Abstract
Background
The Sixth Informatics for Integrating Biology and the Bedside (i2b2) Natural Language Processing Challenge for Clinical Records focused on the temporal relations in clinical narratives. The organizers provided the research community with a corpus of discharge summaries annotated with temporal information, to be used for the development and evaluation of temporal reasoning systems. 18 teams from around the world participated in the challenge. During the workshop, participating teams presented comprehensive reviews and analysis of their systems, and outlined future research directions suggested by the challenge contributions.
Methods
The challenge evaluated systems on the information extraction tasks that targeted: (1) clinically significant events, including both clinical concepts such as problems, tests, treatments, and clinical departments, and events relevant to the patient's clinical timeline, such as admissions, transfers between departments, etc; (2) temporal expressions, referring to the dates, times, durations, or frequencies phrases in the clinical text. The values of the extracted temporal expressions had to be normalized to an ISO specification standard; and (3) temporal relations, between the clinical events and temporal expressions. Participants determined pairs of events and temporal expressions that exhibited a temporal relation, and identified the temporal relation between them.
Results
For event detection, statistical machine learning (ML) methods consistently showed superior performance. While ML and rule based methods seemed to detect temporal expressions equally well, the best systems overwhelmingly adopted a rule based approach for value normalization. For temporal relation classification, the systems using hybrid approaches that combined ML and heuristics based methods produced the best results.
Keywords: clinical language processing, sharedtask challenges, temporal reasoning, natural language processing, medical language processing
Introduction
Understanding the clinical timeline is crucial in determining a patient's diagnosis and treatment. Narrative provider notes from electronic health records frequently detail important information on the temporal ordering of events in a patient's clinical timeline. Temporal analysis of the clinical narrative is therefore a prime target for developing automated natural language processing (NLP) techniques that allow computerized systems to access, reason about, and sequence the clinical events in a patient's record. Such techniques potentially enable or facilitate tracking disease status, monitoring treatment outcomes and complications, discovering medication side effects, etc.
While temporal information extraction and text based temporal reasoning in the clinical domain have attracted some recent attention,1–3 it has generally lagged behind similar work in the general English domain due to the lack of publicly available annotated corpora. In this paper, we present the 2012 Informatics for Integrating Biology and the Bedside (i2b2) Challenge on NLP for Clinical Records, which focuses on temporal relations. In particular, we describe the challenge tasks, data, and evaluation metrics. We then provide an overview of the systems developed for the challenge. We finally outline future directions for clinical temporal relations research that emerged in preparation of the shared task as well as in the analysis and evaluation of the challenge contributions.
In the 2012 i2b2 Challenge, 310 discharge summaries were annotated for temporal information. The challenge focused specifically on the identification of clinically relevant events in the patient records, and the relative ordering of the events with respect to each other and with respect to time expressions included in the records. This task was broken down into two steps, each corresponding to a separate track: (1) extraction of events and time expressions and (2) identification of temporal relations. We also established an end to end track that combined both steps to evaluate state of the art in system performance in temporal information extraction. Eighteen teams participated in the temporal relations challenge (see online supplementary appendix table 1). The results of the challenge were presented in a workshop that i2b2 organized in co-sponsorship with the American Medical Informatics Association (AMIA), at the Fall Symposium of AMIA in 2012.
The remainder of this paper is organized as follows: the section ‘Related work’, describes related work on temporal reasoning and temporal information extraction. The section ‘Data’, presents an overview of the annotation effort. The section ‘Methods’, outlines the methods, including the challenge tracks, inter-annotator agreement, and system evaluation metrics, and gives an overview of the systems in each track. The section ‘Results and discussion’, presents the results and analysis of system performance for each track, and finally, the section ‘Conclusions’, outlines some conclusions and future directions.
Related work
The 2012 i2b2 Challenge builds upon the annotation efforts from the past i2b2 challenges since 2007. The past challenges produced a series of annotation efforts which created ‘layered’ linguistic annotation over a set of clinical notes. These efforts included de-identification of private health information,4 document classification tasks of smoking5 and obesity status,6 extracting information related to medication,7 clinical concept extraction, clinical assertion classification,8 and coreference resolution.9 The 2012 i2b2 Challenge extends these annotations with a temporal layer to encourage the development of better temporal reasoning systems over clinical text using NLP techniques.
Advancement in natural language temporal reasoning research requires corpora annotated with temporal information. In the general domain, one of the better known efforts in text based temporal analysis annotation is the TimeBank corpus which contains 183 news articles and uses the TimeML annotation schema.10 11 TimeML annotation guidelines were used in three recent temporal analysis evaluation tasks in the SemEval competitions, TempEval-1, TempEval-2, and TempEval-3.12–14 As a result of these efforts, the TimeML annotation guidelines, which have been under development since 2002 and served as the basis of the current ISO-TimeML standard,10 have stabilized and reached maturity. TimeML denotes events, time expressions, and temporal relations by the EVENT, TIMEX3, and TLINK tags, respectively.
