Abstract
Objective
To provide a natural language processing method for the automatic recognition of events, temporal expressions, and temporal relations in clinical records.
Materials and Methods
A combination of supervised, unsupervised, and rule-based methods were used. Supervised methods include conditional random fields and support vector machines. A flexible automated feature selection technique was used to select the best subset of features for each supervised task. Unsupervised methods include Brown clustering on several corpora, which result in our method being considered semisupervised.
Results
On the 2012 Informatics for Integrating Biology and the Bedside (i2b2) shared task data, we achieved an overall event F1-measure of 0.8045, an overall temporal expression F1-measure of 0.6154, an overall temporal link detection F1-measure of 0.5594, and an end-to-end temporal link detection F1-measure of 0.5258. The most competitive system was our event recognition method, which ranked third out of the 14 participants in the event task.
Discussion
Analysis reveals the event recognition method has difficulty determining which modifiers to include/exclude in the event span. The temporal expression recognition method requires significantly more normalization rules, although many of these rules apply only to a small number of cases. Finally, the temporal relation recognition method requires more advanced medical knowledge and could be improved by separating the single discourse relation classifier into multiple, more targeted component classifiers.
Conclusions
Recognizing events and temporal expressions can be achieved accurately by combining supervised and unsupervised methods, even when only minimal medical knowledge is available. Temporal normalization and temporal relation recognition, however, are far more dependent on the modeling of medical knowledge.
Keywords: Natural Language Processing; Clinical Informatics; Medical Records Systems, Computerized
Introduction
With the ever-growing importance and utilization of electronic medical records (EMR), there is increasing demand for techniques to reconstruct the patient's clinical history automatically from natural language text. Central to this is the ability to extract clinically relevant events and place them on a timeline. This would enable automated reasoning about a patient’s conditions. Causal relationships are difficult to establish without temporal relationships. Furthermore, without temporal information, one cannot distinguish between a condition that occurred years ago and one that has occurred within a patient’s current hospital visit, and therefore whether it might be related to other symptoms the patient is experiencing.
The 2012 Informatics for Integrating Biology and the Bedside (i2b2) shared task1 focuses on the recognition of temporal relations between events and temporal expressions in clinical documents. It follows the TimeML standard for representing events (Event), temporal expressions (Timex3), and the relations between them (Tlink).
This article presents three distinct methods for resolving these three tasks. These methods are characterized by the central role of supervised techniques, while additionally using temporal normalization rules and unsupervised word clustering from large, unlabeled corpora. Moreover, instead of manually selecting the best features for the supervised methods employed in each task, we rely on a flexible method automatically to select the optimal subset of features from a large set of features.
Background
Temporal information is used in clinical documents to ground events chronologically. However, as with all natural language, temporal information may be ambiguous and requires pragmatic reasoning to be fully grounded. Consider the following sentence from a clinical progress note:
Cardiovascular stable, significant hypertension was noted on 9/7/93 at 5:10 a.m. and therefore 10 cc per kilo albumin was given.
This sentence discusses a change in a patient's medical condition and the response that was taken by the hospital. In order to represent this response formally, several steps are necessary. First, the relevant medical events must be noted. Second, temporal expressions must be extracted and grounded on a timeline. Finally, relations must be detected between these events and temporal expressions in order to enable chronological reasoning about the clinical note.
Temporal relation recognition has been well studied in non-clinical settings. Evaluations for this task on newswire include the TempEval-12 and TempEval-23 challenges, as well as an upcoming TempEval-34 challenge. The interest in these tasks has generated numerous automatic methods5–17 for recognizing events, temporal expressions, and the temporal relations between them. These automatic methods have taken a variety of different approaches: (1) rule-based classifiers incorporating real-world knowledge; (2) supervised classifiers (eg, support vector machines (SVM), maximum entropy, and conditional random fields (CRF)) using common natural language processing (NLP) features; and (3) joint inference-based classifiers, such as Markov Logic.15 Many methods use some combination of these, such as a supervised classifier and a limited set of rules. Our work most closely falls under the second category, as we present a largely supervised approach. We differ from these previous methods, however, by focusing on how to create a flexible method that is customizable to changes in the data, as methods for temporal information extraction in clinical text need to be robust to changes in data (different hospitals, doctors, electronic medical records (EMR) systems, etc).
Previous i2b2 tasks have studied important elements in the processing of clinical text, such as extracting medication information;18 identifying medical concepts, assertions, and relations between concepts;19 and recognizing co-reference relations between medical concepts.20 The most relevant of these to the 2012 i2b2 shared task is the recognition of clinical concepts performed in the 2010 task. The three types of medical concepts in that task (Problems, Treatments, and Tests) form the core subset of the medical events studied in the 2012 task.
Task description
The 2012 i2b2 shared task1 adapts the TimeML21 temporal annotation standard for use on clinical data, providing 310 annotated documents (190 development, 120 test). The i2b2 standard contains three primary annotations:
Event: any situation relevant to the patient's clinical timeline. Events have one of six types: (1) Problem (eg, disease, injury); (2) Treatment (eg, medication, therapeutic procedure); (3) Test (eg, diagnostic procedure, laboratory test); (4) Clinical_dept (ie, the event of being transferred to a department); (5) Evidential (eg, report, show); and (6) Occurrence (ie, all other events). In addition, Events have a polarity (Positive, Negative) and modality (Factual, Conditional, Possible, Proposed).
