Abstract
Background
Temporal information detection systems have been developed by the Mayo Clinic for the 2012 i2b2 Natural Language Processing Challenge.
Objective
To construct automated systems for EVENT/TIMEX3 extraction and temporal link (TLINK) identification from clinical text.
Materials and methods
The i2b2 organizers provided 190 annotated discharge summaries as the training set and 120 discharge summaries as the test set. Our Event system used a conditional random field classifier with a variety of features including lexical information, natural language elements, and medical ontology. The TIMEX3 system employed a rule-based method using regular expression pattern match and systematic reasoning to determine normalized values. The TLINK system employed both rule-based reasoning and machine learning. All three systems were built in an Apache Unstructured Information Management Architecture framework.
Results
Our TIMEX3 system performed the best (F-measure of 0.900, value accuracy 0.731) among the challenge teams. The Event system produced an F-measure of 0.870, and the TLINK system an F-measure of 0.537.
Conclusions
Our TIMEX3 system demonstrated good capability of regular expression rules to extract and normalize time information. Event and TLINK machine learning systems required well-defined feature sets to perform well. We could also leverage expert knowledge as part of the machine learning features to further improve TLINK identification performance.
Keywords:
Introduction
Temporal resolution for events and time expressions in clinical notes is crucial for an accurate summary of patient history, better medical treatment, and further clinical study. Discovery of a temporal relation starts with extracting medical events and time information and aims at building a temporal link (TLINK) between events or between events and time expressions. Clinical practice and research would benefit greatly from temporal expression and relation detection. Therefore, temporal expression and relation discovery in clinical natural language processing (NLP) are timely and inevitable to improve clinical text mining.
This year's i2b2 challenge had three tracks: (1) EVENT/TIMEX3 extraction; (2) End-to-End track—that is, fully automatic EVENT/TIMEX3 extraction and TLINK identification; and (3) TLINK identification. In i2b2, the definitions for EVENT, TIMEX3, and TLINK are as follows:
EVENT is defined as anything that is relevant to the patient's clinical timeline. Events include clinical concepts, such as laboratory tests, medical problems, administered or proposed treatments, diagnoses, patient's complaints, and other concepts. Events also have attributes of type polarity and modality. There are six types of events: test, problem, treatment, clinical_dept, evidential, and occurrence. The first three are categories of clinical concepts (e.g., test: “CT scan”, problem: “headache”, treatment is medications and procedures: “Aspirin” “extubated”). The clinical_dept is to mark the clinical units (e.g., “intensive care unit” “operating room”). The evidential describes information source (e.g., words in “ ” in the followings: CT “shows”, the patient “complained”). The events that do not belong to any of five types above are marked as an occurrence. Polarity refers to whether the event was negated or not. Modality describes whether an event actually occurred or not.
TIMEX3 is phrases that contain time information, specifically date, time, duration, and frequency and has three attributes: type (i.e., date, time, duration, frequency), val (value of the normalized TIMEX3), and mod (i.e., na, approx, more, less, start, end, middle). They follow TIMEX3 tags that are part of the TimeML markup language for temporal and event expression in natural language.1
TLINK is categorized as three types: before, after and overlap. The anchor relation is an Event-Time TLINK and the order relation is an Event-Event TLINK. The i2b2 annotation includes an attribute explicit—i.e., whether TLINK is explicitly determined or not.
The following example shows how to annotate all three categories in the End-to-End track:
“He had chest pain on Feb-13-2012.”
EVENT: chest pain (type=problem)
TIMEX3: Feb-13-2012 (type=date)
TLINK: “chest pain” overlaps with “Feb-13-2012”
We participated in all three tracks. Here, we report our systems and their performance evaluations. All systems were built in Apache Unstructured Information Management Architecture (UIMA) framework (http://incubator.apache.org/uima/) that provides effective management of annotation workflow and easy interface to use existing resources and add new components.
Background
A comprehensive temporal information discovery in clinical text requires medical event extraction, time information, and temporal relation identification. The medical event extraction task was tackled in the 2010 i2b2 clinical NLP challenge.2 This year's event extraction task, however, includes a broader range of medical concepts and all concepts relevant to the patient's clinical timeline.