In the clinical domain, there have been some recent efforts to adapt TimeML annotation guidelines to clinical narratives.3 15 Although these corpora are mostly in pilot stage and size, they have proven the initial success in adopting TimeML style annotations to the clinical domain. In addition, researchers have also explored other alternatives to label temporal information in clinical text. Zhou et al2 proposed a temporal constraint structural representation that translates the temporal relations of events and time expressions in a narrative to temporal interval representation.16 Tao et al17 proposed a web ontology language, CNTRO, to describe temporal relations in clinical narratives. The consensus among these temporal representations3 15 2 17 is that the following elements are the most critical to capture: clinically related events, time expressions with some value normalization, and the temporal relations between entities (events and time expressions). Thus for the 2012 i2b2 Challenge, we modified the TimeML guidelines to emphasize these three aspects: events, temporal expressions, and temporal relations.
Among these three aspects, clinical event extraction is relatively well studied. The existing clinical temporal annotation schemas have a consensus on defining clinically relevant states, procedures, occurrences, and changes as events.3 15 17 Clinical concepts, such as problems, tests, and treatments, fall into these categories. The annotation guidelines for the challenge include all clinical concepts as events. In addition, as a patient's stay in a particular clinical department is also clinically relevant information that can be anchored to the timeline, our guidelines also include clinical departments as events. Clinically relevant events also include evidential words or phrases that indicate the source of information, and other clinically significant activity (such as admission, discharge, transfer, etc), the last of which are categorized as ‘occurrences’. Problems, treatments, and tests had previously been annotated for the 2010 i2b2 Challenge on relation extraction,8 and were re-used in the 2012 i2b2 Challenge on temporal relations.
Although the task of event detection and temporal relation classification for the general and clinical domains demand quite different methods, temporal expression (date, time, duration, and frequency) extraction and normalization in the clinical domain is not much different from that in the general domain, except for the medication dosage and frequency short hand widely used by clinical practitioners. In the general domain, the TempEval 2 shared tasks include a time expression detection track.13 The best performing temporal expression extraction system in TempEval 2 is HeidelTime18 which uses four sets of handcrafted rules to identify and classify temporal expressions.
Temporal relation classification tasks in the 2012 i2b2 Challenge required participants to classify two types of temporal relations: (1) for each event, its temporal relation with the section creation time (EVENT-SectionTime). The creation time of the clinical history section in the discharge summary is the time of admission, and the creation time of the hospital course section is the discharge time (refer to Sun and colleagues19 for more details); (2) temporal relations between events and between events and time expressions. In the general NLP domain, the TempEval Challenges 1 and 212 13 presented similar tasks for the TimeBank corpus of news articles.11 The TempEval Challenges included sub-tasks in which the systems identified each event's temporal relation with the document creation time. The TempEval Challenges further simplified the temporal reasoning task by restricting the potential temporally related entity pairs to the following groups: (a) temporal relations between EVENTs and TIMEX3s within the same sentence; (b) temporal relations between the main EVENTs in adjacent sentences; and (c) temporal relations between two EVENTs where one dominates the other. The results of the TempEval Challenges showed that the temporal relations between EVENTs and document creation time appeared easier to identify than other temporal relations. Further, temporal relations between entities within the same sentence were easier to identify than those between entities from neighboring sentences. In terms of identifying candidate temporal relation pairs, the temporal relation classification track in the 2012 i2b2 NLP Challenge is more complicated than TempEval 1 or 2 in that any two entities in a discharge summary can be a candidate pair to assign temporal relations to. In the medical NLP domain, several review articles exist that summarize and analyze previous work in temporal reasoning in clinical narratives.20 21 However, to our knowledge, until this challenge, there was no temporally annotated clinical corpus on this scale available for the medical informatics research community to access, study, and compare results on.
Data
The 2012 i2b2 temporal relations challenge data include 310 discharge summaries consisting of 178 000 tokens. The records came from Partners Healthcare and the Beth Israel Deaconess Medical Center. Two sections in these discharge summaries, the clinical history and the hospital course, were relatively dense in their characterization of temporal relations. Temporal relations challenge focused on these sections.
Annotation overview
We defined two types of annotations to capture the temporal information presented in the narrative notes:
(1) Events and temporal expressions
Clinically relevant events (denoted by the EVENT tag) were defined to include:
clinical concepts (problems, tests, and treatments, as defined in Uzuner et al8)
clinical departments (such as ‘surgery’ or ‘the main floor’)
evidentials (ie, events that indicate the source of the information, such as the word ‘complained’ in ‘the patient complained about …’), and
occurrences (ie, events that happen to the patient, such as ‘admission’, ‘transfer’, and ‘follow-up’).
Each EVENT has the following attributes: type (as categorized above), polarity (positive or negated events), and modality (indicating whether an event actually happens, is merely proposed, mentioned as conditional, or described as possible).
The temporal expressions, denoted by TIMEX310 11 tags, capturing dates, times, durations, and frequencies,10 11 have three attributes: type (as given above), value, and modifier. The value attribute is a normalized value based on the ISO8601 standard that quantifies the temporal expression.22 The modifier attribute specifies whether a temporal value is exact or not (eg, we can use a modifier value ‘approximate’ to describe a time expression such as ‘several days’). For example, the value field of the TIMEX3 ‘more than 3 h’ is ‘PT3H’ (a period of 3 h) with modifier ‘more’ (ie, ‘more than’).
