Abstract
Objective
To identify the temporal relations between clinical events and temporal expressions in clinical reports, as defined in the i2b2/VA 2012 challenge.
Design
To detect clinical events, we used rules and Conditional Random Fields. We built Random Forest models to identify event modality and polarity. To identify temporal expressions we built on the HeidelTime system. To detect temporal relations, we systematically studied their breakdown into distinct situations; we designed an oracle method to determine the most prominent situations and the most suitable associated classifiers, and combined their results.
Results
We achieved F-measures of 0.8307 for event identification, based on rules, and 0.8385 for temporal expression identification. In the temporal relation task, we identified nine main situations in three groups, experimentally confirming shared intuitions: within-sentence relations, section-related time, and across-sentence relations. Logistic regression and Naïve Bayes performed best on the first and third groups, and decision trees on the second. We reached a 0.6231 global F-measure, improving by 7.5 points our official submission.
Conclusions
Carefully hand-crafted rules obtained good results for the detection of events and temporal expressions, while a combination of classifiers improved temporal link prediction. The characterization of the oracle recall of situations allowed us to point at directions where further work would be most useful for temporal relation detection: within-sentence relations and linking History of Present Illness events to the admission date. We suggest that the systematic situation breakdown proposed in this paper could also help improve other systems addressing this task.
Keywords: Natural Language Processing, Information Extraction, Medical Records, Chronology as Topic, Text Mining
Introduction
The need for automatically extracting information from the text of clinical records is firmly established.1 As the first layers of information extraction (medical entities, negation and modality, some relations between entities) become more mature,2 more complex layers are being challenged. This is the case of temporal information,3–5 encompassing any medical event (occurrence of diseases, treatments, medical encounters, etc), dates, times and other temporal expressions, and relations among these. The i2b2/VA 2012 challenge tasks and corpora6 have created a testbed for the design and evaluation of methods which extract such information from clinical texts.
In this paper we show how existing natural language processing (NLP) components could be reused and adapted during our participation in the challenge for event and temporal expression extraction, and new components designed in complement to address temporal relation extraction. On the one hand, we describe the components we started from and highlight the needed adaptations: lexicons, normalization, features etc. On the other hand, we explore more specifically an issue linked to a prominent feature of many clinical reports, especially in the i2b2/VA 2012 corpus: their clear sectioning into sections including admission date (AD), history of present illness (HPI), hospital course (HC), and discharge date (DD). An intuition shared by many participants of the task was that temporal relations within or across sections have quite different characteristics, as do temporal relations within or across sentences. Hence most participants adopted different strategies to address these different situations.
However, to our knowledge, which breakdown into situations should be designed and how important each situation is in the temporal relation detection process have not been investigated systematically. We therefore decided to push this logic to an extreme by splitting the Tlink space into 57 distinct situations which we define below. We studied the importance of these situations in the training data through an oracle method, and experimentally determined which classifier was best suited for which kind of situation. This enabled us to confirm and refine experimentally the above intuitions, with associated preferred detection methods, and allowed us to outperform by 7.5 F-measure points our official challenge result.
After a short review of related work, we present the methods we used for the detection of events, modality and polarity, temporal expressions (Timex3s), and temporal relations (Tlinks), the results we obtained on the training and test corpora, and discuss directions for future work.
Related work
Concept and Timex3 identification
Clinical information extraction has been the subject of much research (see Meystre et al1 for a review), including in the i2b2/VA 2010 challenge.2 Concept extraction has been addressed by defining rules and gazetteers7 or using linguistic resources obtained from external resources, such as the Unified Medical Language System (UMLS).8–14 Temporal expression identification and the subsequent temporal normalization have been defined and addressed for several years in the Automatic Content Extraction program (ACE) Temporal Expression Recognition and Normalization tasks. Existing systems are mainly rule-based and have made efforts to be extensible.15 16
Temporal relation identification
Temporal relations provide useful information relevant for information extraction and summarization,17 question-answering,18 but they may also help general research purposes such as discourse relations.19 20 While previous work mostly focused on logical aspects,19 21–23 the study of temporal relations has known a renewed interest through NLP applications. Since temporality is deeply entangled in language, a number of studies focus on linguistic features useful for its deciphering. Among the proposed features, we find lexical anchors,24–26 syntactic analysis,19 23 27 28 grammatical information,19 29 and verb semantics.18 19 Notice that these features can be used by both rule-based and machine-learning systems.
Clinical temporal relation extraction has also received a renewed interest, with some methods based on deep language processing30 of event relations and others using machine-learning ranking of start or end times of events.31 The i2b2/VA 2012 challenge corpus6 has created new opportunities to test and evaluate clinical temporal information extraction methods. Most teams participating in the challenge were inspired by the human annotation guidelines and the gold standard document representations, which differentiated section-relative temporal links and other links. For instance, Cherry et al,32 tying for rank #1 for Tlink detection, considered four situations (‘specialists’): local relations (R=0.357), section-relative time (R=0.266), long-distance co-reference (R=0.036), and long-distance before/after (R=0.003). They obtained good precision, but did not compute the reference recall for these situations. Tang et al,33 also tying for rank #1, considered consecutive events, cross-sentence events, and co-reference, then applied Support Vector Machine (SVM) classifiers. They did not show situation-specific gold standard recall either. There is thus a gap in system follow-up and evaluation that we aim to fill.
Challenge corpus description
The training corpus consists of 190 clinical records and the test corpus of 120 files. Participants were asked to identify events (clinical department, evidential, occurrence, problem, test, treatment), temporal expressions (Timex3s: date, duration, frequency, time), and temporal relations (Tlinks), with attributes: ‘type’, ‘polarity’, and ‘modality’ for events, ‘type’ and ‘modifier’ for Timex3s, and ‘type’ for Tlinks. Table 1 shows their distribution; more detail is provided in Sun et al.6
Table 1.