Our event extraction system is based on our previous system in the i2b2 2010 challenge. It uses a conditional random field (CRF) model with a variety of features, including lexical information, natural language components (ie, part of speech, etc), morphological structure, and medical ontology.3
Temporal expression extraction is the first step to resolve temporal relations for any advanced natural language applications, such as text summarization, machine translation, and question answering. Several systems are available for extracting temporal expression. GUTime developed by Georgetown University is an extension of the TempEx tagger,4 which is a temporal tagger based on Perl regular expression. GUTime is now available as part of the TARSQI toolkit.5 HeidelTime6 is a rule-based system that is built in a UIMA framework and performed best for SemEval-2 (http://semeval2.fbk.eu/).7 SUTime8 is also a rule-based system using regular expression and is implemented as one of the annotators in the Stanford CoreNLP pipeline.
In this challenge, we developed MayoTime that adapts the framework of HeidelTime. In MayoTime, we employed icTAKES (http://sourceforge.net/projects/ohnlp/files/icTAKES/) to use basic NLP components rather than using HeidelTime's default one—DKPro Core (http://code.google.com/p/dkpro-core-asl/). icTAKES is an integrated version of cTAKES9 that allows end users to take full advantage of the original cTAKES as well as use the clinical NLP pipeline in more standard project package structure with a simplified installation process. The major functionalities of icTAKES include basic NLP components (eg, sentence parsing, tokenization, term normalization, part-of-speech (POS) tagging, chunking), negation detection, uncertainty resolution, section tagging, clinical concept recognition, drug side-effect identification,10 patient's smoking status classification,11 etc. MayoTime also used adjusted SecTag (http://code.google.com/p/sectag/) in icTAKES to obtain section information. HeidelTime rules, types, and attribute definitions were updated and expanded in MayoTime to cope with the i2b2 definitions.
Similar to the general domain, the task of finding temporal relations between medical events in a clinical note is about finding whether an event is before, after, or overlaps with another event. This process is usually called TLINK detection. Previous studies12 have explored diverse knowledge sources and linguistic information in inferring TLINK among events, including temporal adverbials, tense, aspects,13–15 rhetorical relations,16 pragmatic conventions,17 and background knowledge.18 Mani et al19 employed machine learning models to detect TLINK. Specifically, they trained a MaxEnt model with features such as event class, aspect, modality, tense, and negation as well as event string (a preposition/adverb) and contextual features.
In this TLINK challenge, we used both rules and a combination of rules and machine learning. Most i2b2 discharge summaries consist of two major sections besides admission and discharge headers such as history of present illness and hospital course. The section information can be a strong clue to the order of the two events. For example, laboratory tests are generally performed before diagnosis. Such knowledge can be encoded as simple rules. However, the rules are not accurate in all situations. A machine learning model, when trained well with a variety of features including rules, may improve the performance over a rule-based system. Hence, we used rule-based features in combination with features described by Mani et al19 to train our TLINK model.
Materials and methods
Data
The training set consists of de-identified 190 discharge summaries from Partners Healthcare and Beth Israel Deaconess Medical Center, and has been manually annotated for Events, TIMEX3s, and TLINKs. TLINKs are annotated in all possible pairs. The test set includes 120 discharge summaries from the same sources. For the EVENT/TIMEX3 and End-to-End tracks, the test data are raw text and for the TLINK track, pre-annotated events and TIMEX3s were provided by the organizers.
EVENT extraction
We used icTAKES, which has combined MedTagger20 21 and cTAKES,22 for extracting features, and trained CRF models using MALLET23 for each of the six event types. Features used to train CRF models include raw tokens, MedLex normalized form,24 Unified Medical Language System (UMLS) preferred concept unique identifier string, UMLS semantic type, synonyms, character classes in token, part of speech, negation, prefix (first three characters), and suffix (last three characters). Our MedTagger pipeline consisted of a series of annotators. First, the tokens were syntactically normalized and matched with the MedLex dictionary, which is derived from the UMLS and Mayo clinical notes, to identify the concepts and their semantic type. The next step was to annotate POS tags and classification of the tokens into character classes based on whether they contained alphabet letters, capitalizations, numbers, or Greek numerals. Finally, MedTagger detected polarity using the YTex NegEx algorithm25 and produced the default modality (FACTUAL) for all events. A window of two tokens to the left and one token to the right was used to aggregate features for each token (figure 1).
Figure 1.