Temporal relations
Temporal relations, or temporal links (denoted by the TLINK tag), indicate whether and how two EVENTs, two TIMEX3s, or an EVENT and a TIMEX3 related to each other in the clinical timeline. Possible TLINK types were BEFORE, AFTER, SIMULTANEOUS, OVERLAP, BEGUN_BY, ENDED_BY, DURING, and BEFORE_OVERLAP. Below are some examples of TLINKs, with square brackets indicating EVENT and TIMEX3 connected by a temporal link:
BEFORE: The patient was given stress dose steroids prior to his surgery. ([stress dose steroids] BEFORE [his surgery])
AFTER: Before admission, he had another serious concussion. ([admission] AFTER [another serious concussion])
SIMULTANEOUS: The patient's serum creatinine on discharge date, 2012-05-06, was 1.9. ([discharge date] SIMULTANEOUS [2012-05-06])
OVERLAP: She denies any fevers or chills. ([fevers] OVERLAP [chills])
BEGUN_BY: On postoperative day No 1, he was started on Percocet. ([Percocet] BEGUN_BY [postoperative day No 1])
ENDED_BY: His nasogastric tube was discontinued on 05-26-98. ([His nasogastric] ENDED_BY [05-26-98])
DURING: His preoperative workup was completed and included a normal white count ([a normal white count] DURING [His preoperative workup])
BEFORE_OVERLAP: The patient had an undocumented history of possible atrial fibrillation prior to admission. ([possible atrial fibrillation] BEFORE_OVERLAP [admission])
Annotation effort
The annotation guidelines19 were adapted from TimeML10 to suit clinical data. The intermediate version of the THYME project guidelines was used as a starting point for the adaptation. The discharge summaries utilized for the 2012 Temporal Relations Challenge had previously been used for the 2010 i2b2 Challenge on relation extraction, and already included concepts, assertions, and relations annotations. In the 2011 i2b2 Challenge,9 these discharge summaries had been annotated for coreference resolution. Concepts from the 2010 i2b2 Challenge8 and coreference relations from the 2011 i2b2 Challenge provided a starting point for the 2012 temporal relations annotations. Concepts from the 2010 i2b2 Challenge were included in the EVENTs of the temporal relations challenge. TimeML handles the temporal relation between coreference EVENTs with the ‘IDENTITY’ TLINK type. In our guidelines, as an effort to simply the annotation, given their identical representation in interval algebra, we merged the ‘IDENTITY’ and ‘SIMULTANEOUS’ TLINK types. As a result, the coreference relation between EVENTs from the 2010 i2b2 Challenge were included as the ‘SIMULTANEOUS’ TLINKs in the 2012 corpus. The remaining of the 2012 i2b2 annotation effort included identifying EVENTs that were not annotated in the 2010 annotation, finding and normalizing temporal expressions, and identifying the TLINKs among EVENTs and TIMEX3s. More details on the annotation of the 2012 corpus can be found in Sun et al.19
Data statistics
Our annotations showed that an average note contains 86.6 EVENTs, 12.4 TIMEX3s, and 176 TLINKs. The online supplementary appendix table 2 shows more statistics of the annotated corpus. Our analysis demonstrated that agreement on some TLINKs was low (see section ‘Inter-annotator agreement’ below). In response to this observation, we merged the seven TLINK types as follows: BEFORE, ENDED_BY, and BEFORE_OVERLAP were merged as BEFORE; BEGUN_BY and AFTER were merged as AFTER; and SIMULTANEOUS, OVERLAP, and DURING were merged as OVERLAP. Both unmerged and merged annotations are available at http://i2b2.org/NLP/DataSets.
Inter-annotator agreement
Eight annotators, four of whom have medical background, took part in the annotation task. Each note was dually annotated and then adjudicated by a third annotator. The inter-annotator text span agreements on EVENTs, TIMEX3s, and TLINKs, before and after merge of the TLINKS, are shown in table 1. We report the inter-annotator agreement using both ‘exact matching’ and ‘partial matching’ criteria. For EVENT and TIMEX3s, under ‘exact matching’, an entity span agreement occurs only if the two annotators mark the exact same text span. In contrast, under ‘partial matching’, as long as the text spans that the two annotators marked overlap, it is considered a match. For example, if one annotator marked ‘a severe headache’ as an EVENT and the other marked ‘headache’ as an EVENT, it is considered an agreement under ‘partial matching’ and a disagreement under ‘exact matching’. For TLINK annotation, under ‘exact matching’, a TLINK is considered a match only if both entities’ spans agree under EVENT/TIMEX3 ‘exact matching’ criteria. Under ‘partial match’, a TLINK is considered a match if both entities’ spans agree under EVENT/TIMEX3 ‘partial matching’ criteria.
Table 1.
