Abstract
Objective
Identification of clinical events (eg, problems, tests, treatments) and associated temporal expressions (eg, dates and times) are key tasks in extracting and managing data from electronic health records. As part of the i2b2 2012 Natural Language Processing for Clinical Data challenge, we developed and evaluated a system to automatically extract temporal expressions and events from clinical narratives. The extracted temporal expressions were additionally normalized by assigning type, value, and modifier.
Materials and methods
The system combines rule-based and machine learning approaches that rely on morphological, lexical, syntactic, semantic, and domain-specific features. Rule-based components were designed to handle the recognition and normalization of temporal expressions, while conditional random fields models were trained for event and temporal recognition.
Results
The system achieved micro F scores of 90% for the extraction of temporal expressions and 87% for clinical event extraction. The normalization component for temporal expressions achieved accuracies of 84.73% (expression's type), 70.44% (value), and 82.75% (modifier).
Discussion
Compared to the initial agreement between human annotators (87–89%), the system provided comparable performance for both event and temporal expression mining. While (lenient) identification of such mentions is achievable, finding the exact boundaries proved challenging.
Conclusions
The system provides a state-of-the-art method that can be used to support automated identification of mentions of clinical events and temporal expressions in narratives either to support the manual review process or as a part of a large-scale processing of electronic health databases.
Keywords: clinical text mining, clinical NLP, event extraction, termporal expression extraction, termporal expression normalization
Background
Recent advances in the availability of electronic health records (EHRs) provide an opportunity to improve the quality of clinical care (eg, through large-scale data sharing and integration that can be used to build clinical decision support systems1) and to support medical research (eg, identification of patients with specific conditions to support clinical trials2). While key issues remain in the adoption of EHRs and in managing data confidentiality,3 automated processing of available clinical data is also a major challenge: manual identification of such information is time consuming and often inconsistent and incomplete.4 This is particularly the case with clinical narratives, which are often the primary, preferred, and richest source of patient information. Several efforts have been reported in the area of clinical text mining to bridge the gap between unstructured clinical notes and structured data representation,5–21 including tools such as MetaMap22 23 and KnowledgeMap24 that have been developed to automatically annotate medical concepts in free text, along with systems to identify patient disease status,25–27 medication information,28–30 etc.
The i2b2 Natural Language Processing for Clinical Data challenge series provides a framework for common evaluation of clinical text mining systems. The topic of the 2012 challenge was the identification and linking of mentions of temporal expressions (TEs) (eg, dates, times, durations, and frequencies) and clinically relevant events (eg, patient's problems, tests, treatments) in narratives.31
Extraction of clinical events has recently attracted considerable attention, and was, for example, one of the tasks in the i2b2 2010 challenge.30 A wide variety of approaches (semi-supervised,32 supervised,33 34 hybrid models35 36), features (orthographic, lexical, morphological, contextual, semantic), terminological resources (UMLS,17 18 MedDRA,37 DrugBank38), and heuristic post-processing methods35 were used. The best lenient F score for the extraction of clinical events ranged from 89.80%36 to 92.40%.32
On the other hand, previous research on TE extraction has been mainly focused on the general domain.39 40 The clinical domain has been considered only relatively recently and often as an extension of general systems. For example, Med-TTK41 is built on top of a newswire system (TTK, TARSQI Toolkit42) by modifying and expanding rules developed on a set of 200 clinical narratives. The system identifies mentions of date, time, duration, and frequency TEs (with an overall F score of 85%), but does not provide their normalized values.
In this paper we describe, discuss, and evaluate a system that we have developed as part of our contribution to the i2b2 2012 challenge.
Objective
The aim of the 2012 challenge was to create clinical patient timelines from a set of clinical narratives. The TE extraction task focused on recognition and normalization of TE mentions. Normalization involved assigning three attributes:
value, using the ISO 8601 representation (eg, ‘2012–10–31T09:00’)
type of the TE: Time (eg, ‘the morning of admission’), Date (‘15 May 2007’), Duration (‘4 minutes’), or Frequency (‘PT48H’)
modifier that may be associated with the TE (eg, ‘approx’).
The event extraction task included recognition of instances of Problem (eg, ‘hematoma’), Test (eg, ‘an echocardiogram’), and Treatment (eg, ‘heparin IV’) events. In addition, events included mentions of a Clinical Department (eg, ‘the ER’). Mentions that indicate an evidential source of some specified information (eg, ‘CT [shows],’ ‘the patient [complained]’) are considered Evidential events (note that these can be verbs). Occurrences of all clinically relevant events that occur to the patient but do not belong to other event categories are considered Occurrences (eg, ‘follow up,’ ‘transport,’ etc). In addition to the type of an event, each mention was assigned a modality (Factual, Conditional, Possible, and Proposed) and a ‘polarity’ (ie, negated or not).