Timex3: temporal expressions that enable Events to be chronologically grounded. Timex3s have one of four types (Date, Time, Duration, Frequency) and a modifier (More, Less, Start, Middle, End, Approx, Na). In addition, Timex3 annotations have a value, which is the normalized form of the temporal expression.
Tlink: temporal relations between either Events or Timex3s. For the official evaluation, a simplified set of temporal relations were used (Before, After, and Overlap).
For the example sentence above, the relevant annotations are:
Event{text=“Cardiovascular stable”; type=Occurrence; polarity=Positive; modality=Factual}
Event{text=“significant hypertension”; type=Problem; polarity=Positive; modality=Factual}
Event{text=“albumin”; type=Treatment; polarity=Positive; modality=Factual}
Timex3{text=“9/7/93 at 5:10 a.m.”; type=Time; mod=Na; val=“1993–09–07T05:10”}
- Tlink{from=“Cardiovascular stable”; to=“significant hypertension”; type=Before}
- Tlink{from=“significant hypertension”; to=“9/7/93 at 5:10 a.m.”; type=Overlap}
- Tlink{from=“albumin”; to=“9/7/93 at 5:10 am”; type=After}
From these annotations, we can gather that after an Occurrence (“Cardiovascular stable”), a Problem (“significant hypertension”) emerged around a Time (“9/7/93 at 5:10 a.m.”), after which a Treatment (“albumin”) was administered. To detect Events, Timex3s, and Tlinks automatically, we have designed a large set of features to train classifiers to detect such expressions in clinical records.
Features
Supervised machine learning methods are composed of three primary elements: (1) input features to represent the data; (2) a model to act as the classification function; and (3) a learning process to estimate the parameters of the model. While all three of these elements are important, we have found the proper selection of features to be especially critical for NLP tasks. Importantly, parameter estimation techniques tend to perform poorly when given highly redundant or noisy features. As our goal is to experiment with as many features as possible, we utilize a technique to select the best subset of features automatically. We first give a brief overview of the feature types used in feature selection. We then describe how, from all these potential features, the highest performing subset is automatically selected for each sub-task.
Feature types
In this article, we only provide a high-level overview of the types of features we use due to space limitations. However, we have included supplementary materials (available online only) that give detailed feature descriptions, as well as examples of the feature values on actual data from this task. The list of feature types below is largely organized by the resource that best exemplifies their purpose. Many of the discussed feature types actually correspond to dozens of specific feature types. Additional feature types were considered but discarded. The feature types are:
Common NLP features
These features (eg, bag-of-words, previous token, previous Event type) are simple, largely self-explanatory features common in the NLP literature. They are primarily lexical in nature or rely on specific attributes of the 2012 shared task. Sections ‘event recognition’, ‘temporal expression recognition’ and ‘temporal link recognition’ explain any such feature when necessary.
GENIA features
The GENIA tagger22 is a biomedical text tagger. We use it for part-of-speech tagging, lemmatization, and phrase chunking.
UMLS features
These rely on a lexicon built from the unified medical language system (UMLS) metathesaurus.23 The lexicon contains 4.6 million terms.
Third-party TimeML features
The 2012 i2b2 shared task largely follows the TimeML temporal annotation guidelines. Thus, third-party TimeML systems can provide a useful source of automatic annotations. The four third-party systems we incorporate are: TARSQI,5 HeidelTime,24 SUTime,25 and TERNIP.26
i2b2 concept features
Previous i2b2 tasks evaluated concept extraction, which overlaps significantly with event extraction. We use a pre-existing system27 trained on pre-existing i2b2 data as a source of features.
Quantitative pattern features
These are based on regular expressions that recognize nine different types of entities, largely quantitative in nature: age, date, ICD-9 disease ID, dosage, list element, measurement, (de-identified) name, percent, and time.
Statistical word features
These features use pointwise mutual information to determine automatically the most relevant words for each output class in the data. This acts as a filter to improve the classifier’'s ability to handle bag-of-words style features.
Brown cluster features
Brown clustering28 is an unsupervised technique that has been shown to be effective when used as features in an otherwise supervised setting.29 Brown clustering groups words by their common contexts, based on the distributional hypothesis that similar words occur in similar contexts. We created Brown clusters from 10 different corpora, including the i2b2 data itself, other medical texts, newswire, and Wikipedia.
Relation features
The discourse-level Tlinks are unique within the 2012 i2b2 shared task, as they involve classifying pairs of objects (Events and Timex3s). The relation features leverage this by providing information about how the two arguments relate, what is textually between them, and characteristics of one or both arguments.
In total, close to 300 features were available to the feature selector for each of the sub-tasks. The supplementary materials (available online only) describe the candidate feature types in more detail and provide example feature values of sentences from the i2b2 data.
Feature selection
When all possible features are incorporated into a model, the result is a model that is often too big to fit into memory, too slow to train on a single processor, and—most importantly—performs worse than a model with a few well-chosen features. This problem is traditionally called the curse of dimensionality, but it can typically be described by more practical causes. First, many features are noisy, incomplete, or not well suited for a particular task. Second, features are commonly redundant, expressing only minor differences in the data. Natural language tasks are complex, requiring many different types of information, and encoding this information into features that are both noise free and non-redundant is virtually impossible. The task of feature engineering, therefore, is to determine experimentally the best subset of features. Not only does manual experimentation not scale to hundreds of features, but it commonly results in one-size-fits-all solutions in which multiple classifiers use the same feature set even though those features may be suboptimal for a given task.