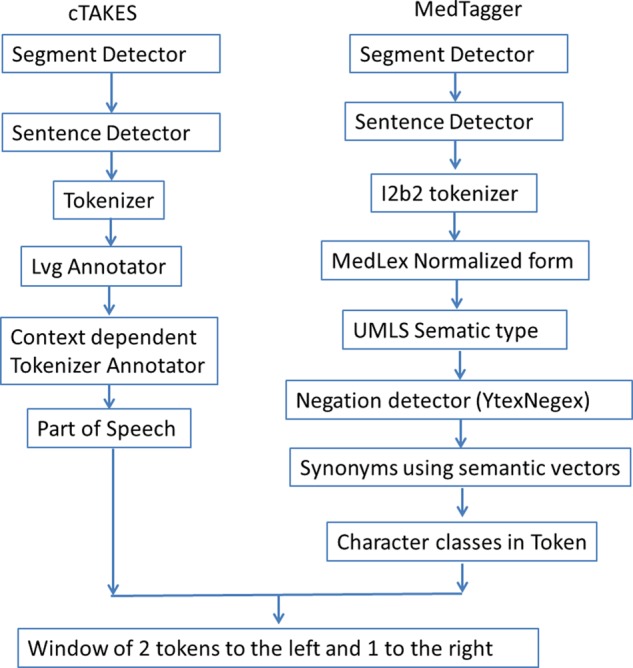
Unstructured Information Management Architecture pipeline for extracting features for event detection. LVG, lexical variant generation; UMLS, Unified Medical Language System.
MALLET was used with a default setting to train the first-order CRF models on the training datasets. We trained separate CRF models for each of the event types. The models for problems, tests, and treatment were trained on a combination of the i2b2 2010 training and test corpora and the i2b2 2012 training corpus. The models for the other events were trained on the i2b2 2012 training corpus (figure 2).
Figure 2.
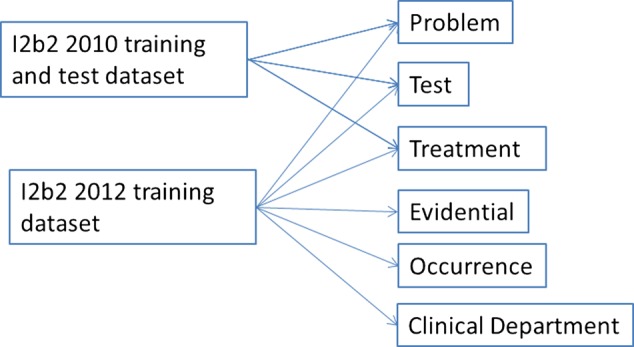
Training sets for conditional random field models for the event types.
TIMEX3 extraction
The i2b2 TIMEX3 has three main attributes (see table 1A for examples): type (date, time, duration, frequency), val (normalized form of TIMEX3), and mod (na, approx, more, less, start, end, middle).
Table 1.
TIMEX3 expression and rules
| Expression | Type | Val | Mod |
|---|---|---|---|
| A. Examples of TIMEX3 expressions (excerpt from the i2b2 annotation guidelines) | |||
| 09/21/2001 day of admission* POD 2* that time* three to four weeks every few days noon 09/17/01 | Date | 2001-09-14 | NA |
| Date | 2005-03-15 | NA | |
| Date | 2005-03-17 | NA | |
| Date | 2005-03-15 | NA | |
| Duration | P3.5W | APPROX | |
| Frequency | RP2D | APPROX | |
| Time | 2001-09-17T12:00 | NA | |
| Type | Rule category | Example | |
|---|---|---|---|
| B. TIMEX3 rule categories and examples | |||
| Date | Day/week/month/year/century | 2005-03-11, Mar 11 2005, Thursday Feb 14, the weekend, March 2005, 2005, 21st century | |
| Independent date | March, Early 2005, the middle of March, the following day, about 1 year ago | ||
| Holidays | Christmas, Easter Sunday | ||
| Relative date | The same day, the day of admission, the operative day, day two of admission, this morning, the night prior | ||
| Exclusion† | march, may, fall | ||
| Duration | Time/day/week/month/year | Less than 60 min, several days, past few weeks, the last three years | |
| Period | A three year period, one-hour course, 3 to 5 minutes | ||
| Constant | Entire hour, overnight, for months, the holiday weekend, the postoperative period | ||
| Exclusion† | About 10 years younger, several days old | ||
| Frequency | Every/each unit | Each day, every Monday, every week, every two days, two days each week | |
| × times | Twice a month, three times a week, 10 times per month | ||
| Specific | On Monday and Wednesday | ||
| Latin abbreviation | p.r.n., bid, q6h, q 6–8h | ||
| Etc | A regular basis many times | ||
| Time | Time stamp | 03/11/2005 15:30, November 24, 2011 1535 GMT | |
| Time point | 3:30 am, 11:30:25 pm, about 9 am Wednesday | ||
| Constant | Midnight, noon | ||
*Referential date—should be referenced to the specific date.