In this paper we focus on the methodologies engineered for the extraction and normalization of TEs and identification of events from clinical narratives.
Materials and methods
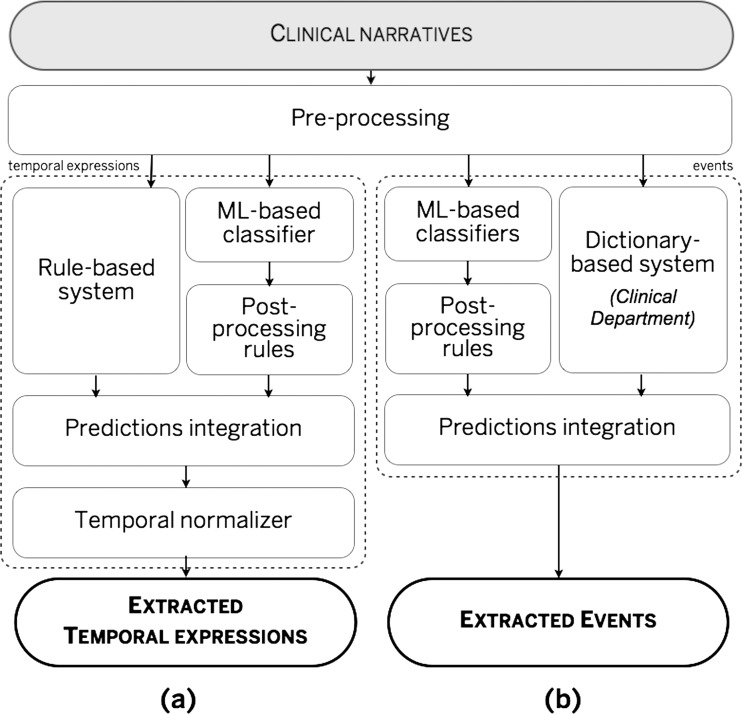
We approached the tasks as named-entity recognition (NER) problems, with the aim to identify relevant text spans and assign required attributes. The system (see figure 1) comprised two tracks: (a) TE identification and normalization, and (b) event recognition. Both tracks start with a common pre-processing step (in order to produce the features for subsequent steps). For identification of TEs we have developed two approaches (a rule-based and a machine learning), which are combined before the TE normalization module. Six separate machine learning (ML) modules were developed for events. Given that target annotations comprise spans of text, we approached the task as a sequence labeling problem and trained a separate conditional random fields43 (CRF) model with a number of shared features. The results from the CRF modules are followed by a set of post-processing rules that are designed to improve the boundaries of the resulting text spans. In addition to CRF models, a dictionary-based module was developed for one of the event classes (Clinical Department).
Figure 1.
The overall system architecture. ML, machine learning.
Data
Dictionaries
We manually crafted a dictionary of temporal terms that included five common types of constituents of TEs: weekdays and months; times of day; spelled-out numbers; medical temporal abbreviations (eg, ‘OBD’); and common TE references (such as ‘previously,’ ‘today,’ etc). This dictionary was used in the feature extraction process for our ML models. In addition, a dictionary of clinical departments was semi-automatically collected using OpenNLP44 NER to automatically extract candidate clinical department names from the i2b2 2010 and 2012 datasets. The candidates were then manually filtered to remove ambiguous terms. This dictionary was used for recognition and classification of mentions of Clinical Departments only.
Annotated corpora
For training, we used the training corpora provided by the i2b2 2012 challenge (190 mention-level annotated narratives). Additionally, the i2b2 2010 challenge corpus comprising 426 narratives annotated with Problem, Test, and Treatment concepts30 was used to support the event recognition track. The methods were tested on the i2b2 2012 test dataset (120 narratives).
Pre-processing
The narratives were first pre-processed (tokenization, sentence splitting, part-of-speech (POS) tagging, chunking) by GATE45 which was used to develop our rule- and dictionary-based modules. Additionally, lexical features for our ML modules were generated by cTAKES, which provided tokens, POS tags, and chunks. Mentions of Problem, Test, and Treatment were pre-generated by the Assertion module of cTAKES. Recognition of other medical entities was done by mapping all nominal chunks using MetaMap. The presence of negation was detected using NegEx,46 and sections (eg, ‘history of present illness,’ ‘hospital course’) within narratives were detected using simple heuristics. We also extracted semantic roles using the Clear Parser semantic role labeller47 module from cTAKES: each token was linked to an associated verb and assigned a role (eg, object, subject) in relation to that verb; verb tokens heading a sentence or sub-sentence were assigned a set of all participants and their roles linked to the verb. For example, the token ‘Thoracentesis’ in the sentence ‘Thoracentesis was performed on 7-12-91.’ would be marked as an object of the verb ‘perform.’