Annotation statistics in percentage on training and test corpora
| Event | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type | Polarity | Modality | |||||||||||
| Clinical department | Evidential | Occurrence | Problem | Test | Treatment | Negative | Positive | Conditional | Factual | Possible | Proposed | ||
| Train | 6.05 | 4.49 | 19.95 | 30.50 | 15.76 | 23.25 | 7.03 | 92.97 | 0.87 | 96.11 | 1.71 | 1.31 | |
| Test | 5.39 | 4.38 | 18.38 | 31.70 | 15.99 | 24.17 | 6.84 | 93.16 | 0.99 | 95.42 | 2.23 | 1.36 | |
|
Timex3 |
Tlink |
||||||||||||
|
Type |
Modifier |
Type |
|||||||||||
| Date | Duration | Frequency | Time | Approximate | End | Middle | More | NA | Start | After | Before | Overlap | |
| Train | 69.38 | 17.19 | 10.52 | 2.91 | 10.98 | 2.41 | 0.08 | 0.30 | 83.83 | 2.41 | 9.56 | 52.36 | 38.08 |
| Test | 67.14 | 18.74 | 10.82 | 3.30 | 9.88 | 1.71 | 0.11 | 0.28 | 86.20 | 1.82 | 9.84 | 54.49 | 35.67 |
Bold face indicates the best results. NA, not applicable.
Methods
Event processing
To identify events, we reused the platforms we developed in previous i2b2/VA challenges: Caramba and Ogmios, described in more detail below. Adaptations included the introduction of new features, relying on tools which we did not use in 2010 (the Charniak McClosky parser, the Brown clustering algorithm), and the construction of new models for the machine-learning part of our systems to deal with the six event categories proposed this year. In the Caramba system, we used a new Conditional Random Field (CRF) engine in place of CRF++.
Event identification
The Caramba system12–14 relies on several tools which allowed us to obtain various features that we fed to Wapiti,34 a CRF classifier implementation:
Document-level features: section id among four sections we defined as follows: AD, DD, HPI, and HC.
- Syntactic features:
- Part-of-speech (POS) tags from the UMLS Specialist Lexicon, the Tree Tagger35 (http://www.ims.uni-stuttgart.de/projekte/corplex/TreeTagger/), and the Charniak McClosky36 biomedical parser (http://stanford.edu/~mcclosky/biomedical.html).
- Chunks from the Charniak McClosky parser and built upon the TreeTagger POS tags.
- Semantic features:
- Supervised features are predefined lexical classes: semantic types and semantic groups from the UMLS (http://www.nlm.nih.gov/research/umls/)37; six event types and other markers provided by both WMatch,38 an analysis engine based upon regular expressions of words, rules and lexicons; and the Ogmios system.39–41
- Unsupervised clusters are created through Brown's algorithm,42 performed over the UMLS Metathesaurus37 terms (multi-words expressions) and over the 2011 i2b2/VA Beth Israel and Partners Healthcare corpora. Clustering was performed with code (http://www.cs.berkeley.edu/~pliang/software/) from Liang's Master's Thesis.43
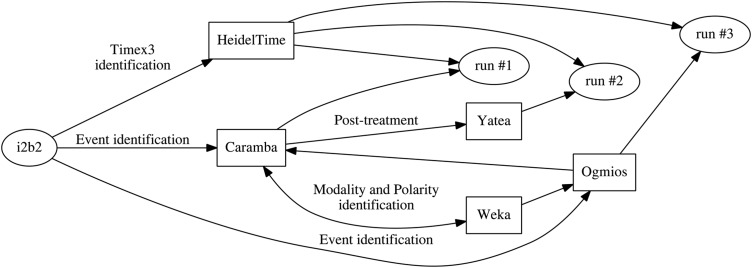
In our experiments on the training corpus, we tested models generating up to 36 million features. Since CRFs may be prone to overfitting (eg, compared to SVMs), we took care to include features that would lead to better generalization: syntactic tags and chunks, and semantic classes of various kinds. The output of the CRF was used as our first submission in both Event/Timex3 and End-to-End tracks (see figure 1).
Figure 1.
Event and Timex3 processing.
To refine the events produced by Caramba, we decided to use the term analysis performed by Yatea,44 a term extraction system. For each multi-word event found both by Yatea and Caramba, we added the boundaries of the event specified by Yatea to the output of Caramba. This was our second submission.
Our third submission in Event/Timex3 and End-to-End tracks was the direct output of the Ogmios platform. The Ogmios system is a configurable linguistic platform dedicated to the processing and annotation of specialized documents in XML and text formats. It integrates NLP tools and, thanks to specific wrappers, it merges information provided by these tools into a homogeneous multilayer structure. Its configuration is similar to that defined for the 2010 i2b2/VA challenge40:
POS tagging is performed with GeniaTagger.45
Event identification relies on the TermTagger Perl module (http://search.cpan.org/~thhamon/Alvis-TermTagger/) and terminological resources.
A specific post-processing fitted to this year's challenge selects and extends previously identified events.
We exploited four terminological sources:
- 316 368 terms from the UMLS37 which belong to several semantic axes related to the involved types of events:
- medical problems
- tests
- treatments
- clinical departments.
243 869 entries from RxNorm46 used for the detection of medication names (treatments).
The available annotations of the 2012 i2b2 training sets.
Additional contextual tags for marking specific contexts for different types of events (pre-problem, pre-test, pre-treatment, post-treatment...).
These resources were manually checked and adapted to increase their coverage. Section titles were also used for event categorization, for instance, the Admission Diagnosis section usually contains medical problems.
A post-processing step first extends to the right the event strings provided that acceptable POS tags are found for this extension, while non-acceptable stopwords stop the extension. Then, it selects the predicted event according to several indicators: (i) in case of competing annotations, the larger event string is preferred; (ii) events identified as non i2b2 concepts (as well, M.D., etc) are rejected; (iii) events occurring in section titles are removed except for tests (Serologies) and occurrences (DD).