In this article, we aim to overcome this feature engineering bottleneck by utilizing automated feature selection for the extraction of temporal information in clinical texts. This flexible framework allows our system to determine automatically the best set of features for each sub-task given (a) a large collection of features, (b) a classifier, (c) a set of labeled data, and (d) a feature selection strategy. The features, as described above, are considered at the type level (eg, previous token) instead of the specific instantiation level (eg, previous token is the). Otherwise, instead of selecting from among hundreds of features, it would be selecting among tens of millions. The classifier is largely a ‘black box’ function: given data and features, return a score. The score is obtained through a fivefold cross-validation on the training data. For sequence classification (eg, Event boundary detection), each fold is evaluated using the F1-measure. For multi-class classification (eg, Timex3 type classification), each fold is evaluated using accuracy. Instead of taking the arithmetic mean of the scores for each fold, as is typical, we use the harmonic mean, which is less susceptible to changes in a single fold, favoring moderate increases in all folds over larger increases in one or two. This results in a more conservative method for accepting new features. Given the ability to test the utility of a single feature set, the feature selection strategy dictates how features will be experimentally chosen.
The feature selection strategy we employ is known as floating forward feature selection,30 sometimes referred to as greedy forward/backward. This greedy algorithm starts with an empty feature set and iterates until it fails to find new features that improve the cross-validation score. Let F be the best known feature set at the current stage of the algorithm. At the start of each iteration (the forward step), for every unused feature g not in F, the feature set F ∪ g is given to the classifier for testing. Let g* be the feature g that corresponds to the best performing feature set. If the score for F ∪ g* is greater than the score for F and some small margin ɛ (we use ɛ = 0.0001), then g* is added to F. If not, the algorithm has failed to find any beneficial features and terminates. If a new feature was found, then (in the backward step), for every currently selected feature f in F, f ≠ g*, the feature set F–{f} is given to the classifier for testing. If some reduced feature set obtains a better score than F, the feature(s) corresponding to the reduced feature set is greedily removed.
In short, the algorithm iteratively adds the best unused feature, and after each feature is added the detrimental features are pruned. Intuitively, the backward step is necessary because, as features are added, redundancies are found. While a feature might have improved the classification performance during the second iteration, that may no longer be true during the ninth iteration. Furthermore, it is common for the first selected feature to be a good predictor, yet have a high level of noise. As more and more features are added, they act as a better combined predictor than the noisy feature, and so it may be pruned. In a manual feature engineering process, such pruning is rarely performed, resulting in diminished classification performance.
The result of this technique is not only a near optimal subset of features, but one that was achieved without manual intervention. This method is largely agnostic to both the classifier (we use both SVMs and CRFs) and data (we report results on eight separate subsets). All the features reported for Event and Timex3 recognition, as well as the section time Tlinks, were chosen by this feature selection technique. Unfortunately, there was insufficient time to incorporate discourse Tlink recognition. While it uses many of the same features, its classification method was based on a separate platform and could not be reliably integrated before the submission deadline. A post-hoc evaluation of discourse Tlinks was conducted, showing small improvements. This evaluation is discussed further in the Results section.
Event recognition
Figure 1 illustrates the architecture of our Event recognition system. First, we identify Event boundaries with a CRF classifier. Then we detect type, modality, and polarity using separate SVM classifiers. In particular, we use the Mallet31 CRF implementation and LibLinear32 SVM implementation. The chosen features for all Event classifiers are shown in table 1.
Figure 1.
Event Recognition Architecture.
Table 1.
Event features
| Boundary features | Type features |
|---|---|
| (E1.1) GENIA previous token lemma | (E2.1) Event text (uncased) |
| (E1.2) GENIA next token part-of-speech | (E2.2) Event last token (uncased) |
| (E1.3) GENIA next token phrase chunk IOB | (E2.3) Previous token |
| (E1.4) GENIA 1-token lemma context | (E2.4) Previous bigram (uncased) |
| (E1.5) UMLS category IOB | (E2.5) Contains punctuation |
| (E1.6) Brown cluster (TREC-1000) | (E2.6) Previous Event type |
| (E1.7) Brown cluster (i2b2–100) | (E2.7) i2b2 Concept type (exact) |
| (E1.8) Brown cluster prefix (XIN-100, 2) | (E2.8) i2b2 Concept type (overlap) |
| (E1.9) Brown cluster prefix (CNA-1000, 6) | (E2.9) TARSQI event polarity |
| (E2.10) UMLS category prefix (5) | |
| (E2.11) Brown cluster prefix (TREC-100, 4) | |
| (E2.12) Brown cluster (TREC-1000) | |
| (E2.13) Brown cluster (Pubmed-100) | |
| (E2.14) Unigram PMI>0 | |
| (E2.15) Sentence Unigram PMI strongest type (sum) | |
| Polarity features | Modality features |
| (E3.1) Event unigrams | (E4.1) Previous token lemma |
| (E3.2) Previous Event polarity | (E4.2) Previous previous token lemma |
| (E3.3) Indexed previous token lemmas | (E4.3) Previous Event modality |
PMI, pointwise mutual information.