Inter-annotator agreement
| EVENT | ||
| Exact match | Partial match | |
| Span | 0.83 | 0.87 |
| Average precision and recall | Kappa | |
| Type | 0.93 | 0.9 |
| Modality | 0.96 | 0.37 |
| Polarity | 0.97 | 0.74 |
| TIMEX3 | ||
| Exact match | Partial match | |
| Span | 0.73 | 0.89 |
| Average precision and recall | Kappa | |
| Type | 0.9 | 0.37 |
| Val | 0.75 | – |
| Mod | 0.83 | 0.21 |
| TLINK | ||
| Exact match | Partial match | |
| Span | 0.39 | – |
| Average precision and recall | Kappa | |
| Type | 0.79 | 0.3 |
Methods
Challenge tracks
The temporal relations challenge included three tracks:
EVENT/TIMEX3 track: This track included the identification of EVENTs and TIMEX3s with their spans and all of their attributes from raw discharge summaries.
TLINK track: In this track, the released inputs included the EVENT tags, with their type, polarity, and modality attributes, and TIMEX3 tags, with their type, value, and modifier attributes. Systems identified the temporal relations between the EVENTs and TIMEX3s.
End-to-End track: This track required the systems to use raw discharge summaries to first find EVENTs and TIMEX3s, and then the TLINKs between them.
We evaluated systems on each of the tracks separately.
Inter-annotator agreement metrics
We calculated the inter-annotator agreement using average precision and recall—that is, by holding one annotation as the gold standard and measuring the precision of the other annotation and then doing the reverse to get the average. More specifically, for EVENT/TIMEX3 span detection, we measured the average of the percentage of entities in one annotation that can be verified in the other annotation. For EVENT/TIMEX3 attributes, we measured the average of the percentage of attributes (in one annotation) of entities that appear in both annotations, which can be verified in the other annotation. For TLINK classification, we measured the average of the percentage of TLINKs in one annotation that can be verified in the closure of the other annotation. Closure takes transitivity of relations into account. For example, if the annotation indicates that EVENT A happens at the same time as EVENT B, and EVENT B happens at the same time as EVENT C, transitive closure (TC) adds the TLINK indicating that EVENT A happens at the same time as EVENT C.
Evaluation metrics
The two subtasks of the EVENT/TIMEX3 track were evaluated independently, with separate scores reported for EVENT and TIMEX3 extraction.
The EVENT extraction task requires detecting the spans of EVENTs and identifying their attributes. We used the F measure, the harmonic mean of precision and recall of the system output against the gold standard to evaluate EVENT span detection performance. For EVENT attributes, we calculated classification accuracy—that is, the percentage of correctly identified EVENT attributes for the EVENTs whose spans are identified correctly. The primary evaluation metric for EVENT extraction is the span F measure. We report the accuracy for type, modality, and polarity attributes for completeness.
 |
The TIMEX3 extraction requires span detection, attribute identification, and value normalization. TIMEX3s values attributes need to be normalized to ISO8601 standards. We used the F measure of the TIMEX3 span detection multiplied by the accuracy of the value field as the primary metric for TIMEX3 extraction evaluation. We report the accuracy of modifier and type attributes separately for completeness.
For the TLINK classification task, F measure was used as the primary evaluation metric. Prior to evaluation, we compute the TC of the TLINKS provided by the system and the TC of the TLINKs found in the gold standard. The precision of the system output is the percentage of system TLINKs that can be verified in the TC of the gold standard TLINKs. Recall of the system output is the percentage of gold standard TLINKs that can be verified in the TC of the system TLINK output. We adapted the TLINK evaluation script for TempEval3 by UzZaman and Allen24 to compute temporal closure in our TLINK evaluation.
For the end to end track, the F measure of TLINK extraction served as the primary evaluation metric. The evaluation scores for EVENTs and TIMEX3s are reported for completeness.
Systems
Eighteen teams took part in the 2012 i2b2 Temporal Relations Challenge. After 2 months of development, the systems were evaluated on held-out test data. Each team submitted up to three system runs per track, and was ranked on their best performing system. Overall, 76 system runs were ranked, covering a wide range of machine learning (ML), rule based, and hybrid approaches.
EVENT/TIMEX3 track
For EVENT span detection, nine of the top 10 systems used conditional random fields (CRFs), a statistical modeling method for sequential data labeling.25 For EVENT attribute classification, most systems used support vector machines (SVM).26 Some teams were able to utilize and improve their concept detection systems from the 2010 i2b2 Challenge for EVENT detection.27 28 The input features for these systems included the Unified Medical Language System,29 the output from TARSQI tool kit,30 and Brown clustering31 of extended text resources.32 Yu-Kai Lin et al33 utilized Wikipedia and MetaMap34 to extract semantic features of medical terms. Other medical domain knowledge tools such as cTAKES35 were also utilized by some of the participants.36 37
All of the top 10 systems incorporated rule based temporal expression detection and/or normalization into their systems. Four of the top 10 systems used hybrid approach combining ML (CRF or SVM) and rules28 32 33 38 39 40 for this purpose. Rules from HeidelTime,18 the best performing rule based TIMEX3 detection system from TempEval2,13 were adopted by some teams.27 33 41 Other third party temporal expression taggers, SUTIME42 and GUTIME,30 were utilized by others.38 Despite some success in utilizing existing systems, most teams developed their own frequency detection and normalization components.