Event modality and polarity attributes
To predict the modality and polarity of events, we used machine learning through the Weka toolbox (http://www.cs.waikato.ac.nz/ml/weka/).47 48 We chose to build two different models, one for modality and another for polarity. We experimented with several algorithms on the training corpus, using 10-fold cross validation. Features were the four left and right tokens surrounding the identified event. Among our experiments, the Random Forest algorithm, based on multiple decision trees, performed best: F=0.994 on modality and 0.987 on polarity. Our models were only trained on Caramba outputs. For the test, we used these models on the outputs of the event identification stage (using either Caramba or Ogmios) to predict modality and polarity.
Timex3 identification
We studied and tested several existing Timex identification systems and found that HeidelTime15 had the best combination of performance and adaptability. While this system proposes a good coverage of the temporal expressions used in general-language documents, it needed to be adapted to the clinical domain. We added:
Medical and especially clinical temporal expressions, such as post-operative day #, b.i.d., day of life, qac.
A preprocessing step to identify the AD of each document. We considered it as the document creation date and provided it to HeidelTime to compute the reference time of relative expressions such as 2 days later.
Post-processing normalizations of the temporal expressions to fit i2b2 requirements: normalization of durations towards approximative numerical values rather than towards the undefined ‘X’-value; and external computation for some durations and frequencies due to limitations in HeidelTime's internal arithmetic processor, etc.
Temporal relation identification
Situations
To help assign, as summarized from the guidelines, ‘all Tlinks necessary to create a clinical timeline’, we divided the Tlink detection problem according to different situations. Given two events or Timex3s (henceforth collectively named ‘EVTs’ (events)) that might be linked through a temporal relation, quite different situations arise depending on whether they are of the same kind (eg, Event–Event) or not (Event–Timex3), in the same sentence or paragraph or not, etc. We identified the following dimensions, which define a total of 56 situations (plus one, ‘OTHER’, for the remaining cases):
Sections of the source and target EVTs: AD, DD, HPI, HC.
Element types of the source and target EVTs: Timex3 or Event.
Distance between EVTs: same sentence SS, adjacent sentences in same section S1, more distant sentences.
Number of Timex3s between EVTs: no Timex3 NTB or at least one Timex3.
Each combination of these dimensions defines a situation for which a distinct method can be applied. For instance, TIMEX3-EVENT-DD-HC codes a situation where we consider a Timex3 in the DD section (ie, the DD itself) and an Event in the HC section (see figure 2). Given the set of EVT pairs found in the training corpus for a situation, we experimented with a set of classifiers to assign each pair one of the four classes {BEFORE, AFTER, OVERLAP, NIL}, the latter meaning ‘no Tlink’. This is equivalent to defining an initial decision tree (with features based on the above four dimensions), after each leaf of which another classifier is applied. For instance, a baseline decision for TIMEX3-EVENT-DD-HC could be that the Event occurred BEFORE the DD. A question which naturally arises in this context is: which of these situations are most important to address, and with which methods?
Figure 2.
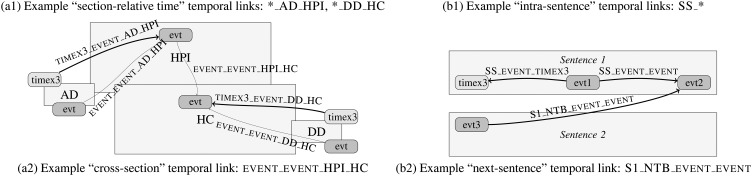
Example situations: (A) related to section time; (B) intra-sentence and next-sentence.
This is first an evaluation problem. Although one can train and test classifiers with cross-validation, the recall and precision measured this way by usual classifier machinery concern individual Tlinks. To take into account temporal inference, the TempEval-type evaluation provided by the organizers’ program must be used. Therefore, all our evaluations of Tlinks use the default settings of the official evaluation program, that is, original against closure mode. Second, the gold standard is organized by document, not according to our situations, which means that the recall reported for a given situation is always fairly low, since a situation never covers all Tlinks in a document. We therefore computed an oracle recall (OR) for each situation in the following way. For each situation, we identified which Tlinks of the gold standard files were relevant for this situation, and we prepared new files containing only these Tlinks. Evaluating these new files against the full gold standard files obtains a perfect precision (no error has been introduced into these files) and a recall which is the best recall that can be obtained in this situation (since these files contain all the expected TLinks for this situation): this is what we call the OR for this situation. This is the ideal recall that should be reached by a classifier trained for this situation. Initial experiments showed us that we did not obtain gains when considering those situations which had an OR lower than 0.01; we thus focused on those above that threshold.
Features
We used the following two sets of features to represent a candidate pair of EVTs (the top three of each set were used in our challenge submissions):
- Intra-EVT:
- Text of the EVTs (if among the 50 most frequent).
- All EVT annotations (modality, polarity, subtypes).
- For events, a subcategorization into surgery, states, punctual events, locations, follow-up events, or others, based on a lexical study of the development set.
- EVT tokens (if occurring at least 10 times in the training corpus).
- We also tested Brown clusters of EVT tokens and combinations with temporal prepositions, but this did not improve over the above tokens.
- Inter-EVT:
- Distance between the EVTs.
- Temporal or other prepositions between the EVTs.
- Number of other EVTs in between.
- For pairs of events located in the same sentence, syntactic dependencies obtained with the Charniak McClosky parser,36 as converted into Stanford dependencies.
- We interpreted initial better results of decision trees with the above features as indicating that conjoined features are meaningful predictors of Tlinks. Therefore we added several such conjunctions, beginning with the triples <source-event-type, syntactic-dependency, target-event-type>, where source- and target-event types are the i2b2/VA 2012 categories: clinical department, evidential, occurrence, problem, test, treatment; date, duration, frequency, time.
NIL relations and transitive closure
The definition of the task implies that manual annotations are incomplete: some positive instances of temporal relations can be inferred (through transitive closure) from those provided in the reference annotations of the training set. Thus they should not be considered as negative examples. We therefore ran transitive closure over all training Tlinks, using the Sputlink system,49 and all resulting Tlinks were used as positive instances. The cross-product of EVTs of each document was used to generate a full set of candidate Tlinks, from which positive instances were removed to form the set of negative instances. On the contrary, transitive closure was not performed when applying the trained classifiers to the development or test data.