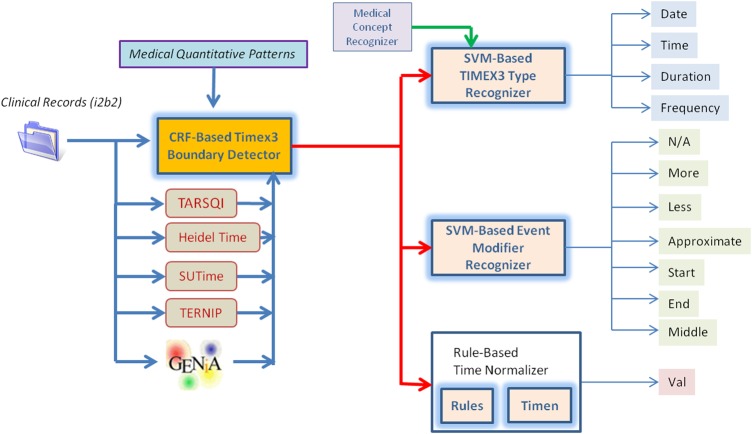
Figure 2.
Timex3 Recognition Architecture.
For boundary detection, the feature selector primarily chose features based on GENIA and Brown clusters. GENIA lemmas, parts of speech, and phrase chunks were all found to be important Event indicators. Four different Brown cluster features were chosen from four different corpora. The two medical corpora are likely to be useful for medical words, while the two Gigaword sub-corpora cluster more general words.
For type classification, the feature selector chose a diverse set of feature types, indicating the wide variety of ways in which Event types may be discovered. The lexical features (E2.1 and E2.2) capture the common Events from the training data (eg, noted, administered), while features E2.3 and E2.4 suggest the importance of an Event's context. Feature E2.6, the previous Event's type, was chosen because often Events of the same type occur together, such as in a list. While the UMLS feature (E2.10) is clearly useful, as UMLS is organized into a taxonomy compatible with the event types, it is interesting that several Brown cluster features (E2.11–13) were also chosen. These features indicate that the Brown clusters are able to differentiate between event types, justifying the distributional hypothesis.
For polarity and modality classification, the feature selector chose much smaller sets of features. Both include a feature indicating the previous Event's classification. Note again that these features were chosen automatically. Although it may seem logical, the decision to use the previous event's type/polarity/modality for each of the respective sub-tasks is based entirely on the data (via feature selection) without any manual intervention. For instance, if the previous event's polarity had been useful for modality classification, it would have been included by the feature selector. Thus this automatic method still produces a logical feature set for each task.
Temporal expression recognition
The architecture of our temporal expression recognition method is shown in figure 2. Similar to Event annotation, a CRF classifier recognizes Timex3 boundaries and two SVM classifiers determine the type and modifier. A rule-based method then normalizes the Timex3. The chosen features for all Timex3 classifiers are shown in table 2.
Table 2.
Timex3 features
| Boundary features | Type features |
|---|---|
| (T1.1) Current token (numbers replaced with ‘0’) | (T2.1) Timex3 unigrams (uncased) |
| (T1.2) Next token | (T2.2) Timex3 text (uncased) |
| (T1.3) 6-character token prefix | (T2.3) Previous token |
| (T1.4) 4-character token suffix | (T2.4) GENIA coarse part-of-speech |
| (T1.5) Quantitative pattern IOB | |
| (T1.6) GENIA next token lemma | |
| (T1.7) GENIA previous token lemma | Mod features |
| (T1.8) GENIA 1-token part-of-speech context | (T3.1) Timex3 unigrams (uncased) |
| (T1.9) GENIA 4-token lemma context | (T3.2) Previous token (uncased) |
| (T1.10) GENIA 4-token phrase chunk context | (T3.3) Contains uppercase |
| (T1.11) GENIA 6-token phrase chunk context | (T3.4) GENIA coarse part-of-speech |
| (T1.12) Third-party Timex3 IOB (all systems) | (T3.5) GENIA part-of-speech trigrams (replace verbs) |
| (T1.13) Third-party Timex3 IOB (HeidelTime) | |
| (T1.14) Third-party Timex3 IOB (TERNIP) |
For boundary detection, the feature selector chose a combination of lexical and syntactic features, along with utilizing third-party Timex3 systems. The basic text feature (T1.1) generalizes the text by replacing all non-zero digits with a zero. For instance, both 10:05 am and 11:14 am become 00:00 am. The prefix (T1.3) and suffix (T1.4) features provide a functionality similar to lemmatization. The quantitative patterns (T1.5) contain regular expressions for times and dates, so it is not surprising they prove useful for detecting Timex3s. Unlike Events, the third-party methods were quite useful for detecting Timex3s. HeidelTime (T1.13) and TERNIP (T1.14) in particular were chosen in addition to the combination of all systems (T1.12).
For type and modifier classification, the feature selector largely chose lexical features, including a bag-of-words (T2.1, T3.1), a complete Timex3 span (T2.2), and a previous token (T2.3, T3.2) feature. Lexical cues are good indicators of the type of Timex3, words like am indicates Time, and daily indicates Frequency. Beyond the lexical features, the coarse-grained part-of-speech (T2.4, T3.4) feature was chosen to generalize the Timex3's syntactic category. Feature T3.3 indicates whether the Timex3 contains any uppercase characters. The final feature (T3.5) is a part-of-speech trigram replace feature, returning the parts of speech of all three-word sequences within the Timex3, but replacing verbal parts of speech with the verb itself.