TLINK track
In the TLINK classification track, there was a larger variation in ML methods from maximum entropy (MaxEnt), Bayesian, and SVM to CRF, explored by the participants. Some teams also incorporated heuristics and rule based components in their systems. For example, Cherry et al43 separated the task into four sub-tasks: anchoring EVENTs against the admission/discharge date (EVENT to section time TLINKs), detecting relations between EVENTs/TIMEX3s within the same sentence (intra-sentence TLINKs), assigning ‘OVERLAP’ relation to EVENTs in different sentences (cross sentence TLINKs), and determining causal relation induced TLINKs. Tang et al27 used heuristics in selecting candidate entity pairs to assign TLINKs to. Chang et al44 integrated the results from a rule based TLINK extraction component that looks for sentence internal, cross sentence, and section time related TLINKS, and a MaxEnt component that first detects candidate pairs and then assigns TLINK types to them. Nikfarjam et al45 also utilized a rule based component alongside their SVM based system. Some teams divided the tasks into even more specific sub-tasks for improved performance.28 41
Results and discussion
EVENT extraction
Performance of the top 10 systems on EVENT extraction is shown in the first section of table 2. The average F measure of the 10 teams is 0.8716, with an SD of 0.0296. Compared with the average F measure of 0.9009 with an SD of 0.0119 in the 2010 i2b2 clinical concept extraction task, this year's EVENT detection appears to be more challenging. This is due to the addition of three new EVENT types: evidential, occurrence, and clinical department. In particular, the evidential and occurrence EVENT types seem more difficult to detect than other EVENT types. Nevertheless, the best result of EVENT extraction (0.9166) is very close to the best result from the 2010 concept extraction task (0.9244).
Table 2.
System results for EVENT, TIMEX, and TLINK tracks
| Organization | Span F measure | Type accuracy | Polarity accuracy | Modality accuracy | Method | ||
|---|---|---|---|---|---|---|---|
| EVENT | |||||||
| Beihang University; Microsoft Research Asia, Beijing; Tsinghua University | 0.92 | 0.86 | 0.86 | 0.86 | CRF | ||
| Vanderbilt University | 0.9 | 0.84 | 0.85 | 0.83 | CRF + SVM | ||
| The University of Texas, Dallas | 0.89 | 0.8 | 0.85 | 0.84 | CRF+SVM | ||
| The University of Texas, Dallas—deSouza | 0.88 | 0.71 | 0.85 | 0.05 | CRF | ||
| University of Arizona, Tucson | 0.88 | 0.73 | 0.79 | 0.8 | CRF+SVM+NegEx | ||
| University of Novi Sad, Novi Sad, Serbia; University of Manchester | 0.87 | 0.82 | 0.79 | 0.82 | CRF+dictionary based | ||
| Siemens Medical Solutions | 0.86 | 0.71 | 0.78 | 0.77 | CRF+MaxEnt | ||
| MAYO Clinic | 0.85 | 0.76 | 0.75 | 0.76 | CRF | ||
| LIMSI–CNRS; INSERM; STL CNRS; LIM&BIO | 0.83 | 0.8 | 0.84 | 0.85 | CRF+SVM | ||
| University of Illinois at Urbana-Champaign | 0.83 | 0.74 | 0.75 | 0.77 | Integer Quadratic Program | ||
| Organization | Primary score– Value F-measure | Span F measure | Type accuracy | Value accuracy | Modifier accuracy | Method | |
| TIMEX3 | |||||||
| MAYO Clinic | 0.66 | 0.9 | 0.86 | 0.73 | 0.86 | Regular Exp | |
| Beihang University; Microsoft Research Asia, Beijing; Tsinghua University | 0.66 | 0.91 | 0.89 | 0.72 | 0.89 | CRF+SVM+rule based | |
| University of Novi Sad, Novi Sad, Serbia; University of Manchester | 0.63 | 0.9 | 0.85 | 0.7 | 0.83 | Rule based | |
| Vanderbilt University | 0.61 | 0.87 | 0.85 | 0.7 | 0.85 | Rule based+HeidelTime | |
| University of Arizona, Tucson | 0.61 | 0.88 | 0.81 | 0.69 | 0.8 | HeidelTime+CRF | |
| The University of Texas, Dallas | 0.55 | 0.89 | 0.78 | 0.62 | 0.79 | CRF+SVM+rule based | |
| Siemens Medical Solutions | 0.53 | 0.89 | 0.86 | 0.6 | 0.8 | SUTime | |
| The University of Texas, Dallas—deSouza | 0.53 | 0.89 | 0.78 | 0.59 | 0.79 | GUTime+CRF+rule base | |
| Bulgarian Academy of Sciences; American University in Bulgaria; University of Colorado School of Medicine | 0.49 | 0.8 | 0.72 | 0.61 | 0.71 | Regular Exp | |
| LIMSI–CNRS; INSERM; STL CNRS; LIM&BIO | 0.45 | 0.84 | 0.75 | 0.54 | 0.72 | HeidelTime | |
| Organization | F measure | Precision | Recall | Method | |||
| TLINK | |||||||
| Vanderbilt University | 0.69 | 0.71 | 0.67 | Rule based pair selection+CRF+SVM | |||
| National Research Council Canada | 0.69 | 0.75 | 0.64 | MaxEnt+SVM+rule based | |||
| Beihang University; Microsoft Research Asia, Beijing; Tsinghua University | 0.68 | 0.66 | 0.71 | SVM | |||
| Arizona State University | 0.63 | 0.76 | 0.54 | SVM+rule-based | |||
| The University of Texas, Dallas—deSouza | 0.61 | 0.54 | 0.72 | CRF | |||
| University of California, San Diego; Department of Veterans Affairs, Tennessee Valley Healthcare System | 0.59 | 0.65 | 0.54 | MaxEnt/Bayes | |||
| Academia Sinica; National Taiwan University; Institute For Information Industry; Yuan Ze University | 0.56 | 0.57 | 0.56 | Rule based+MaxEnt | |||
| The University of Texas, Dallas | 0.56 | 0.48 | 0.66 | SVM | |||
| LIMSI–CNRS; INSERM; STL CNRS; LIM&BIO | 0.55 | 0.51 | 0.59 | SVM | |||
| Brandeis University | 0.43 | 0.34 | 0.59 | MaxEnt | |||
CRF, conditional random field; MaxEnt, maximum entropy; SVM, support vector machines.