Different classifiers
The decision tree classifier J48, an implementation of the C4.5 decision tree learning algorithm,50 as implemented in the Weka toolbox,47 48 obtained the best results globally, without the EVT token features. As explained above, we also evaluated the individual results of different classifiers (Naïve Bayes, Decision Tree, SVM (LibSVM), k nearest neighbors, logistic regression (aka MaxEnt), Random Forest) for the main situations. We selected the best classifier for each situation and applied the combination of the winners. We also tested voting combinations of classifiers, using average confidence as the combination operator. All classifiers were used through Weka.
Contradictions
Tlinks predicted by a classifier might contradict previously predicted Tlinks. To address this issue, when applying each model, we applied Verhagen's link merging algorithm49: we sorted the predicted Tlinks in descending order of confidence and added them in this order: for each one, its consistency with the current Tlink set was computed, including transitive closure on BEFORE and AFTER relations. In case of inconsistency, the predicted Tlink was discarded. This greedy approach was also applied at the level of situations, since each situation was processed in turn in descending order of its precision on the training set.
Rule-based decisions
Some Tlinks can be identified directly based on a few rules before any classifier: Admission (resp. Discharge) events and dates OVERLAP (this is the most frequent case of co-reference), and Admission is BEFORE Discharge.
Evaluation and discussion
Evaluation metrics are described in the overview paper of the challenge.6
Track 1: Event/Timex3 detection
In the event identification task, our best submission achieved a 0.8307 global F-measure using the Ogmios system (run #3, see table 2) and ranked #9/14 (top=0.92). Ogmios also obtained a higher precision (0.7812) while Caramba achieved a better recall (0.9576), probably due to our choice of features geared towards generalization.
Table 2.
Aggregated scores on the Event/Timex3 task
| Run | Precision | Recall | Average P&R | F-measure | Type | Polarity | Modality |
|---|---|---|---|---|---|---|---|
| Event | |||||||
| #1 | 0.6526 | 0.9576 | 0.7764 | 0.7762 | 0.8607 | 0.9098 | 0.9142 |
| #2 | 0.6339 | 0.9567 | 0.7627 | 0.7625 | 0.8598 | 0.9089 | 0.9139 |
| #3 | 0.7812 | 0.8869 | 0.8307 | 0.8307 | 0.7993 | 0.8401 | 0.8463 |
| Type | Val | Modifier | |||||
| Timex3 | |||||||
| #1 | 0.8605 | 0.8170 | 0.8382 | 0.8382 | 0.7495 | 0.5363 | 0.7203 |
| #2 | 0.8605 | 0.8170 | 0.8382 | 0.8382 | 0.7495 | 0.5363 | 0.7203 |
| #3 | 0.8611 | 0.8170 | 0.8385 | 0.8385 | 0.7478 | 0.5357 | 0.7203 |
Italics indicates the best results.
Polarity and modality detection was only performed on Caramba output. For Ogmios output events, if the event was found by both systems, we used the modality and polarity attributes computed for Caramba's event. If an event was only identified by Ogmios, we gave it the default modality and polarity values (ie, factual and positive). In consequence, the modality and polarity scores are better in runs #1 and #2 (Caramba) than in run #3 (Ogmios). These scores are on a par with the best systems (0.86). Unfortunately, their distribution in the test corpus was such that a better score would have been obtained had we given the majority attribute values to each event (0.93 for polarity and 0.95 for modality).
In the Timex3 identification sub-task, the HeidelTime algorithm performed well (F=0.8385 on Ogmios output, ranking #10/14, top=0.91). We noticed that some Timex3s were incorrectly normalized, for example, ‘x’ characters instead of a correct date. More work on normalization is thus needed.
Track 2: End-to-End Tlink detection
In the End-to-End task, our best submission was also run #3 (based on Ogmios) and achieved a 0.4932 F-measure for temporal links (see table 3), ranking #5/7 (top=0.63). It should be much improved by our new Tlink detection results obtained after the challenge.
Table 3.
Aggregated scores on the End-to-End task (Tlink evaluation)
| Run | Precision | Recall | Average P&R | F-measure |
|---|---|---|---|---|
| Tlink | ||||
| #1 | 0.5551 | 0.4114 | 0.4900 | 0.4726 |
| #2 | 0.5511 | 0.4166 | 0.4908 | 0.4745 |
| #3 | 0.6155 | 0.4115 | 0.5175 | 0.4932 |
Italics indicates the best results.
