparova D, et al. Using semantic search to reduce cognitive load in an electronic health record. 2011 IEEE 13th International Conference on e-Health Networking, Applications and Services. 13–15 Jun 2011, Columbia, MO, USA [Google Scholar]
- 23.Natarajan K, Stein D, Jain S, et al. An analysis of clinical queries in an electronic health record search utility. Int J Med Inform 2010;79:515–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanauer DA. EMERSE: the Electronic Medical Record Search Engine. AMIA Annu Symp Proc 2006:941. [PMC free article] [PubMed] [Google Scholar]
- 25.Gregg W, Jirjis J, Lorenzi NM, et al. StarTracker: an integrated, web-based clinical search engine. AMIA Annu Symp Proc 2003:855. [PMC free article] [PubMed] [Google Scholar]
- 26.Chang CCLC. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2011;2:1–27 [Google Scholar]
- 27.Sing TSO, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics 2005;21:3940–1 [DOI] [PubMed] [Google Scholar]
- 28.Friedman C, Hripcsak G. Natural language processing and its future in medicine. Acad Med 1999;74:890–5 [DOI] [PubMed] [Google Scholar]
- 29.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. J Biomed Inform 2002;35:222–35 [DOI] [PubMed] [Google Scholar]