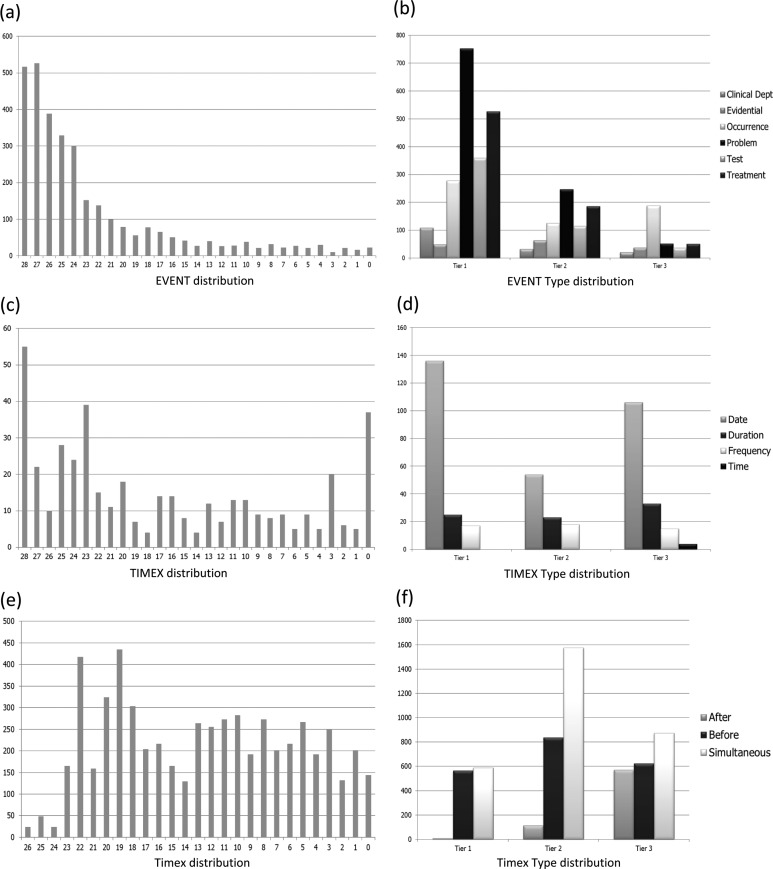
In order to better understand system strengths and weaknesses, we randomly selected 25% of the test data from this year's challenge and analyzed system outputs in this sample. A total of 28 submissions from the top 10 teams were included in this analysis. Figure 1A shows the distribution of EVENT counts against the number of submissions in which the EVENT is correctly identified. The x axis shows the number of submissions that correctly identified the EVENT and the y axis shows the EVENT count in the corresponding bar. For example, the first bar indicates that 517 EVENTs in the sample records were correctly identified by all 28 submissions. Figure 1A shows that about 64% of all EVENTs in the sample set were correctly discovered by most systems (24 or more systems out of the total 28 systems); about 24% of the EVENTs were recognized by some systems (15 or more out of the 28 systems); and only less than half of the submissions correctly identified the remaining 12% of the EVENTs. The EVENT type distribution of the above three groups is shown in figure 1B.
Figure 1.
System result analysis. (A) EVENT distribution. (B) EVENT type distribution. (C) TIMEX distribution. (D) TIMEX type distribution. (E) Timex distribution. (F) Timex type distribution.
Most of the easier identifiable EVENTs were clinical concepts— that is, problems, treatments, and tests. In particular, spelled out clinical concepts such as ‘hypothyroidism’ and ‘the cardiology department’ are better detected than acronyms, such as ‘UCx’ and ‘ACD’. Coreference of clinical concepts in pronoun form also proves to be challenging to detect. Evidential and occurrence EVENTs appear to be more difficult to identify. Among these EVENTs, high frequency evidential and occurrence EVENTs, such as ‘report’, ‘admission’, and ‘discharge’ are among the best detected group, while less frequently appearing EVENTs, such as ‘dried’ and ‘plateaued’, are less effectively detected.