Extraction and normalization of temporal expressions
This module aims to identify and normalize TEs in pre-processed clinical narratives. The TE identification component accepts plain-text narratives and produces the spans of text recognized as TEs. We developed two identification modules: one based on rules and one ML-based. The results of both modules were integrated and passed to the normalization component, which provided the type, value, and modifier for the identified TEs.
The rule-based module
The rule-based module was developed using GATE. A total of 65 rules were engineered containing literal expressions derived from initial collocation extraction of TEs in the training data. The rule set is made up of (a) JAPE45 macros which defined a set of recurring literals and symbols (eg, temporal modifiers, weekdays, name of months, temporal medical abbreviations, etc) and (b) JAPE rules which combine macros and JAPE grammar for rule formalism. The effectiveness of rules (in terms of precision, recall, and F score) was analyzed on the training data to identify those that could have a positive effect on precision, recall, or F score.
The CRF-based module
The CRF-based module used token-level features that included the token's own properties and context features of the neighboring tokens (the experiments on the training data showed that two tokens each side provided the best performance). We used the inside-outside (I-O) annotation. The following features were engineered for each token:
Lexical features included the token itself, its lemma, and POS tag, as well as lemmas and POS tags of the surrounding tokens. Each token was also assigned features from its associated chunk (phrase): the type of phrase (nominal, verbal, etc), tense and aspect (if the phrase was verbal), the location of the token within the chunk (beginning or inside), and the presence of negation as returned by NegEx.
Domain features capture mentions of specific clinical/healthcare concepts. All nominal chunks were fed to MetaMap and the returned UMLS semantic class was used as a feature for all tokens within that particular chunk. In the case of multiple semantic classes returned by MetaMap, we concatenated them alphabetically and used the resulting string as a hybrid semantic class. Additionally, mentions of Problem, Test, and Treatment (as generated by cTAKES) were assigned to the token.
Semantic role features model dependencies between the token and associated verb, following the approach of Llorens et al.48 Each token is assigned the role, the verb, and their combination (eg, ‘object+perform’) in order to capture particular verb–role preferences.
Section type feature represents the section type in which the token appeared.
TE features represent five features that indicated the presence of the five common types of constituents of TEs in a given token (see the temporal dictionary mentioned in the Data section above).
An example of a complete feature vector for the CRF model is given in the online supplementary data. The results of the CRF-based tagging were post-processed to adjust the boundary/scope of token-level tags (eg, including determiners and pronouns where appropriate) and remove obvious false positives (eg, single character predictions such as ‘/’ or ‘a’).
The results of both identification modules were integrated. In cases of overlap, the union at the token level is taken: for example, consider the segment ‘starting at 9am of the morning of admission;’ if our rule-based method tagged the segment ‘9am of the morning’ and our CRF model annotated ‘morning of admission,’ the final integrated result will be ‘9am of the morning of admission.’
Temporal expression normalization module
The temporal expression normalization module has three components:
A rule-based extractor of key reference dates within the clinical pathway (namely, time of admission, discharge, operation, transfer) uses a set of regular expressions to extract and associate these main clinical events to a date. The rules are based on the proximity of specific keywords (eg, ‘operative,’ ‘hospital,’ ‘discharge,’ ‘operation’) and their direction (is the event mention before or/and after these keywords).
A rule-based utterance time selector pairs each TE with a reference time by analyzing its component words. For example, the expression ‘the day after the admission’ will be paired with the date of the admission, where the expression ‘postoperative day 2’ will be paired with the operation date. In the case of ambiguous expressions (such as ‘that time,’ ‘that period’), the module used the time assigned to the preceding TE. Otherwise, for all other TEs, the default reference time (admission) was used.
Clinical NorMA, a rule-based clinical TE normalizer, provides the value, type, and (optional) modifier to identified TEs. It extends a pre-existing open-source general-domain normalizer.49 To each TE and its associated reference time (from the utterance time selector), Clinical NorMA applies dictionary-driven regular expressions (83 general domain and 66 rules specifically designed for the clinical domain) to identify the value and type of the TE. The TE modifier is set only if a specific syntactic expression is triggered, for example, ‘in [number] or [number] days’. If the modifier has not been assigned using such expressions, the modifier (MOD) component checks for the presence of trigger words (eg, ‘approximately,’ ‘several,’ ‘nearly’). These triggers have been mined from the training corpus by applying a feature selection algorithm based on mutual information. Finally, the post-processing component applies additional rules that correct systematic errors or provide default values (eg, the substitution of the undefined number of days in ‘PXD’ with a default value, which was set as 3 for the i2b2 challenge).