Track 3: Tlink detection on ground truth events
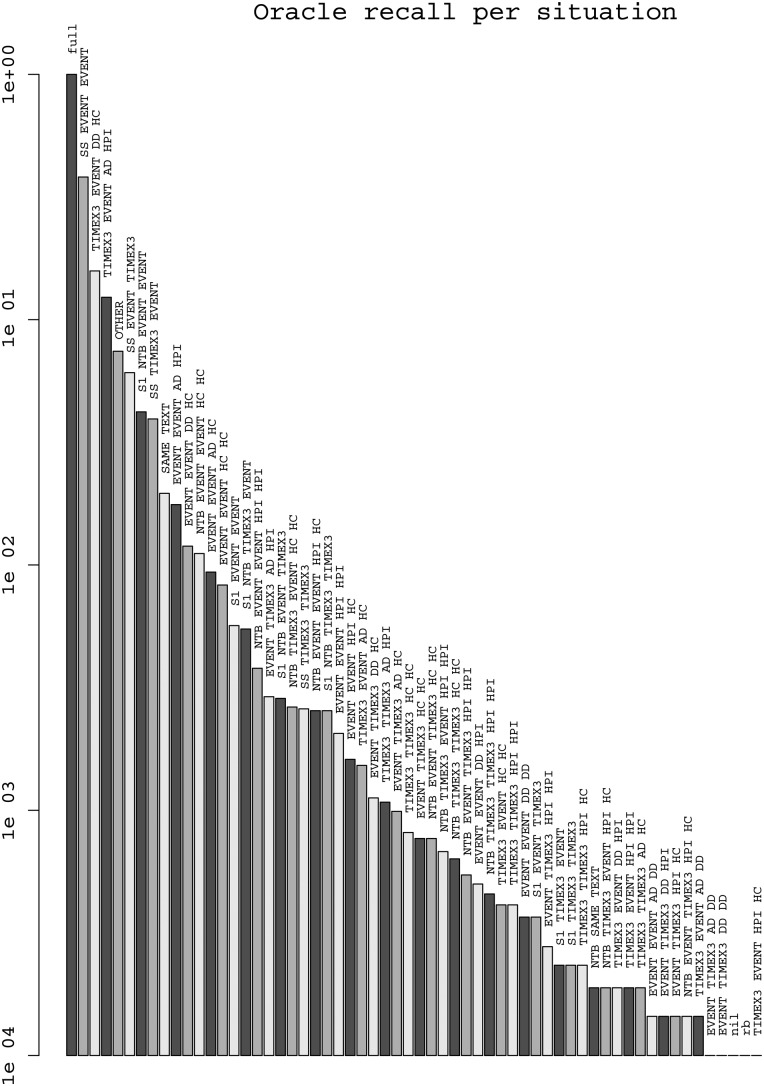
Figure 3 plots the OR of each situation on the training corpus in decreasing order on a semi-logarithmic scale. The most contributing situations (with OR>0.01) include three intra-sentence (SS-*) situations (total OR=0.48), four ‘section-relative times’ (*-AD-HPI, *-DD-HC: total OR=0.31), a next-sentence situation (S1−NTB−EVENT−EVENT: OR=0.04), co-reference (SAME−TEXT: OR=0.02), and longer distance events in HC (NTB−EVENT−EVENT−HC−HC: OR=0.01). We focused on the first eight of these, leaving aside for now 0.17pt of recall (including 0.7 for diverse OTHER situations). We quickly noticed that EVENT−TIMEX3−AD−HPI could obtain a good precision and relative recall and added it to the set.
Figures 3.
Oracle recall of each situation on the training corpus in decreasing order on a semi-logarithmic scale.
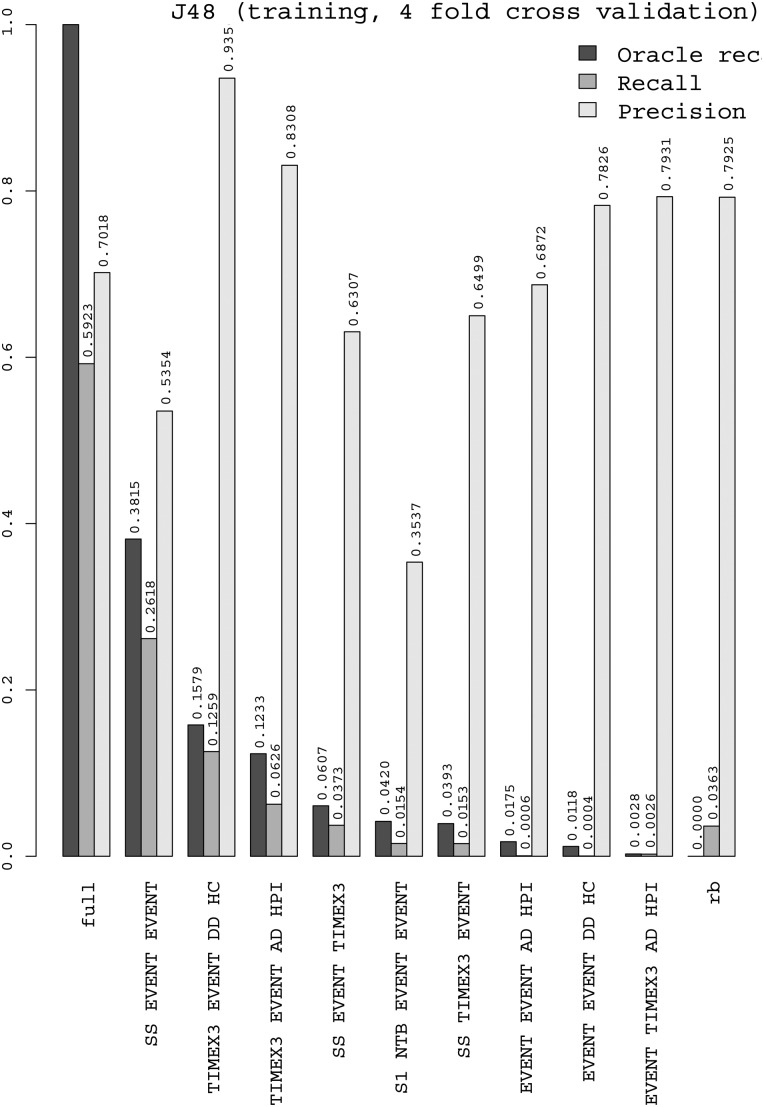
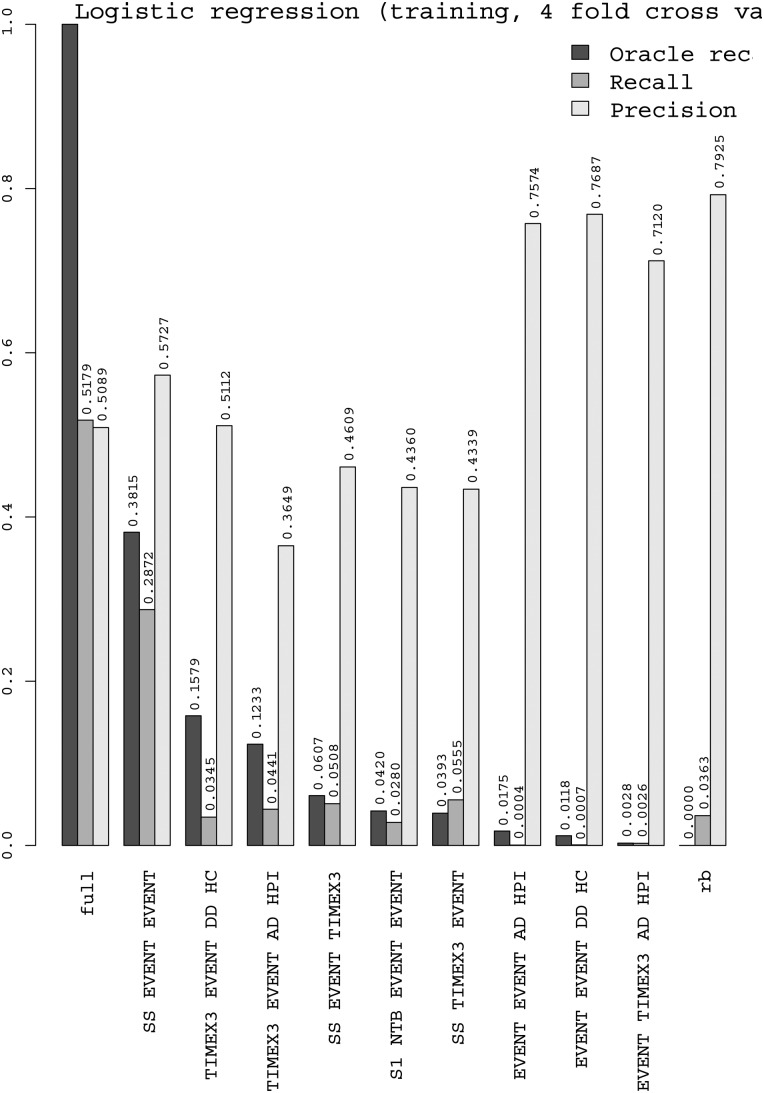
Figures 4 and 5 show the performance of the J48 decision-tree and logistic regression classifiers on each of the nine situations, plus rb (rule-based decisions) and full (total). The precision of J48 is generally good (but less on intra-sentence), and thanks to the OR we can see that its recall is good on TIMEX3–EVENT–DD–HC and moderate on SS–EVENT–EVENT and TIMEX3–EVENT–AD–HPI. The distance to OR is still large for these two situations (resp. 10pt for Logistic and 6pt for J48), which makes them first candidates to explore in further work. Logistic regression generally has lower precision; it has better recall than J48 on intra-sentence and lower on section-relative times. We explain this by the capacity of decision trees to build feature conjunctions, useful for these situations, which logistic regression cannot do by itself. We also tested an SVM with LibSVM, but it had a longer training time and did not rank first in any of the situations where we tested it. Random Forest, with token features, seems to be more robust to situations with a smaller number of instances.
Figure 4.
Performance of the J48 decision tree classifier, on each of the 9 top situations, plus rb (rule-based decisions) and full (total).
Figure 5.
Performance of the logistic regression classifier on each of the 9 top situations, plus rb (rule-based decisions) and full (total).