TIMEX3 extraction
The second section of table 2 displays the system scores for the TIMEX3 extraction task. The primary score, the product of value field accuracy and the F measure, averaged for the top 10 teams, is 0.5701, with an SD of 0.0721. The best system achieved a span F measure of 0.9003 and a value accuracy of 0.7291.
Figure 1 (C, D) shows the distributions of TIMEX3 count and TIMEX3 types against the number of submissions that correctly identified and normalized them. We found that well formatted calendar dates— for example, ‘5/24/2001’ and ‘Jan 14, 2002’—are easy to identify and normalize. Calendar dates make up the majority of the date TIMEX3s identified correctly by most systems. The date TIMEX3s that turn out to be challenging to recognize are relative dates, such as ‘the next morning’ and ‘hospital day number five’. The results for the duration and frequency TIMEX3s follow the same pattern—that is, well formatted, absolute durations and frequencies are easier to identify, while durations and frequencies without standard formats which require inference to normalize are more challenging.
TLINK extraction
The last section of table 2 displays the system scores for the TLINK task. The average F measure of the top ten teams is 0.5997, with an SD of 0.0822. The best system achieved a TLINK extraction F measure of 0.6932.
Analysis of the 25% sample records shows that the recognition of EVENT to section time TLINKs in general is easier than the recognition of other types of TLINKs (EVENT-EVENT, EVENT-TIMEX3, TIMEX3-TIMEX3, and TIMEX3-EVENT); 45.87% of the TLINKs in the sample gold standard records anchor EVENTs to section time. On average, 19.98 submissions out of the total 28 submissions correctly identified EVENT to section time TLINKs while only 11.92 out of 28 submissions correctly identified the other type of temporal relations. Among the non-section time TLINKs, EVENT-TIMEX3 and EVENT-EVENT relations were easier to detect, while TIMEX3-TIMEX3 and TIMEX3-EVENT were more challenging. Further analysis of the TLINKs suggests that this is likely due to the fact that many of the TIMEX3-TIMEX3 and TIMEX3-EVENT temporal relations involve the anchoring of relative dates and durations, a problem that is consistent with a similar issue in TIMEX3 extraction.
For EVENT to section time TLINKs, performance figures for the three TLINK types—BEFORE, AFTER, and OVERLAP—are fairly similar. Each EVENT to section time TLINK of type BEFORE is recognized on average by 19.59 submissions out of 28. Each AFTER TLINK is recognized on average by 20.12 out of 28 systems. The OVERLAP section time TLINKs is recognized on average by 18.46 submissions. Among the non-section time TLINKs, the OVERLAP TLINKs, which comprise 59.22% of all non-section time TLINKs, are recognized best, with an average of 14.44 out of 28 submissions recognizing each such TLINK. BEFORE relations, which account for 26.04% of all non-section time TLINKs, and AFTER relations, which account for 14.73% of all non-section time TLINKs, are recognized on average by 9.47 and 6.14 systems, respectively (table 3).
Table 3.
TLINK task—system performance analyzed by TLINK entity type
| % in all TLINKs | Average No of correct submissions (%) | |
|---|---|---|
| EVENT—section time TLINKs | 45.87 | 19.98 |
| Non–section time TLINKs | ||
| EVENT-EVENT | 42.07 | 11.87 |
| EVENT-TIMEX3 | 9.64 | 12.78 |
| TIMEX3-TIMEX3 | 0.80 | 8.04 |
| TIMEX3-EVENT | 1.62 | 10.62 |
End to end track
The results of the end-to-end track are shown in table 4.
Table 4.
Top 7 systems in end to end track
| Primary score (F measure of TLINK) | F measure EVENT span | F measure of TIMEX3 value | |
|---|---|---|---|
| Vanderbilt University | 0.6278 | 0.9011 | 0.8607 |
| Beihang University; Microsoft Research Asia, Beijing; Tsinghua University | 0.5924 | 0.9166 | 0.9098 |
| The University of Texas, Dallas | 0.5258 | 0.8933 | 0.8907 |
| The University of Texas, Dallas—deSouza | 0.5126 | 0.8835 | 0.8886 |
| LIMSI–CNRS; INSERM; STL CNRS; LIM&BIO | 0.4932 | 0.8307 | 0.8385 |
| MAYO Clinic | 0.3741 | 0.8548 | 0.8999 |
| University of Novi Sad, Novi Sad, Serbia; University of Manchester | 0.3448 | 0.8611 | 0.8607 |
Conclusions
In the i2b2 2012 Temporal Relation NLP Challenge, we created a clinical temporal relation corpus that includes clinical EVENTs, temporal expressions, and temporal relations. Eighteen teams from all over the world participated and achieved very encouraging performance in all tracks of the challenge. The results of the challenge provide some necessary guidance for understanding the current state of the art in clinical temporal analysis. It also reveals some of the remaining problems in each of the component tasks:
Events. Clinically relevant events that proved hardest to detect for the majority of systems were acronyms and anaphoric expressions, suggesting that better coreference resolution and acronym handling may improve the results.