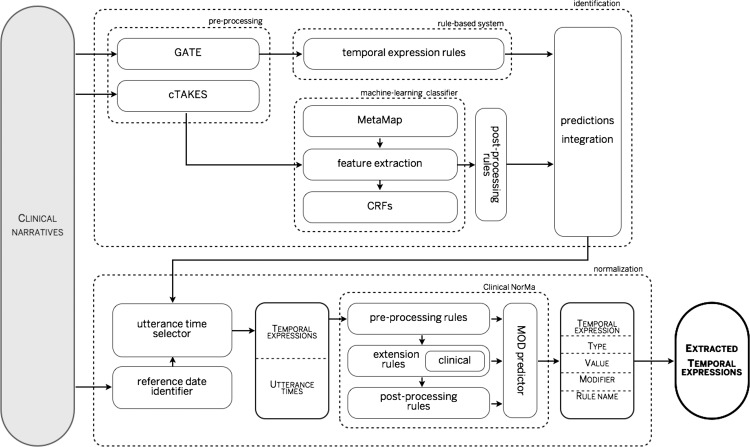
Figure 2 summarizes the architecture for the identification and normalization of TEs.
Figure 2.
Temporal expression extraction and normalization architecture. CRF, conditional random field.
Extraction of event mentions
This module aims at the extraction of event mentions. Apart from Clinical Department, all other event types were identified using CRFs only. The mentions of Clinical Departments were identified using both a manually-curated dictionary (see the Data section above) and a CRF module, which were integrated at the token level (like the TEs above).
The event CRF models were trained on relevant (type-specific) subsets of the training data and they all shared a number of feature groups (see table 1 for a summary). However, the Evidential and Occurrence types relied on additional feature groups as their scopes were not as focused as the scopes of other four event classes. We therefore added three additional feature groups for these event types:
Frequency of the token annotated as Occurrence or Evidential in the training set, with the aim to help the model resolve confusion between them.
Co-occurring events: An analysis of the training data revealed that mentions of these two event types correlate with the presence of other events in the same sentence. For example, the verb is 'noted' often annotated as Evidential if it is preceded with a Problem event (eg, ‘<Problem>Oral cyanosis and shallow respirations </Problem> were <Evidential>noted</Evidential>’). We therefore decided to include predictions from Problem, Test, Treatment, and Clinical Department modules as features for the CRF models of the Evidential and Occurrence categories. The resulting tags of the Evidential model were also used as features in the Occurrence CRF model. We note that therefore the CRFs were run in a particular order: the models for Treatments, Tests, Problems, and Clinical Departments were run in parallel; the outputs of these models were then used as features for the Evidential CRF, whose predictions were used as a feature in the Occurrence model.
Expanded lexical features: The initial experiments also revealed that the lexical variability of the Occurrence class was high, so an additional feature was added to indicate if a token is a typical Occurrence unigram. These unigrams were derived manually from a list of the 500 most frequent unigrams associated with this category (as obtained from the training data); after removing ambiguous terms, the list comprised 289 unigrams. The feature was also considered for the Evidential events, but associated words were heavily context dependent and thus not useful.
Table 1.
Groups of features used in the CRF models
| Entity type | Lexical features | Domain features | Semantic role features | Section type feature | Temporal expression features | Frequency features | Co-occurring event features | Expanded lexical features |
|---|---|---|---|---|---|---|---|---|
| Problem | X | X | X | X | X | X | ||
| Test | X | X | X | X | X | X | ||
| Treatment | X | X | X | X | X | X | ||
| Clinical department | X | X | X | X | X | X | ||
| Evidential | X | X | X | X | X | X | X | X |
| Occurrence | X | X | X | X | X | X | X | X |
| Temporal expressions | X | X | X | X | X | X |
CRF, conditional random fields.
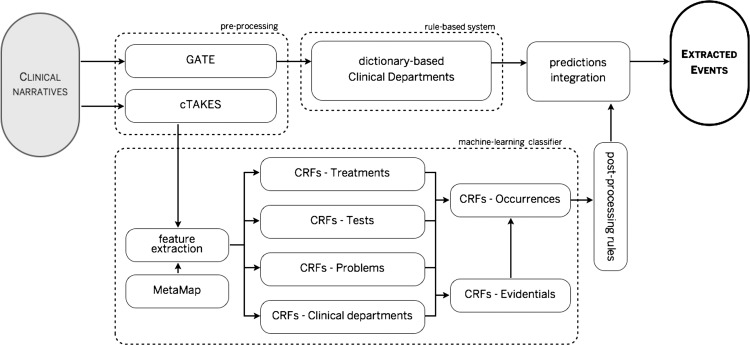
All CRF-based results were post-processed in the same way as TEs. Figure 3 summarizes the architecture developed for extraction of event mentions.
Figure 3.
Event extraction architecture. CRF, conditional random field.