Our Timex3 normalization (val) method is rule based, combining custom-built rules with TIMEN,33 an open-source temporal normalizer. In combination with TIMEN, we use two types of rules: (1) pre-TIMEN rules that fully normalize temporal expressions, and (2) post-TIMEN rules that correct common TIMEN mistakes or annotation differences between TimeML and the i2b2 annotations. Many of these rules require other Timex3s to be normalized already, so the procedure is performed in a loop: if a needed normalization is unavailable, the Timex3 is skipped and normalization is attempted at the next iteration.
The pre-TIMEN rules are generally either simple regular expressions, or knowledge-intensive procedures. The regular expressions parse Timex3s such as twice daily or 12-14-2005. The knowledge-intensive procedures attempt to capture discourse and document-level knowledge and normalize time expressions that refer to important events such as the admission date, discharge date, or date of a major operation (eg, postoperative day #5). For this last example, our method will search the document for types of operations (eg, surgeries, transplants) and determine the relevant operation. The number of postoperative days is added to the relevant operation's date to obtain the Timex3's val. Similar knowledge-intensive rules are used to normalize the relative hospital day and the admission/discharge dates.
The post-TIMEN rules largely target differences between how TIMEN normalizes dates and how the i2b2 annotations were made. One illustrative example is the word several, such as in ‘several months’, in which TIMEN leaves a placeholder variable (PXM), but the i2b2 defines several as 3 (P3M).
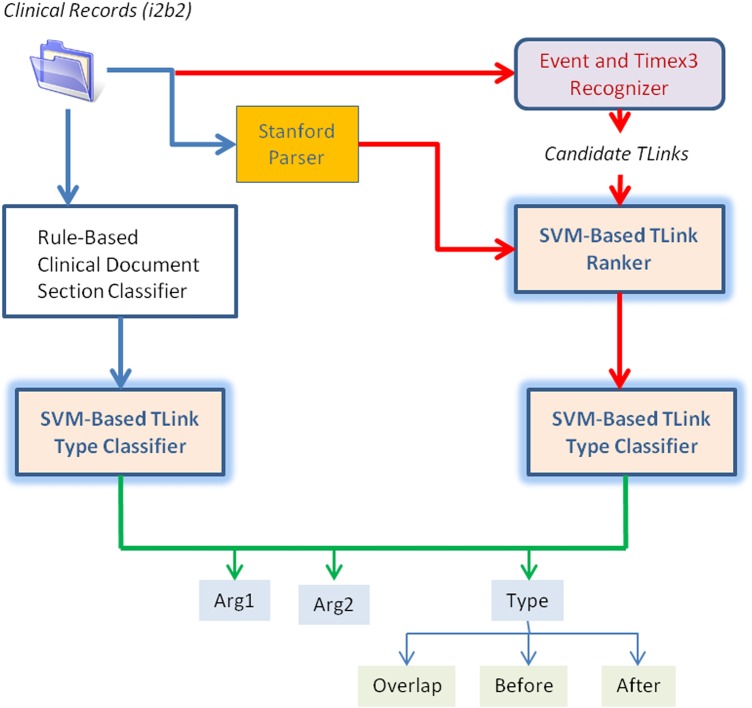
Temporal link recognition
The architecture of our temporal link recognition method is shown in figure 3. We separate the detection of Tlinks between Events and Timex3s in the discourse (document text) from the detection of section time Tlinks. For discourse Tlinks, we use a SVM-ranker to detect links between Events and Timex3s (for simplicity, we refer to them collectively as ets), and a multi-class SVM classifier to determine the Tlink type. For section time Tlinks, we use a rule to connect each Event to either the admission or discharge date, then employ a multi-class SVM classifier to determine the Tlink type. All Tlink features are shown in table 3.
Figure 3.
Tlink Recognition Architecture.
Table 3.
Tlink features
| Discourse detection features | Section time features |
|---|---|
| (DL.1) Has intermediate event | (SL.1) et unigrams |
| (DL.2) Same sentence | (SL.2) et sentence unigrams |
| (DL.3) Dependency path between arguments | (SL.3) GENIA previous token part of speech (SL.4) GENIA part of speech (replace verbs) |
| (DL.4) First argument text | |
| (DL.5) Second argument text | |
| (DL.6) First argument sentence unigrams | |
| (DL.7) Second argument sentence unigrams | |
| (DL.8) Argument order | |
| (DL.9) et Type sequence | |
| (DL.10) et Class sequence |
For discourse Tlinks, we first generate candidate Tlinks for an et by considering all ets in the current and previous sentence, ignoring ets in the current sentence that occur after the given et. We then employ an SVM-ranker by scoring all candidate Tlinks, ranking them by the confidence score. The SVM-ranker is trained using the same process for generating candidate Tlinks. Our ranker allows for tuning how many Tlinks to consider: by increasing the number of accepted Tlinks, recall is increased at the cost of precision. During experiments on the training set, it was found that using only the top-ranked Tlink maximized F1-measure. A multi-class SVM classifier then chooses the Tlink type (Before, After, Overlap) using the same feature set. Features DL.1 and DL.2 are proximity features, as most Tlinks are links between nearby ets. Feature DL.3 uses the dependency parse34 to determine syntactic relationships between ets (eg, direct object, prepositional object). Feature DL.9 uses each et's type (eg, Treatment-Date), while feature DL.10 uses the class of the et (eg, Event-Timex3), allowing the classifier to learn argument type/class distributions and which types/classes are compatible in a Tlink.