The two runs we submitted for this last task were anterior to our systematic study of situations, and included 20 approximately ordered situations (see table 4), ranking #9. Different tradeoffs of recall and precision were obtained by varying the proportion of NIL relations in the training set. Adding new features and the above-mentioned management of NIL relations, voting, and assigning different classifiers to different situations all improved the F-measure. Then the systematic study of situations, guided by the OR, allowed us to focus on situations with the best relative recall, while optimizing precision by ordering classifiers by descending precision. All our studies were performed on the training corpus using fourfold cross validation to avoid overfitting. Thanks to these precautions, all improvements observed when optimizing on the training corpus were reflected by improvements on the test corpus, bringing it to the level of rank #5.
Table 4.
Aggregated scores on the Tlink task
| #S | Classifier | Training corpus (4-fold c-v) | Test corpus | Delta | ||||
|---|---|---|---|---|---|---|---|---|
| P | R | F | P | R | F | |||
| 9 | Sel[P] (tokens) | 0.711 | 0.628 | 0.667 | 0.655 | 0.594 | 0.623 | −0.044 |
| 9 | C | 0.652 | 0.663 | 0.658 | 0.602 | 0.631 | 0.616 | −0.042 |
| 9 | j48 | 0.702 | 0.592 | 0.642 | 0.661 | 0.555 | 0.603 | −0.039 |
| 9 | rf (tokens) | 0.661 | 0.591 | 0.624 | 0.628 | 0.557 | 0.590 | −0.034 |
| 9 | rf | 0.624 | 0.573 | 0.598 | 0.580 | 0.549 | 0.564 | −0.034 |
| 9 | nb (tokens) | 0.569 | 0.581 | 0.575 | 0.390 | 0.520 | 0.445 | −0.129 |
| 9 | logistic | 0.509 | 0.518 | 0.513 | 0.345 | 0.470 | 0.398 | −0.115 |
| 9 | nb | 0.478 | 0.537 | 0.506 | 0.331 | 0.454 | 0.383 | −0.123 |
| 20 | Sel[F] | 0.601 | 0.615 | 0.608 | ||||
| 20 | vote | 0.654 | 0.549 | 0.597 | ||||
| 20 | j48 | 0.644 | 0.532 | 0.583 | ||||
| 20 | LibSVM | 0.659 | 0.511 | 0.576 | ||||
| 20 | rf | 0.589 | 0.522 | 0.553 | ||||
| 20 | #1 j48 | 0.511 | 0.589 | 0.547 | ||||
| 20 | #2 j48 | 0.494 | 0.602 | 0.543 | ||||
Italics indicates the best results.
The vote (by average predictions) combined J48, LibSVM, Random Forest, k nearest neighbors, and Naïve Bayes. Tokens, including the tokens feature. The 9 situations are precisely ordered in descending precision, the 20 situations are approximately ordered. #1 and #2 were our official runs. Data in empty cells are unavailable.
c-v, cross-validation; Delta, difference in F-measure between test corpus and training corpus; F, F-measure; nb, Naïve Bayes; P, Precision; R, Recall; rf, Random Forest; #S, number of Situations; Sel, Selection of different classifiers for different situations, optimizing Precision [P] or F-measure [F].