†Exclude even if matched by the other rules.
For TIMEX3 annotation we developed MayoTime that was adapted from publicly available HeidelTime.6 MayoTime finds TIMEX3 through an externalized pattern matching rules based on regular expression, which provides an easy interface for users to customize the system. MayoTime also used adjusted SecTag to obtain section information, which is crucial to infer relative time information—for example, ‘day of admission/discharge’ ‘hospital day’ ‘this/that time’, etc.
The HeidelTime has different types and attribute categories from those of i2b2. In MayoTime, we adjusted them according to the i2b2 definitions as follows: (1) map the type ‘set’ in HeidelTime to ‘frequency’ in i2b2; (2) adjust ‘value’ definitions to be the same as for i2b2 (eg, ‘once a day’: P1D in HeidelTime to RP1D in i2b2); (3) map different ‘mode’ expressions to the i2b2 standard (eg, ‘more than’ in HeidelTime to ‘more’ in i2b2).
Following this process, we derived a set of TIMEX3 patterns and rules accordingly to the i2b2 TIMEX3 definitions. Our basic principles for each type of extraction are described as follows:
Date: It is the most dominant TIMEX3 type in i2b2. There are basically two categories for date description—explicit (eg, 09/29/1993, 2015-11-13, etc.) and relative (eg, ‘the day of admission’ ‘hospital day three’ ‘this time’, etc). Most date information is explicit but there are some relative date expressions. MayoTime catches explicit date expressions directly through rules and regular expressions. The relative dates are set by referring to the anchor date and computing the elapsed date, if necessary. For example:
“The day of admission/discharge”: refers to the admission/discharge date in the beginning of the discharge summaries.
“Postoperative day one”, “hospital day three”, “day of life two”: anchors to the admission date and adds the elapsed number of days. To correctly find the postoperative date the actual operation date should be used. However, in our system we used admission date as an anchor date to simplify the process unless we saw specific evidence (see details at the end of this section).
“The day” “this/that time”: the ‘gold standard’ annotations are not consistent to annotate these cases—that is, some mentions are annotated but some are not. The principle we used is that: (1) if the previous date is in the same section, the system uses it; (2) if the previous date is in a different section, the system uses the admission or discharge date depending on the section information—that is, if a given date is in the “history of present illness” section, the system uses admission date; if a given date is in the “hospital course” section, the system uses discharge date.
We also performed the post-processing to remove frequent false-positive cases, such as incorrect month information ‘fall’, ‘may’, etc.
Frequency: Most frequency expressions come with indication words (eg, ‘each’ ‘every’ ‘x’ ‘times’) or Latin abbreviations (eg, b.i.d., p.r.n., q6h, q day, etc). We compiled a set of indication words. The i2b2 frequency values may consist of the number of repeats and the interval (ie, R[# repeats][duration]). MayoTime first catches the frequency unit and frequency number and then compute the corresponding duration. For example, for ‘twice a month’: MayoTime first finds frequency unit ‘month’ (M) and frequency number ‘2’, next it computes duration by 1/frequency number, which is 1/2= 0.5 in this example, and finally, it generates the i2b2 frequency format RP0.5M.
Duration: This is temporal expressions that specify a period of time. The duration value is represented by P[period][unit]. For example, ‘for 10 days’ is represented as ‘P10D’, ‘for half a year’ is represented as ‘P0.5Y.’ HeidelTime rules were expanded accordingly to the i2b2 representation.
Time: The i2b2 time format is an extension of ISO 8601, such as [hh]:[mm]:[ss]. If time expressions occur with date expression, it over-rides date and follows the format [YYYY]-[MM]-[DD]T[hh]:[mm]:[ss]. MayoTime first anchors a specific date using section information and relative date, if available, and annotates time expressions.
We noticed that the operation date can be inconsistently treated as either postoperation day (POD) 0 or POD 1 in different notes. We used a heuristic to determine how the operation date is recorded in a specific note. For example, if the patient is discharged on POD 7 and we know the discharge date, then it is clear how the operation date is recorded.
There are about 300 rules in MayoTime TIMEX3 extraction. The detailed rules are categorized and described with examples in table 1B.