Finally, each of the recognized events was checked with NegEx to determine polarity. For modality, lexical clue-based rules were explored during the development phase, but produced no significant improvement as compared to setting this attribute to Factual for every recognized event (95% of all events in the training data were Factual).
Results
Extraction and normalization of temporal expressions
The main evaluation metric for the TE recognition task was the product of the F score calculated with the lenient matching strategy (requiring that the system output overlaps the gold standard) and the accuracy obtained for the value attribute. Three different results were officially evaluated, based on the way temporal extractions were identified (the normalization module was always applied in full):
run 1: only TEs identified by the rules optimized for F score;
run 2: the union of recall-optimized rule-based predictions and CRF-generated tags;
run 3: only TEs identified by the rules optimized for precision.
The optimization has been performed on the training set with respect to the lenient matching strategy. Table 2 provides the results: run 2 provided the best F score (90.08%) along with the highest recall (91.54%). The strict evaluation scores (requiring an exact match between the system output and gold standard) were significantly lower (by 10–12% for the F score) indicating that both the ML and the rule-based approaches would benefit from a better method of boundary adjustment. The normalization scores were also highest in run 2 (type: 84.73%, value: 70.44%, modifier: 82.75%). We note that this is a state-of-the-art result as our run 2 was a top ranked outcome of the 2012 challenge (there were no significant differences between the top three runs, coming from three different teams). Compared to the results on the training data, there was some drop in the strict F score values (see online supplementary data), in particular for the rule-based runs, but overall lenient F score and normalization results were comparable with around a 1% difference between them (in some cases, the test results were even better).
Table 2.
Temporal expression extraction: micro-averaged results on the test data (120 narratives, 1820 temporal expressions)
| Identification | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strict matching | Lenient matching | Normalization | ||||||||
| P (%) | R (%) | F (%) | P (%) | R (%) | F (%) | Type (%) | Value (%) | Modifier (%) | ||
| Run 1 | 78.03 | 78.41 | 78.22 | 89.23 | 89.62 | 89.42 | 83.30 | 69.73 | 81.98 | |
| Run 2 | 77.03 | 79.62 | 78.30 | 88.68 | 91.54 | 90.08 | 84.73 | 70.44 | 82.75 | |
| Run 3 | 79.85 | 77.09 | 78.45 | 90.38 | 87.25 | 88.79 | 80.88 | 67.91 | 79.67 | |
F, F score; P, precision; R, recall.
Extraction of event mentions
The event extraction task was evaluated using the F score calculated with the lenient matching strategy, averaged across all the annotations in the test corpus. Accuracy was used to evaluate polarity and modality attributes. Two different results were officially evaluated, different only in how mentions of Clinical Departments were identified:
run 1 targeted precision by choosing Clinical Department predictions based on dictionary matches only;
run 2 targeted recall, so Clinical Department predictions included the union of CRF- and dictionary-based tags.
The mentions of other event types were identical in both runs. Table 3 provides the results: overall, run 2 gave better results, with the better F score (87.29%), recall (85.32%), and accuracies: polarity (79.45%) and modality (81.53%). Nonetheless, as expected, better precision (89.64%) was achieved in run 1. The strict F scores were 8% lower in both submissions, indicating again that determining the right boundaries for token-level recognized events was challenging. When compared to the training data (see online supplementary data), the system seems to have generalized well as there was even a slight increase in F score (around 1%) as compared to the training data.
Table 3.
Event extraction: micro-averaged results on the test data (120 narratives, 13 593 events)
| Strict matching | Lenient matching | Attributes | ||||||
|---|---|---|---|---|---|---|---|---|
| P (%) | R (%) | F (%) | P (%) | R (%) | F (%) | Polarity (%) | Modality (%) | |
| Run 1 | 82.05 | 77.05 | 79.71 | 89.64 | 84.66 | 87.08 | 78.81 | 80.08 |
| Run 2 | 81.74 | 78.05 | 79.85 | 89.35 | 85.32 | 87.29 | 79.45 | 81.53 |
F, F score; P, precision; R, recall.
Discussion
Temporal expression recognition
The results indicate that textual spans that represent TEs can be identified with an F score of 90%, with no significant differences between rule-based and integrated models (as expected, the rule-based runs showed slightly better precision; the CRF model on its own showed lower performance, with an F score of 86.70%). Common errors include mentions of clinical findings that follow date patterns or ambiguous mentions (such as ‘now’). The online supplementary data provide a detailed error analysis.
Temporal expression normalization
The normalization results vary for different attributes: while type and modifier have reasonable accuracies (84.73% and 82.75%, respectively), the value attribute proved challenging (70.44%). This is expected given that the value prediction asked for complete identification of a TE, whereas the other two attributes provide categorization-like values. We note that in some cases the normalizer failed to correctly distinguish between Date and Duration (and less frequently between Time and Duration). This is mostly due to wrong boundaries inherited from the TE recognition (eg, omission of an important preposition like ‘three days’ (date or duration) versus ‘every three days’ (frequency)). A detailed error analysis is provided in the online supplementary data.