We started to attempt the exploration of situations with lower OR, but so far increases in recall have not compensated associated decreases in precision. Investigating more features should be the next avenue. Failing to use word features for logistic regression also probably hindered its performance. Token features helped the classifiers which could handle the larger number of features, and we assume that a stronger Maximum Entropy classifier should be able to do so too.
Conclusion
In this paper, we presented the experiments we made to process the temporal relations between clinical events and temporal expressions as part of our participation in the 2012 i2b2/VA challenge.
We reused and adapted the Caramba and Ogmios platforms we developed during the 2010 and 2011 i2b2/VA editions to detect clinical events (F=0.83). These platforms integrate both rules and machine-learning processes. We identified event modality and polarity with Random Forest models (F=0.91), and temporal expressions through HeidelTime with adaptations to deal with clinical document specificities (F=0.84). Temporal relation detection relied on a rule-based division into a set of situations which were then handled by specific classifiers. Exhaustive enumeration of positive and negative instances at training time, token, syntax-based and combined features, voting, and oracle-recall-driven management of situations enabled us to improve our challenge system by 7.5pt of F-measure (F=0.62).
There are still many directions for enhancements. The CRF-based Caramba and rule-based Ogmios systems have complementary strengths. This raises the question of the best way to combine a data-driven system with a non-trainable system. Better normalization of Timex3s should not be too complex. For temporal relations, designing specific additional features for each identified prominent situation should be the natural path, together with switching to more scalable classifier implementations. Instead of a greedy decision procedure, a global decision procedure could be designed which would examine the graph of all predicted temporal relations, including conflicting ones, together with their prediction confidence, and would select a globally consistent sub-graph with a best overall prediction score. The characterization of the OR of situations allowed us to point at directions where further work would be most useful for temporal relations: within-sentence relations and linking HPI events to the AD. We believe that this OR should also help other teams addressing this task to identify their needs better.
Acknowledgments
This project was supported by Informatics for Integrating Biology and the Bedside (i2b2) award number 2U54LM008748 from the NIH/National Library of Medicine (NLM), by the National Heart, Lung and Blood Institute (NHLBI), and by award number 1R13LM01141101 from the NIH NLM. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NLM, NHLBI, or the National Institutes of Health.
Footnotes
Contributors: CG, NG, TH, SR, XT, and PZ jointly designed the experiments. NG and TH built the Ogmios system; CG, SR, and PZ built the Caramba system; XT and PZ built the Tlink detection system. All authors contributed to drafting the manuscript; CG, TH, and PZ revised the manuscript; all authors read and approved the final manuscript.
Funding: This work was supported by ANR grant number ANR-12-CORD-0007-03 (Accordys project) and has been partly done within the framework of the Quaero project funded by Oseo, French State Agency for Research and Innovation.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008:128–44 [PubMed] [Google Scholar]
- 2.Uzuner O, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harkema H, Setzer A, Gaizauskas R, et al. Mining and modelling temporal clinical data. Proceedings of the UK e-Science All Hands Meeting 2005, Nottingham, UK, 2005: 507–14 [Google Scholar]
- 4.Zhou L, Melton GB, Parsons S, et al. A temporal constraint structure for extracting temporal information from clinical narrative. J Biomed Inform 2006;39:424–39 Epub 29 Aug 2005 [DOI] [PubMed] [Google Scholar]
- 5.Savova G, Bethard S, Styler W, et al. Towards temporal relation discovery from the clinical narrative. AMIA Annu Symp Proc 2009:568–72 [PMC free article] [PubMed] [Google Scholar]
- 6.Sun W, Rumshisky A, Uzuner O. Evaluating Temporal Relations in Clinical Text: 2012 i2b2 Challenge. J Am Med Inform Assoc 2013;20:806–13 [DOI] [PMC free article] [PubMed]
- 7.Mykowiecka A, Marciniak M, Kupść A. Rule-based information extraction from patients’ clinical data. J Biomed Inform 2009;42:923–36 [DOI] [PubMed] [Google Scholar]
- 8.Friedman C, Shagina L, Lussier Y, et al. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc 2004;11:392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Wu YFB. Identifying important concepts from medical documents. J Biomed Inform 2006;39:668–79 [DOI] [PubMed] [Google Scholar]
- 10.Denecke K. Semantic structuring of and information extraction from medical documents using the UMLS. Methods Info Med 2008;47:425–34 [DOI] [PubMed] [Google Scholar]
- 11.Lindberg DA, Humphreys BL, McRay AT. The unified medical language system. Methods Info Med 1993;32:281–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grouin C, Ben Abacha A, Bernhard D, et al. CARAMBA: concept, assertion, and relation annotation using machine-learning based approaches. Proceedings of i2b2/VA Workshop, Washington, DC, 2010 [Google Scholar]
- 13.Minard AL, Ligozat AL, Ben Abacha A, et al. Hybrid methods for improving information access in clinical documents: concept, assertion, and relation identification. J Am Med Inform Assoc 2011;18:588–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grouin C, Dinarelli M, Rosset S, et al. Coreference resolution in clinical reports. The LIMSI participation in the i2b2/VA 2011 challenge. Proceedings of i2b2/VA Workshop, Washington, DC, 2011 [Google Scholar]
- 15.Strötgen J, Gertz M. Temporal tagging on different domains: challenges, strategies, and gold standards. Proceedings of LREC, Istanbul, Turkey, 2012 [Google Scholar]
- 16.Chang AX, Manning CD. SUTime: a library for recognizing and normalizing time expressions. Proceedings of LREC, Istanbul, Turkey, 2012 [Google Scholar]
- 17.Mani I. Recent developments in temporal information extraction. In: Nicolov N, Mitkov R, Proceedings of RANLP'03. Amsterdam, The Netherlands/Philadelphia, PA: John Benjamins, 2004: 45.– [Google Scholar]
- 18.Pustejovsky J, Knippen R, Littman J, et al. Temporal and event information in Natural Language Text. Language Resources and Evaluation. 2005;39:123–64. [Google Scholar]
- 19.Lascarides A, Asher N. Temporal interpretation, discourse relations and commonsense entailment, linguistics and philosophy. Linguist Philos 1993;16:437–93, Kluwer Academic Publishers, Dordrecht, Holland [Google Scholar]
- 20.Wang R, Zhang Y. Recognizing textual entailment with temporal expressions in natural language texts. Proceedings of IWSCA, Incheon, South Korea, 2008: 109–16 [Google Scholar]
- 21.Reichenbach H. Elements of symbolic logic. New York: The Macmillan Co, 1947 [Google Scholar]
- 22.Prior AN. Tense logic and the logic of earlier and later. In: Prior AN. Papers on time and tense. Oxford, UK: Oxford University Press, 1968: 117–38 [Google Scholar]
- 23.Song F, Cohen R. Tense interpretation in the context of narratives. Proceedings of AAAI, Anaheim, CA, 1991: 131–6 [Google Scholar]
- 24.Mani I, Wilson G. Robust temporal processing of news. Proceedings of ACL, Hong Kong 2000: 69–76 [Google Scholar]
- 25.Schilder F, Habel C. From temporal expressions to temporal information: semantic tagging of news messages. Proceedings of Workshop on Temporal and Spatial Information Processing, Toulouse, France, 2001: 65–72 [Google Scholar]
- 26.Pustejovsky J, Belanger L, Castaño J, et al. NRRC Summer Workshop on Temporal and Event Recognition for QA Systems. 2002.