TLINK identification
The TLINK detection consists of two main tasks. The first is to link events and/or time expressions based on rules and the second task is to assign a TLINK type, either by rules or the combination of rules and machine learning technique.
Rule-based framework
The annotations for TLINK events include relations within a single sentence/clause and beyond. As a preliminary investigation, we designed a rule-based system for extracting TLINK relations and classifying the relation between them as BEFORE, OVERLAP, and AFTER. The rules for extraction and classification of TLINK events are centered on two broad principles.
TLINK extraction within a single sentence
We outlined some of the rules that extract TLINK events in a single sentence, which were designed after manually examining the annotations in the training corpus. These rules operate within clausal/sentence boundaries to achieve high precision. We also show an example sentence that matches each of the rules.
EVENT1 WORD{0,2} (and|as well as|which is|that is|for) WORD{0,2} EVENT2 => OVERLAP (EVENT1, EVENT2)
Eg) “He was also started on intravenous Ticarcillin for possible gram negative pneumonia”
EVENT WORD{0,2}(on|within) D{0,2} TIMEX” => OVERLAP (EVENT, TIMEX)
Eg) “He was diuresed with intravenous Lasix to which he responded with at least a 2L urine output on the first day”
EVENT1 VERB(PAST) EVENT2 => BEFORE (EVENT1, EVENT2)
Eg) “Coronary angiography revealed occlusion of the right coronary artery proximally with insignificant plaquing in the left anterior descending artery and circumflex arteries.”
EVENT1 WORD* after WORD{0,3} EVENT2 => AFTER(EVENT1, EVENT2)
Eg) “It appears from the Discharge summary that the patient was placed on Unasyn 3 grams intravenously every six hours after his ERCP.”
TLINK extraction beyond a single sentence
We also implemented rules to relate and classify events and time expressions, which occur beyond sentence boundaries. These rules are based on either knowledge derived from the inherent structure of the clinical note or on sound linguistic discourse principles such as co-reference. These rules significantly improve the recall of the system. We briefly discuss some of the rules below:
If TIMEX3s occur within hospital course section then they are automatically classified as BEFORE to discharge date.
If TIMEX3s occur within admission section, then they are automatically classified as BEFORE to admission date unless the sentence refers to the admission process (such as presents with pneumonia).
If two events co-refer as determined by the lexical match and semantic match sieves,26 they are considered as OVERLAP.
Between two TIMEX3s, we used the measure of absolute difference between them (when available) in order to designate the TLINK as OVERLAP, BEFORE, or AFTER.
Machine learning framework
In the machine learning framework, we used a supervised model that requires potential TLINK pairs as an input and produces TLINK labels. In the i2b2 definition, TLINK is a time relation (ie, before, after, overlap) between ‘any’ combination of pairs from Event and TIMEX3. This leads to a lot of potential TLINK pairs; however, there exist a large number of negative pairs (ie, pairs without any TLINK) that outnumber TLINK pairs. Thus, it is impractical to train a machine learner on all the potential pairs. Also, the imbalanced class distribution due to the preponderance of negative pairs would degrade the classification performance. To avoid this problem, we used a simple approach to select potential TLINK pairs: (1) pairs from our previous rules (in Rule-based framework); (2) any possible pairs within the same sentence. The union of (1) and (2) will be our potential pairs. These potential pairs were used in both the training and test phases.
We use three sources for feature generation. First, rule-based TLINK labels determined by the rules from rule-based framework section. Second, attributes extracted from the Clinical Narrative Temporal Relation Ontology (CNTRO)27 28—that is, concept definitions about time (eg, temporal instant, temporal interval) and relations between the time concepts (eg, prior to, before, earlier, after, until, subsequent, overlap, during, etc). CNTRO, as domain ontologies, have been used in information technology for providing semantic definitions of a particular domain, which enable automated agents to perform queries intelligently and infer new knowledge. Third, features listed in the following were also found to be discriminative for detecting temporal relations.
Section head: admission, history, hospital course, discharge
LVG: normalized time stamps with lexical variant generator
POS: part of speech of involved events, medical events
Adjacent events: events close to involved events, including their types, POS and event names
Coreferring events: events coreferring with current events
Relative temporal relations: temporal relations found among current events and other events
Context: some adverbs or prepositions before or after involved events
Modality of events: FACTUAL, POSSIBLE, PROPOSED and CONDITIONAL
Polarity of events: NEGATION, POSITIVE, NEUTRAL
Time phrases: time-related words
Tense: PAST, PRESENT, FUTURE, PERFECT, FINISHED
Event type: OCCURRENCE, PROBLEM, EVIDENTIAL, TREATMENT, TEST, CLINICAL_DEPT
TIMEX3 type: DATE, TIME, DURATION, FREQUENCY
Section headings are important clues for TLINK. For example, most events in the history section occur before admission, and events in the hospital course occur after admission and before discharge. Normalized time stamps make temporal expressions more consistent. POS can help distinguish events of different types.