For section time Tlink recognition, we first apply a rule-based classifier to determine whether an Event should be related to the admission or discharge date. This looks at the name of the section containing the Event, mapping sections related to patient history to the admission date, and sections related to the hospital course to the discharge date. For section time Tlink type classification, the feature selector chose lexical and part-of-speech features. Features SL.1 and SL.2 are lexical features for the Event and its sentence, respectively. Feature SL.3 largely captures tense information for the verb immediately before the Event (eg, ‘experiencing [pain]’ might indicate that pain Overlaps the admission date). Finally, feature SL.4 is similar to feature T3.5 with the exception that the entire Event span is used instead of trigrams.
Results
The official results are shown in table 4. Our submission ranked third (of 14) in Event detection, sixth (of 14) in Timex3 detection, eighth (of 12) in Tlink detection with gold input, and third (of 7) on end-to-end Tlink detection. Event and Timex3 boundaries were evaluated with precision, recall, and F1-measure using exact, overlapping, and partial credit schemes. Event type, modality, and polarity were judged on their accuracy using the gold Events and Timex3s found via the overlap scheme. Tlinks were judged on precision, recall, and F1-measure using two separate schemes. First, the links in the system output are directly compared against those in the gold output (‘original’). Second, the links are compared using a graph-based technique that incorporates a temporal closure (‘tempeval’), meaning that equivalent relations (eg, A before B, B after A) are considered equal.35 The primary metric for measuring Events was the type score. The primary metric for measuring Timex3s was the val score (the temporal normalization). The primary metric for measuring Tlinks is the tempeval score.
Table 4.
Submission results for 2012 i2b2 shared task
| Event results | Timex3 results | ||||||
|---|---|---|---|---|---|---|---|
| Boundary | Exact | Partial | Overlap | Boundary | Exact | Partial | Overlap |
| P | 0.7984 | 0.8543 | 0.9102 | P | 0.8309 | 0.8911 | 0.9513 |
| R | 0.7692 | 0.8231 | 0.8770 | R | 0.7319 | 0.7846 | 0.8374 |
| F1 | 0.7835 | 0.8384 | 0.8933 | F1 | 0.7783 | 0.8345 | 0.8907 |
| Accuracy | Accuracy | ||
|---|---|---|---|
| Type | 0.8045 | Type | 0.7813 |
| Polarity | 0.8507 | Val | 0.6154 |
| Modality | 0.8417 | Mod | 0.7879 |
| Tlink results (gold Event and Timex3s) | Tlink results (end-to-end) | ||||
|---|---|---|---|---|---|
| Original | Tempeval | Original | Tempeval | ||
| P | 0.5292 | 0.4846 | P | 0.5174 | 0.4848 |
| R | 0.5400 | 0.6614 | R | 0.5133 | 0.5745 |
| F1 | 0.5346 | 0.5594 | F1 | 0.5154 | 0.5258 |
| Comparison to 2012 i2b2 submissions | ||||||
|---|---|---|---|---|---|---|
| F1-measure | Rank | Teams | Mean | Median | Max | |
| Event | 0.8933 | 3 | 14 | 0.7915 | 0.8569 | 0.9166 |
| Timex3 | 0.5481 | 6 | 14 | 0.4732 | 0.5292 | 0.6564 |
| Tlink | 0.5594 | 8 | 12 | 0.5535 | 0.5759 | 0.6932 |
| End-to-end | 0.5258 | 3 | 7 | 0.4958 | 0.5126 | 0.6278 |
The primary metric for each task is the Event type, the Timex3 val, and the TLink tempeval score.
The single greatest point of loss for Events is boundary detection. Our analysis shows that our approach frequently overlaps the gold Events, but leaves off or adds an extra modifier, hence the much higher overlap score. Many errors resulted from annotation inconsistency, but we feel there is still room for improvement through more advanced sequence classification. For instance, semi-Markov CRFs36 allow for the encoding of phrase-level features during token-level sequence classification. For Timex3, the most difficult task is normalization (val), as many temporal expressions require a significant amount of discourse knowledge to be normalized. One potential solution is the creation of new NLP tasks specifically targeted at recognizing the critical elements of a clinical note, such as the primary diagnosis and treatment (eg, a surgery is far more likely to be the primary treatment than a pain killer). Systems trained on the data annotated for such a task can be used by the normalizer as a source of discourse-level knowledge. For instance, to normalize ‘post-operative day 5’, one needs to identify the date of the actual operation. While this could theoretically be done with co-reference, discourse-level knowledge (such as the patient's primary diagnosis and treatment) are commonly needed clinical facts, and could probably be performed as document-level tasks. Finally, our Tlink classification has a significant amount of difficulty due to the depth of semantic and pragmatic understanding it requires. A Tlink may be the result of an explicit expression of a temporal relation in text, it may be based entirely on clinical knowledge of how concepts relate, or it could be based on an understanding of the overall discourse. This could also be aided by greater clinical knowledge extraction, as the annotators commonly linked Events to a primary treatment or diagnosis for temporal comparison. Based on post-hoc analysis of other Tlink methods, it seems clear that architectural choices are very important for this task. For instance, it would be useful to separate single-sentence Tlink classification from inter-sentence classification, or use modules that are intended for specific types of Tlinks based on medical knowledge.