Event recognition
The type-specific lenient evaluation results are given in table 4. The best F scores were achieved for frequent, well-defined event types, such as Problem (91.38%), Test (91.11%), and Treatment (89.26%). This result showed that CRF models generalized well with an abundance of training data (the additionally used 2010 dataset) and benefited from the use of terminological processing (cTAKES, MetaMap). For example, when the 2010 dataset is removed, the F scores drop notably for Problem (by 21%), Test (19%), and Treatment (20%) (data not shown). We note that these results are in line with the top performing systems in the i2b2 2010 challenge (the lenient F score ranges from 89.80%36 to 92.40%32).
Table 4.
Event recognition: per category performance on the test data (run 2, lenient matching)
| Event type | Frequency | P (%) | R (%) | F (%) |
|---|---|---|---|---|
| Problem | 4309 | 95.24 | 87.82 | 91.38 |
| Treatment | 3285 | 95.68 | 83.65 | 89.26 |
| Occurrence | 2499 | 63.43 | 66.91 | 65.12 |
| Test | 2173 | 95.05 | 87.48 | 91.11 |
| Clinical department | 732 | 76.02 | 83.61 | 79.64 |
| Evidential | 595 | 64.99 | 75.80 | 69.98 |
F, F score; P, precision; R, recall.
The results for the Clinical Department type were a surprise, as they proved much more lexically variable and ambiguous (eg, ‘Wound care’) than was indicated by both the training and 2010 data. The context-dependant Evidential type showed moderate results (69.98%), indicating that capturing the right context for annotations needed improvement. Finally, the Occurrence category (any clinically related event that could not be classified as any of the other categories) was too broad and underspecified for an ML method to generalize, and thus resulted in a lower F score (65.12%). The online supplementary data provide an error analysis and describe the impact that particular groups of features have on performance.
The accuracy of the polarity identification (79.45%) was around 10% lower when compared to the results typically obtained on manually input data (Goryachev et al50), which is expected given that the input was from our recognition component. Our assumption that setting the modality to Factual for each event mention would provide reasonably high results was correct (the percentage of Factual modality in the test data was consistent with the training). The drop in accuracy (81.53%) can be explained by the imperfect event spans returned by the recognition module.
Conclusion
This paper presents and evaluates various approaches to the extraction of clinically relevant events and TEs from clinical narratives, as part of our participation in the i2b2 2012 challenge. The methodology relies on combining rule-based approaches with feature-rich ML, which includes morphological, lexical, syntactic, semantic, and domain-specific features. The rule-based components were designed to handle the recognition and normalization of TEs and Clinical Departments, while CRF models were trained for all event and temporal recognition tasks.
The hybrid temporal recognition and normalization system provides state-of-the-art results with a micro F score of over 90% for lenient matching, and accuracies of 84.73% (type), 70.44% (value), and 82.75% (modifier) for the TE normalization. Clinical event extraction showed good performance with a micro F score of 87.29% (lenient). The well-scoped classes (such as Problem, Treatment, and Test) showed very good performance (F score of 90%), whereas unfocused and context-dependent categories (eg, Clinical Department, Occurrence, Evidential) proved to be challenging. Our study also revealed that the use of additional annotated corpora can indeed benefit the models, relaxing the need for specific terminological information.
While performance based on lenient matching was good, the most challenging part remains deciding the right boundaries of mentions (strict F scores were 78% and 80% for TE and event mentions, respectively). In addition, future work needs to explore new methods and features to capture context-dependent mentions and model unfocused categories.
A comparison to the agreement between the human annotators (89% for TEs and 87% for event recognition) indicates that the quality of the system's performance is comparable to what can be expected from manual efforts, and thus can be used either as a pre-processing step for a manual review process or as a part of a large-scale processing of electronic health databases.
The methods described here are packed in the TERN and CliNER tools, which are freely available at http://gnode1.mib.man.ac.uk/hecta.html
Acknowledgments
AK would like to acknowledge the Serbian Ministry of Education and Science for funding his stay at the University of Manchester (June–August 2012).
Footnotes
Contributors: AK developed the machine learning models and the overall system architecture, and drafted the manuscript; AD developed the rule-based temporal recognition models, collected the Clinical Department dictionary, and drafted the manuscript; MF developed the clinical temporal expression normalization component and drafted the manuscript; JAK performed critical revision of the manuscript and gave final approval; GN was the team leader in the i2b2 challenge, supervised the development of the whole system, and performed critical revision of the manuscript.