Building a machine learning TLINK detection system
Figure 3 shows the overall architecture of the TLINK detection system. It is composed of the collection reader, analysis engines, and Common Analysis System (CAS) consumers. The collection reader is designed to read data. The aggregate rule generator generates diverse rules and relevant annotations. All previous annotations are saved in UIMA CAS, allowing feature extractions to be accomplished online also. We employed MALLET pipe data structure to pass and deliver feature vectors. We evaluated several machine learning models (such as MaxEnt, NaïveBayes, etc) during the training phase, and decided to use LibSVM as it produced the best results. In LibSVM, C=31 and g=0.0925 were found to be the optimal parameter settings, obtained by running a cross-validation grid search. To reduce the bias leaning towards features with larger numbers, feature values were normalized between −1 and 1.
Figure 3.
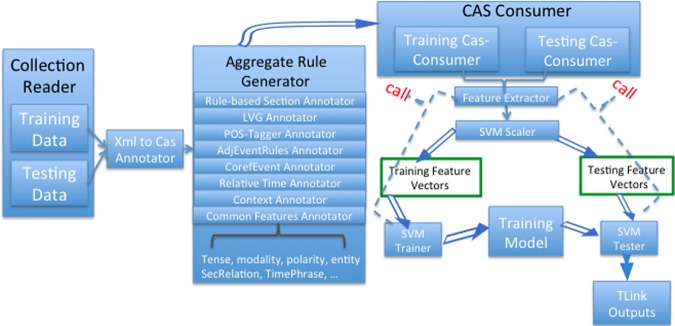
The workflow of Unstructured Information Management Architecture-based TLINK detector. CAS, Common Analysis System; LVG, lexical variant generation; POS, part of speech; SVM, support vector machines.
Results
EVENT
Table 2 summarizes the results of the evaluation on the test dataset. We note that the recall of the system was far lower than the precision. It may be possible to further improve the F-measure by optimizing the recall–precision trade-off.
Table 2.
Event evaluation on the test set
| Span match | Recall | Precision | Average P&R | F-measure |
|---|---|---|---|---|
| A. Annotation evaluation | ||||
| Exact | 0.7256 | 0.8648 | 0.7896 | 0.7891 |
| Overlap | 0.8037 | 0.9482 | 0.8702 | 0.8700 |
| B. Attribute accuracy | Accuracy |
| Type | 0.7677 |
| Polarity | 0.7625 |
| Modality | 0.7689 |
TIMEX3
The most dominant type of TIMEX3 is ‘date.’ In the training set, date covers about 73% of the whole TIMEX3 expression, followed by duration (16%), frequency (8%), and time (3%). We implemented three runs; run 1 used the comprehensive rule sets with exact matching, run 2 used the comprehensive rule sets allowing lower-case pattern matching, and run 3 used compacted rule sets for date allowing lower-case pattern matching—that is, excluded two-digit month and year information (ie, mm/yy, mm-yy), season information (eg, summer, winter), day information (eg, earlier yesterday, Monday), and approximate year information (eg, mid-1990s, early 1990s). Table 3 shows the evaluation performance of all runs on the test set. All three runs produced similar F-measures but run 2 had a slightly higher F-measure wing to higher recall. The accuracy scores for all runs were similar.
Table 3.
TIMEX3 evaluation on the test set
| Evaluation measure | Run 1 | Run 2 | Run 3 |
|---|---|---|---|
| Precision (P) | 0.8794 | 0.8791 | 0.8789 |
| Recall (R) | 0.9214 | 0.9225 | 0.9165 |
| Average P&R | 0.8999 | 0.9002 | 0.8972 |
| F-measure | 0.8999 | 0.9003 | 0.8973 |
| Type | 0.8593 | 0.8604 | 0.8533 |
| Val | 0.7313 | 0.7313 | 0.7302 |
| Modifier | 0.8555 | 0.8566 | 0.8538 |
TLINK
We report the runs for the hybrid and the rule-based system in table 4.
Table 4.