We performed a post-hoc evaluation of feature selection for discourse Tlinks, as we were unable to integrate automated feature selection into the task before the submission. The automatically chosen feature set includes DL.1, DL.2, DL.4, DL.5, DL.6, and DL.9 from table 3. In addition, two more features were chosen: the words between the arguments (if they are within five tokens), and whether or not either argument is the first et in the sentence. This second feature is useful as annotators were more likely to include the first Event/Timex3 in a sentence as part of a Tlink for both intra and inter-sentence relations. This slightly smaller feature set results in a minimal gain of half a point (0.005) in F1-measure, which would have improved our Tlink rank from eighth to seventh. Such a small gain further indicates that the Tlink task requires important architectural considerations, while the Event and Timex3 tasks can achieve good results with straightforward machine learning classifiers combined with feature selection.
To evaluate further the contribution of various types of features, we have performed a simple feature test on our Event boundary classifier (there is insufficient space for a detailed test of all nine classifiers). We place the features for Event boundary recognition into one of three categories: (1) GENIA (four features); (2) UMLS (one feature); and (3) Brown Clusters (four features). We also include a test of all candidate (almost 300) features. The results for these experiments are shown in table 5. These experiments show that GENIA features perform best, followed by Brown Clusters. Curiously, the UMLS feature performs poorly and significantly hurts the performance of the GENIA features, but improves the performance when all three feature sets are considered by the classifier. We believe the most likely cause of this is that the UMLS lexicon is noisy: it contains many terms that correspond to gold Events, but it is also missing many types of events (such as evidential and Occurrence). A system with only the GENIA features is insufficient to handle this noise, but a system with all three sets of features is sufficiently robust to incorporate the UMLS feature. It is thus less likely that the UMLS feature would have been added to the feature set using manual feature selection, as there are cases in which it hurts the overall score. Instead, our flexible feature selection framework allows for it to be automatically incorporated when judged beneficial in the context of other features. By comparison, using nearly 300 features (instead of the nine selected) performs worse by 1.3 points of F1-measure, despite requiring significantly more memory and taking almost 17 times as long to evaluate (120 min vs 7 min on a 3 GHz server).
Table 5.
Feature experiments for Event boundary recognition
| Features | P | R | F |
|---|---|---|---|
| GENIA only | 0.7547 | 0.6849 | 0.7181 |
| UMLS only | 0.3708 | 0.1422 | 0.2055 |
| Brown clusters only | 0.7268 | 0.6741 | 0.6995 |
| GENIA+UMLS | 0.6262 | 0.5055 | 0.5594 |
| GENIA+Brown clusters | 0.7932 | 0.7657 | 0.7792 |
| UMLS+Brown clusters | 0.7390 | 0.6923 | 0.7148 |
| All selected features | 0.7984 | 0.7692 | 0.7835 |
| All candidate features | 0.7929 | 0.7497 | 0.7706 |
Conclusion
We have presented our approach for the 2012 i2b2 shared task. We combine a variety of methods, including supervised learning (with CRFs and SVMs), unsupervised learning (based on clustering), and rule-based methods (centered on knowledge acquisition). Our methods achieved excellent results in the recognition of medical events and temporal expressions, and they performed well at normalizing temporal expressions and detecting temporal relations. For future work, we would like to incorporate additional rules to handle less frequent temporal expressions, especially those unique to clinical documents, more advanced sequence learning methods to improve on CRFs.
Footnotes
Collaborators: KR acted as the primary designer and implementer of the research, and drafted the initial manuscript. BR contributed to the design, implementation, and analysis, as well as assisting in the drafting of the manuscript. SMH contributed to the design and analysis, as well as assisting in the drafting of the manuscript.
Funding: The authors received no specific grant from any funding agency in the public, commercial or not-for-profit sectors. The 2012 i2b2 shared task was supported by Informatics for Integrating Biology and the Bedside (i2b2) award number 2U54LM008748 from the NIH/National Library of Medicine (NLM), by the National Heart, Lung and Blood Institute (NHLBI), and by award number 1R13LM01141101 from the NIH NLM. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NLM, NHLBI, or the National Institutes of Health.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: All data used are published, publicly accessible, or part of the i2b2 challenge data (which will become publicly available in 1 year).