Funding: This work was partially supported by PhD scholarships from the University of Manchester and UK EPSRC (to AD, MF) and the following projects: "Development of new information and communication technologies, based on advanced mathematical methods, with applications in medicine, telecommunications, power systems, protection of national heritage and education" (III44006) and "Infrastructure for Technology Enhanced Learning in Serbia" (III47003) (the Serbian Ministry of Education and Science, to AK, GN); ‘Linked2Safety: A next-generation, secure linked data medical information space for semantically-interconnecting electronic health records and clinical trials systems advancing patients safety in clinical research’ project (EU FP7 ICT, contract 288328, to JAK, GN); ‘A study using techniques from clinical text mining to compare the narrative experiences of patients with medulloblastoma with factors identified from their hospital records’ (The Christie Paediatric Oncology Charitable Fund, to JAK, GN, AD). Health eResearch Centre (HeRC) is funded by a consortium of ten UK government and charity funders, led by the Medical Research Council (MRC). The i2b2 challenge is supported by Informatics for Integrating Biology and the Bedside (i2b2) award number 2U54LM008748 from the NIH/National Library of Medicine (NLM), by the National Heart, Lung, and Blood Institute (NHLBI), and by award number 1R13LM01141101 from the NIH NLM. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NLM, NHLBI, or the National Institutes of Health.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Wagholikar KB, Maclaughlin KL, Henry MR, et al. Clinical decision support with automated text processing for cervical cancer screening. J Am Med Inform Assoc 2012;19:833–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGregor JI, Brooks CJ, Chalasani P, et al. The Health Informatics Trial Enhancement Project (HITE): Using routinely collected primary care data to identify potential participants for a depression trial. Trials 2010;11:39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrández O, South B, Shen S, et al. BoB, a best-of-breed automated text de-identification system for VHA clinical documents. J Am Med Inform Assoc 2013;20:77–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogren PV, Savova GK, Buntrock JD, et al. Building and evaluating annotated corpora for medical NLP systems. AMIA Annual Symposium Proceedings; 2006:1050. Washington, DC, USA. [PMC free article] [PubMed] [Google Scholar]
- 5.Sager N, Friedman C, Chi E, et al. The analysis and processing of clinical narrative. MedInfo 1986;2:1101–5 [Google Scholar]
- 6.Sager N, Friedman C, Lyman M. Medical language processing: computer management of narrative data. Reading, MA: Addison-Wesley, 1987 [Google Scholar]
- 7.Hripcsak G, Friedman C, Alderson PO, et al. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med 1995;122:681–8 [DOI] [PubMed] [Google Scholar]
- 8.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hripcsak G, Austin JH, Alderson PO, et al. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology 2002;224:157–63 [DOI] [PubMed] [Google Scholar]
- 10.Haug PJ, Koehler S, Lau LM, et al. Experience with a mixed semantic/syntactic parser. Proceedings of the Annual Symposium on Computer Application in Medical Care 1995:284–8. New Orleans, LA, USA [PMC free article] [PubMed] [Google Scholar]
- 11.Fiszman M, Chapman WW, Evans SR, et al. Automatic identification of pneumonia related concepts on chest x-ray reports. Proceedings of AMIA Symposium; 1999:67–71. Washington, DC, USA [PMC free article] [PubMed] [Google Scholar]
- 12.Haug PJ, Christensen L, Gundersen M, et al. A natural language parsing system for encoding admitting diagnoses. Proceedings AMIA Annual Fall Symposium; 1997:814–18. Nashville, TN, USA [PMC free article] [PubMed] [Google Scholar]
- 13.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng QT, Goryachev S, Weiss S, et al. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak 2006;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spackman KA, Campbell KE, Cote RA. SNOMED-RT: a reference terminology for health care. Proceedings AMIA Annual Fall Symposium; 1997:640–4. Nashville, TN, USA [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization ICD -10 classifications of mental and behavioural disorder: clinical descriptions and diagnostic guidelines. Geneva: World Health Organization, 1992 [Google Scholar]
- 17.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system. Methods Inf Med 1993;32:281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Q, Cimino JJ. Automated knowledge extraction from the UMLS. Proceedings of AMIA Symposium; 1998:568–72. Orlando, FL, USA [PMC free article] [PubMed] [Google Scholar]
- 19.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol 2005;6:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith CL, Goldsmith CA, Eppig JT. The mammalian phenotype ontology as a tool for annotating, analyzing, and comparing phenotypic information. Genome Biol 2005;6:R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheuermann RH, Ceusters W, Smith B. Toward an ontological treatment of disease and diagnosis. Summit on Translational Bioinformatics; 2009:116–20. San Francisco, CA, USA [PMC free article] [PubMed] [Google Scholar]