TLINK evaluation on the test set
| Methodology | Recall | Precision | Average P&R | F-measure |
|---|---|---|---|---|
| Hybrid | 0.5249 | 0.5502 | 0.5382 | 0.5373 |
| Rule-based | 0.3681 | 0.5873 | 0.4779 | 0.4526 |
Discussion
Events used in this i2b2 temporal resolution challenge were broader than conventional event definitions in the 2010 i2b2 medical concept definition.2 In this study, Events can be any verb or adjective/adverb phrases that are relevant to the patient's clinical timeline (eg, reports, follow-up, awake, very well). In order to capture conventional events (ie, clinical concepts) and other types of events, well-defined expert knowledge and ontologies might be useful features in a machine learning model to further improve performance. We experimented with a variety of features to improve the performance of the system. Lexical features, semantic types, and POS tags, were useful as compared with other features. We did not use post-processing rules to refine the system output, which might have further improved performance. Also, there was a considerable room to optimize for the F-measure by improving recall–precision trade-off, since the recall was far lower than the precision.
MayoTime produced the best performance in i2b2 challenge teams and was able to find most TIMEX3. We believe that manually curated comprehensive rules to cover most TIMEX3 expressions, heuristic rules to catch relative date and time, and the systematic value normalization process produce this high performance. The explicit time expressions were easy to find, but relative time expressions, which require inference or reasoning, were often difficult to catch. Inconsistency of relative time annotation in the gold standard makes the problem even more difficult. For example, about half of the date expressions ‘this/that time’ were annotated in the gold standard but the rest were not. Some frequency TIMEX3 annotations require more reasoning to obtain the value attribute correctly. For example, ‘p.o. on Monday, Wednesday, and Friday (val=RP48h)’ ‘2 puffs q6h (val=R2P6H).’ Our current system does not consider those cases.
The hybrid TLINK system outperformed the rule-based one by 8% in F-measure. The hybrid approach produced balanced precision and recall while the rule-based system produced comparatively high precision but low recall. The rule-based approach identifies specific sets of TLINK pairs, leading to higher precision but much lower recall than the hybrid approach. It was observed that temporal features including CNTRO attributes and section information improved the performance by about 10%. However, added coreference resolution did not show significant improvement, perhaps owing to the prevalence of false-positive coreferences links. POS and tense features also did not contribute much to performance enhancement.
A few observations have been noticed for future improvements. TLINKs generated by the system can be expanded. For example, if the system generates ‘A before B’ and ‘B before C,’ then we can add ‘A is before C.’ It may be helpful to use intra-sentence TLINK types as features of inter-sentence TLINK. Finally, assigning different weights to the rule-based features in machine learning training might improve the precision.
Conclusion
A comprehensive temporal information extraction and classification system was designed and tested in the i2b2 2012 NLP challenge. A rule-based approach could be effectively applied to extract time expression patterns. The machine learning approach was attempted for Events and TLINK tasks, as these annotations require a variety of factors. We could also take advantage of knowledge engineering into a machine learning part, such as simple TLINK rules as part of machine learning features. We plan to release MayoTime, our TIMEX3 system, as an open source.
Acknowledgments
The 2012 i2b2 challenge was supported by Informatics for Integrating Biology and the Bedside (i2b2) award number 2U54LM008748 from the NIH/National Library of Medicine (NLM), by the National Heart, Lung, and Blood Institute (NHLBI), and by award number 1R13LM01141101 from the NIH NLM. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NLM, NHLBI, or the National Institutes of Health.
Footnotes
Contributors: All authors conceived and designed the experiments. SS, HL, KBW, DL, SRJ, and RKE participated in data analysis and system development. SS, KBW, DL, and SRJ wrote the first draft of the manuscript. All authors contributed to the writing of the manuscript and revisions. All authors reviewed and approved the final manuscript.
Funding: This study was made possible by National Science Foundation ABI:0845523, National Institute of Health R01LM009959A1, Strategic Health IT Advanced Research Projects (SHARP) Program (90TR002) administered by the Office of the National Coordinator for Health Information Technology, NLM 1K99LM011389.
Competing interests: None.