References
- 1.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 challenge overview. J Am Med Inform Assoc 2013;20:806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Verhagen M, Gaizauskas R, Schilder F, et al. The TempEval challenge: identifying temporal relations in text. Language Resour Eval 2009;43:161–79 [Google Scholar]
- 3.Verhagen M, Sauíi R, Caselli T. James pustejovsky. SemEval-2010 task 13: TempEval-2. Proceedings of the 5th International Workshop on Semantic Evaluation; 2010:57–62 [Google Scholar]
- 4.UzZaman N, Llorens H, Allen JF, et al. TempEval-3: Evaluating Events, Time Expressions, and Temporal Relations. Ann Arbor, Michigan, USA: Association for Computational Linguistics. arXiv.1206.5333v1, 2012
- 5.Verhagen M, Mani I, Saurí R, et al. Automating temporal annotation with TARSQI. Proceedings of the 43rd Annual Meeting of the Association for Computational Linguistics; Demo Session, 2005 [Google Scholar]
- 6.Bethard S, Martin JH. CU-TMP: Temporal relation classification using syntactic and semantic Features. Proceedings of the 4th International Workshop on Semantic Evaluation (SemEval); 2007:129–32 [Google Scholar]
- 7.Badulescu A, Srikanth M. LCC-SRN: LCC's SRN system for semEval 2007 task 4. Proceedings of the 4th International Workshop on Semantic Evaluation (SemEval); 2007:215–18 [Google Scholar]
- 8.Cheng Y, Asahara M, Matsumoto Y. NAIST.Japan: temporal relation identification using dependency parsed tree. Proceedings of the 4th International Workshop on Semantic Evaluation (SemEval); 2007:245–8 [Google Scholar]
- 9.Hepple M, Setzer A, Gaizauskas R. USFD: preliminary exploration of features and classifiers for the tempEval-2007 task. Proceedings of the 4th International Workshop on Semantic Evaluation (SemEval); 2007:438–41 [Google Scholar]
- 10.Grover C, Tobin R, Alex B, et al. Edinburgh-LTG: tempEval-2 system description. Proceedings of the 5th International Workshop on Semantic Evaluation (SemEval); 2010:333–6 [Google Scholar]
- 11.Strotgen J, Gertz M. HeidelTime: High quality rule-based extraction and normalization of temporal expressions. 2010:321–4
- 12.Kolomiyets O M-F Moens. KUL: recognition and normalization of temporal expressions. Proceedings of the 5th International Workshop on Semantic Evaluation (SemEval), Association for Computational Linguistics, Uppsala, Sweden, 2010:325–8 [Google Scholar]
- 13.Boro ES. ID 392:TERSEO + T2T3 transducer. A systems for recognizing and normalizing TIMEX3. Proceedings of the 5th International Workshop on Semantic Evaluation; 2010:317–20 [Google Scholar]
- 14.Llorens H, Saquete E, Navarro B. TIPSem (English and Spanish): evaluating CRFs and semantic roles in TempEval-2. Proceedings of the 5th International Workshop on Semantic Evaluation (SemEval); 2010:284–91 [Google Scholar]
- 15.UzZaman N, Allen J. TRIPS and tRIOS system for TempEval-2: extracting temporal information from text. Proceedings of the 5th International Workshop on Semantic Evaluation (SemEval); 2010:276–83 [Google Scholar]
- 16.Derczynski L, Gaizauskas R. USFD2: annotating temporal expresions and TLINKs for TempEval-2. Proceedings of the 5th International Workshop on Semantic Evaluation (SemEval); 2010:337–40 [Google Scholar]
- 17.Vicente-Díez MT, Moreno-Schneider J, Mart´inez P. UC3M system: determining the extent, type and value of time expressions in TempEval-2. Proceedings of the 5th International Workshop on Semantic Evaluation (SemEval); 2010:329–32 [Google Scholar]
- 18.Uzuner Ö, Imre S, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uzuner Ö, South B, Shen S, et al. uVall. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uzuner Ö, Bodnari A, Shen S, et al. Evaluating the state of the art in coreference resolution for electronic medical records. J Am Med Inform Assoc 2011; 18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pustejovsky J, Castano J, Ingria R, et al. TimeML: Robust specification of event and temporal expressions in text. IWCS-5 Fifth International Workshop on Computational Semantics; 2003 [Google Scholar]
- 22.Tsuruoka Y, Tateishi Y, Kim J-D, et al. Developing a robust part-of-speech tagger for biomedical text. Proceedings of the 10th Panhellenic Conference on Informatics; 2005:382–92 [Google Scholar]
- 23.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods of Information in Medicine. 199332:281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strotgen J, Gertz M. Multilingual and cross-domain temporal tagging. Lang Resour Eval 2012. http://link.springer.com/article/10.1007/s10579-012-9179-y/fulltext.html [Google Scholar]
- 25.A Chang, C X Manning D. SUTime: A library for recognizing and normalizing time expressions. Proceedings of the Eighth International Conference on Language Resources and Evaluation; 2012 [Google Scholar]
- 26.Christopher Northwood TERNIP: temporal expression recognition and normalisation in Python [PhD thesis]. University of Sheffield, Sheffield, UK, 2010 [Google Scholar]
- 27.Roberts K, Sanda M. A flexible framework for deriving assertions from electronic medical records. J Am Med Inform Assoc 2011;18:568–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown PF, deSouza PV, Mercer RL, et al. Class-based n-gram models of natural language. Comput Linguist 1992;18:467–79 [Google Scholar]
- 29.Turian J, Ratinov L, Bengio Y. Word representations: a simple and general method for semi-supervised learning. Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics; 2010:384–94 [Google Scholar]
- 30.Pudil P, Novovičova J, Kittler J. Floating search methods in feature selection. Pattern Recogn Lett 1994;15:1119–25 [Google Scholar]
- 31.McCallum AK. 2002. MALLET: A Machine Learning for Language Toolkit. http://mallet.cs.umass.edu. [Google Scholar]
- 32.Fan R-E, Chang K-W, Hsieh C-J, et al. LIBLINEAR: a library for large linear classification. J Mach Learning Res 2008;9:1871–4 [Google Scholar]
- 33.Llorens H, Derczynski L, Gaizauskas R, et al. TIMEN: an open temporal expression normalisation resource. Proceedings of the Eighth International Conference on Language Resources and Evaluation; 2012 [Google Scholar]
- 34.de Marneffe M-C, MacCartney B, Manning C, et al. Generating typed dependency parses from phrase structure parses. Proceedings of the Fifth International Conference on Language Resources and Evaluation; 2006 [Google Scholar]
- 35.Naushad N, Allen JF. Temporal evaluation. Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics (Short Papers); 2011 [Google Scholar]
- 36.Sarawagi S, Cohen WW. Semi-Markov conditional random fields for information extraction. Proceedings of the 18th Annual Conference on Neural Information Processing Systems; 2004:1185–92 [Google Scholar]