- 22.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances . J Am Med Inform Assoc 2010;17:229–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aronson AR. Effective mapping of biomedical text to the UMLS metathesaurus: the MetaMap program. Proceedings of AMIA Symposium; 2001:17–21. Washington, DC, USA [PMC free article] [PubMed] [Google Scholar]
- 24.Denny JC, Miller RA, Johnson KB, et al. Development and evaluation of a clinical note section header terminology. AMIA Annual Symposium Proceedings; 2008;156–60. Washington, DC, USA [PMC free article] [PubMed] [Google Scholar]
- 25.Uzuner Ö. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc 2009;16:561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solt I, Tikk D, Gál V, et al. Semantic classification of diseases in discharge summaries using a context-aware rulebased classifier. J Am Med Inform Assoc 2009;16:580–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H, Spasić I, Keane J, et al. A text mining approach to the prediction of a disease status from clinical discharge summaries. J Am Med Inform Assoc 2009;16:596–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spasic I, Sarafraz F, Keane JA, et al. Medication information extraction with linguistic pattern matching and semantic rules. J Am Med Inform Assoc 2010;17:532–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun W, Rumshisky A, Uzuner Ö. Evaluating Temporal Relations in Clinical Text: 2012 i2b2 Challenge Overview. J Am Med Inform Assoc 2013;20:806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Bruijn B, Cherry C, Kiritchenko S, et al. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc 2011;18:557–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurulingappa H, Hofmann-Apitius M, Fluck J. Concept identification and assertion classification in patient health records. Proceedings of the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data; Boston, MA, USA, 2010 [Google Scholar]
- 34.Patrick JD, Nguyen DH, Wang Y, et al. A knowledge discovery and reuse pipeline for information extraction in clinical notes. J Am Med Inform Assoc 2011;18:574–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang M, Chen Y, Liu M, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc 2011;18:601–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang N, Afzal Z, Singh B, et al. Using an ensemble system to improve concept extraction from clinical records. J Biomed Inform 2012;45:423–8 [DOI] [PubMed] [Google Scholar]
- 37.Merrill GH. The MedDRA paradox. AMIA Annual Symposium Proceedings; 2008:470–4. Washington, DC, USA [PMC free article] [PubMed] [Google Scholar]
- 38.Wishart DS. DrugBank and its relevance to pharmacogenomics. Pharmacogenomics 2008;9:1155–62 [DOI] [PubMed] [Google Scholar]
- 39.Verhagen M, Gaizauskas R, Schilder F, et al. SemEval-2007 task 15: TempEval temporal relation identification. Proceedings of the 4th International Workshop on Semantic Evaluations; Prague, Czech Republic: Association for Computational Linguistics, 2007:75–80 [Google Scholar]
- 40.Verhagen M, Saurí R, Caselli T, et al. SemEval-2010 task 13: TempEval-2. Proceedings of the 5th International Workshop on Semantic Evaluation; Los Angeles, CA: Association for Computational Linguistics, 2010:57–62 [Google Scholar]
- 41.Reeves RM, Ong FR, Matheny ME, et al. Detecting temporal expressions in medical narratives. Int J Med Inform 2013;82:118–2. [DOI] [PubMed]
- 42.Verhagen M, Pustejovsky J. Temporal processing with the TARSQI toolkit. 22nd International Conference on Computational Linguistics: Demonstration Papers (COLING ‘08). Association for Computational Linguistics; Stroudsburg, PA, USA, 2008:189–92 [Google Scholar]
- 43.Lafferty JD, McCallum A, Pereira FCN. Conditional random fields: probabilistic models for segmenting and labeling sequence data. ICML; 2001: 282–9 [Google Scholar]
- 44.The Apache Software Foundation http://opennlp.apache.org/ (accessed 3 Jan 2013).
- 45.Cunningham H, Maynard D, Bontcheva K, et al. GATE: a framework and graphical development environment for robust NLP tools and applications. Proceedings of the 40th Anniversary Meeting of the Association for Computational Linguistics (ACL'02); 2002. Philadelphia, PA, USA [Google Scholar]
- 46.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 47.Choi JD, Palmer M. Transition-based semantic role labeling using predicate argument clustering. Proceedings of the ACL 2011 Workshop on Relational Models of Semantics. RELMS ‘11; Stroudsburg, PA, USA, 2011:37–45 [Google Scholar]
- 48.Llorens H, Saquete E, Navarro B. TIPSem (English and Spanish): evaluating CRFs and semantic roles in TempEval-2. Proceedings of the 5th International Workshop on Semantic Evaluation; Uppsala, Sweden, 2010:284–91 [Google Scholar]
- 49.Filannino M. Temporal expression normalisation in natural language texts, CoRR 2012; abs/1206.2010, 2012
- 50.Goryachev S, Sordo M, Zeng QT, et al. Implementation and evaluation of four different methods of negation detection. Technical report, DSG, 2006 [Google Scholar]