- 27.Filatova E, Hovy EH. Assigning time-stamps to event-clauses. Proceedings of ACL Workshop on Temporal and Spatial Reasoning, Toulouse, France, 2001 [Google Scholar]
- 28.Mani I, Schiffman B, Zhang J. Inferring temporal ordering of events in news. Proceedings of HLTC, Edmonton, Canada, 2003 [Google Scholar]
- 29.Lapata M, Lascarides A. Learning sentence-internal temporal relations. J Artif Intell Res 2006;27:85–117 [Google Scholar]
- 30.Jung H, Allen J, Blaylock N, et al. Building timelines from narrative clinical records: initial results based-on deep natural language understanding. Proceedings of BioNLP, Portland, OR, 2011 [Google Scholar]
- 31.Raghavan P, Fosler-Lussier E, Lai AM. Learning to temporally order medical events in clinical text. Proceedings of 50th ACL, Jeju Island, South Korea, 2012;2:70–4 [Google Scholar]
- 32.Cherry C, Zhu X, Martin J, et al. A la Recherche du Temps Perdu-Extracting Temporal Relations from Medical Text in the 2012 i2b2 NLP Challenge. J Am Med Inform Assoc 2013. Published Online First 23 March 2013 [DOI] [PMC free article] [PubMed]
- 33.Tang B, Wu Y, Jiang M, et al. A hybrid system for temporal information extraction from clinical text. J Am Med Inform Assoc 2013;20:828–35 [DOI] [PMC free article] [PubMed]
- 34.Lavergne T, Cappé O, Yvon F. Practical very large scale CRFs. Uppsala, Sweden, 2010: 504–13 [Google Scholar]
- 35.Schmid H. Probabilistic part-of-speech tagging using decision trees. Proceedings of International Conference on New Methods in Language, Manchester, UK, 1994: 44–9 [Google Scholar]
- 36.McClosky D, Charniak E, Johnson M. Automatic domain adaptation for parsing. Proceedings of HLTC, Los Angeles, CA, 2010 [Google Scholar]
- 37.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32:267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galibert O. Approches et méthodologies pour la réponse automatique à des questions adaptées à un cadre interactif en domaine ouvert. PhD thesis, Université Paris-Sud, Orsay, 2009 [Google Scholar]
- 39.Hamon T, Grabar N. Linguistic approach for identification of medication names and related information in clinical narratives. J Am Med Inform Assoc 2010;17:549–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamon T, Périnet A, Nobécourt J, et al. Linguistic and semantic annotation for information extraction and characterization. Proceedings of i2b2/VA Workshop, Washington, DC, 2010 [Google Scholar]
- 41.Hamon T, Grabar N. Concurrent linguistic annotations for identifying medication names and the related information in discharge summaries. Proceedings of i2b2/VA Workshop, San Francisco, CA, 2009 [Google Scholar]
- 42.Brown PF, Della Pietra VJ, de Souza PV, et al. Class-based n-gram models of natural language. Comput Linguist 1992;18:467–79 [Google Scholar]
- 43.Liang P. Semi-supervised learning for natural language. Master's thesis, MIT, 2005 [Google Scholar]
- 44.Aubin S, Hamon T. Improving term extraction with terminological resources. In: Advances in NLP (5th International Conference on NLP, FinTAL), Turku, Finland, 2006 [Google Scholar]
- 45.Tsuruoka Y, Tateishi Y, Kim JD, et al. Developing a robust part-of-speech tagger for biomedical text. Proceedings of Advances in Informatics—10th Panhellenic Conference on Informatics, LNCS; Volos, Greece: Springer, 2005: 382–92 [Google Scholar]
- 46.National Library of Medicine, Bethesda, Maryland RxNorm, a standardized nomenclature for clinical drugs 2009. http://www.nlm.nih.gov/research/umls/rxnorm/docs/index.html (accessed 2 Apr 2013).
- 47.Hall MA, Frank E, Holmes G, et al. The WEKA data mining software: an update. SIGKDD Explor Newsl 2009;11:10–18 [Google Scholar]
- 48.Witten IH, Frank E, Hall MA. Data mining: practical machine learning tools and techniques. 3rd edn Amsterdam, The Netherlands: Morgan Kaufmann, 2011 [Google Scholar]
- 49.Verhagen M, Pustejovsky J. Temporal processing with the TARSQI Toolkit, Manchester, UK, 2008: 189–92 [Google Scholar]
- 50.Quinlan R. C4.5: Programs for machine learning. San Francisco, CA: Morgan Kaufman Publishers, 1993 [Google Scholar]