Ethics approval: i2b2 data use agreement.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Pustejovsky J, Castano J, Ingria R, et al. TimeML: Robust specification of event and temporal expressions in text. New Directions in Question Answering In AAAI Spring Symposium on New Directions in Question-Answering, Stanford, CA, 2003:28–34
- 2.Uzuner O, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torii M, Wagholikar K, Liu H. Using machine learning for concept extraction on clinical documents from multiple data sources. J Am Med Inform Assoc 2011;18:580–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mani I, Wilson G. Annual Meeting on Association for Computational Linguistics, New Brunswick, NJ, 2000:69–76
- 5.Verhagen M, Pustejovsky J. Temporal processing with the TARSQI toolkit. International Conference on Computational Linguistics, Manchester, UK 2008:189–92
- 6.Strötgen J, Gertz M. HeidelTime: High quality rule-based extraction and normalization of temporal expressions. International Workshop on Semantic Evaluation. Uppsala Sweden, 2010:321–4
- 7.Verhagen M, Sauri R, Caselli T, et al. SemEval-2010 task 13: TempEval-2. International Workshop on Semantic Evaluation. Uppsala Sweden 2010:57–62
- 8.Chang AX, Manning CD. SUTIME: a library for recognizing and normalizing time expressions. International Conference on Language Resources and Evaluation (LREC 2012), Istanbul, Turkey, 2012:3735–40 [Google Scholar]
- 9.Savova G, Masanz J, Ogren P, et al. Mayo Clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sohn S, Kocher J-PA, Chute CG, et al. Drug side effect extraction from clinical narratives of psychiatry and psychology patients. J Am Med Inform Assoc 2011;18(Suppl 1):144–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sohn S, Savova GK. Mayo Clinic Smoking Status Classification System: Extensions and Improvements. AMIA Annual Symposium; 2009, San Francisco, CA: 2009:619–23 [PMC free article] [PubMed] [Google Scholar]
- 12.Kehler A. Resolving temporal relations using tense meaning and discourse interpretation. Formalizing the dynamics of information. Stanford: CSLI Publications, 2000 [Google Scholar]
- 13.Passonneau RJ. A computational model of the semantics of tense and aspect. Comput Linguist 1988;14:44–60 [Google Scholar]
- 14.Webber BL. Tense as discourse anaphor. Comput Linguist 1988;14:61–73 [Google Scholar]
- 15.Hwang CH, Schubert LK. Tense Trees as the fine structure of discourse. Annual Meeting on Association for Computational Linguistics, Newark, DE, 1992:232–40
- 16.Lascarides A, Asher N. Temporal interpretation, discourse relations and commonsense entailment. Linguist Philos 1993;16:437–93 [Google Scholar]
- 17.Kamp H, Reyle U. From discourse to logic: introduction to modeltheoretic semantics of natural language, formal logic and discourse representation theory. The Netherlands: Kluwer Academic Dordrecht, 1993 [Google Scholar]
- 18.Mani I, Pustejovsky J. Temporal discourse models for narrative structure. ACL Workshop on Discourse Annotation, Barcelona, Spain, 2004:57–64
- 19.Mani I, Verhagen M, Wellner B, et al. Machine learning of temporal relations. Annual Meeting on Association for Computational Linguistics. Sydney, Australia, 2006:753–60
- 20.Wagholikar K, Torii M, Jonnalagadda S, et al. Feasibility of pooling annotated corpora for clinical concept extraction. AMIA Summits Transl Sci Proc 2012;2012:38. [PMC free article] [PubMed] [Google Scholar]
- 21.Wagholikar K, Torii M, Jonnalagadda S, et al. Pooling annotated corpora for clinical concept extraction. J Biomed Semantics 2013. Published Online First [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCallum, Andrew Kachites.2002. MALLET: A Machine Learning for Language Toolkit. http://mallet.cs.umass.edu.
- 24.Liu HWS, Li D, Jonnalagadda S, et al. Towards a semantic lexicon for clinical natural language processing Annual Symposium of American Medical Informatics Association; Chicago: 2012 [Google Scholar]
- 25.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 26.Jonnalagadda SR, Li D, Sohn S, et al. Coreference analysis in clinical notes: a multi-pass sieve with alternate anaphora resolution modules. J Am Med Inform Assoc 2012;19:867–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao C, Solbrig HR, Chute CG. CNTRO 2.0: a harmonized semantic web ontology for temporal relation inferencing in clinical narratives. AMIA Summits Transl Sci Proc 2011:64–8 [PMC free article] [PubMed] [Google Scholar]
- 28.Tao C, Wei WQ, Solbrig HR, et al. CNTRO: a semantic web ontology for temporal relation inferencing in clinical narratives. AMIA Annu Symp Proc 2010:787–91 [PMC free article] [PubMed] [Google Scholar]