Time expressions. Relative time normalization remains a challenging problem for most systems, indicating that context aware temporal expression understanding requires further research.
Temporal relations. Identification of candidate entity pairs, as well as relative time anchoring—both prerequisites for a full scale temporal reasoning system—are presently not well addressed by the current state of the art. Future research on these topics may help advance the accuracy of temporal reasoning in the clinical domain.
Acknowledgments
The authors would like to thank the organizing committee members of the 2012 i2b2 Challenge, including Peter Szolovits, Lynette Hirschman, Cheryl Clark, John Aberdeen, Martha Palmer, Guergana Savova, and James Pustejovsky, for their valuable advice and kind help in organizing this shared task challenge.
Correction notice: This article has been corrected since it was published Online First. Reference 26 has been corrected.
Ethics approval: The data use portion of this work was approved by the institutional review boards at the Massachusetts Institute of Technology, Partners Healthcare, and SUNY Albany.
Contributors: WS is the primary author. WS played the key role in the preparation of the 2012 i2b2 Natural Language Processing (NLP) Challenge, which included leading the data analysis and managing the annotation and preparation of the 2012 corpus, under the guidance of AR. AR and WS developed annotation guidelines and evaluation strategies for the 2012 i2b2 NLP Challenge. AR and OU are the primary organizers of the 2012 i2b2 NLP Challenge, offered insights and guidance from data preparation to result analysis, and provided substantial edits to the manuscript. OU led the 2012 i2b2 Challenge Workshop.
Funding: This project was supported by Informatics for Integrating Biology and the Bedside (i2b2) award No 2U54LM008748 from the NIH/National Library of Medicine (NLM), by the National Heart, Lung, and Blood Institute (NHLBI), and by award No 1R13LM01141101 from the NIH NLM. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NLM, NHLBI, or the National Institutes of Health.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The data developed for the challenge were made available to the challenge participants and will be made available to the community at large a year after the challenge.
References
- 1.Harkema H, Setzer A, Gaizauskas R, et al. Mining and modelling temporal clinical data. Nottingham, UK: Proceedings of the UK e-Science All Hands Meeting, Vol 2005; 2005:507–14 [Google Scholar]
- 2.Zhou L, Melton GB, Parsons S, et al. A temporal constraint structure for extracting temporal information from clinical narrative. J Biomedical Inform 2006;39: 424–39 [DOI] [PubMed] [Google Scholar]
- 3.Savova G, Bethard S, Styler W, et al. Towards temporal relation discovery from the clinical narrative. San Francisco, CA, USA: AMIA Annual Symposium Proceedings, Vol 2009 American Medical Informatics Association, 2009:568. [PMC free article] [PubMed] [Google Scholar]
- 4.Uzuner Ö, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007;14:550–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uzuner Ö, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008;15:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uzuner Ö. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc 2009;16:561–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18: 552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uzuner O, Bodnari A, Shen S, et al. Evaluating the state of the art in coreference resolution for electronic medical records. J Am Med Inform Assoc 2012;00 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauri R, Littman J, Knippen B, et al. TimeML annotation guidelines, V.1.2.1, 2005. Available at: http://www.timeml.org/timeMLdocs/AnnGuide14.pdf
- 11.Pustejovsky J, Hanks P, Sauri R, et al. The timebank corpus. Proceeding of Corpus Linguistics, Lancaster University, UK, 2003, 647–56.
- 12.Verhagen M, Gaizauskas R, Schilder F, et al. Semeval-2007 task 15: tempeval temporal relation identification. Prague, Czech Republic: Proceedings of the 4th International Workshop on Semantic Evaluations 2007:75–80
- 13.Verhagen M, Sauri R, Caselli T, et al. SemEval-2010 task 13: TempEval-2. Uppsala, Sweden: Proceedings of the 5th International Workshop on Semantic Evaluation. Association for Computational Linguistics, 2010:57–62
- 14.UzZaman N, Llorens H, Allen J, et al. TempEval-3: evaluating events, time expressions, and temporal relations. arXiv preprint arXiv:12065333 2012. Available at http://arxiv.org/pdf/1206.5333.pdf
- 15.Galescu L, Blaylock N. A corpus of clinical narratives annotated with temporal information. Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium. IHI '12. New York, NY, USA: ACM, 2012:715–20 [Google Scholar]
- 16.Vilain M, Kautz H. Constraint propagation algorithms for temporal reasoning. Philadelphia, PA, USA: Proceedings of the Fifth National Conference on Artificial Intelligence; 1986:377–82
- 17.Tao C, Wei WQ, Solbrig HR, et al. CNTRO: a semantic web ontology for temporal relation inferencing in clinical narratives. Washington DC, USA: AMIA Annual Symposium Proceedings; Vol 2010 American Medical Informatics Association, 2010:787. [PMC free article] [PubMed] [Google Scholar]
- 18.Strotgen J, Gertz M. HeidelTime: high quality rule-based extraction and normalization of temporal expressions. Proceedings of the 5th International Workshop on Semantic Evaluation; 2010:321–4
- 19.Sun W, Rumshisky A, Uzuner O, et al. Annotating temporal information in clinical narratives. 2013 (submitted) [DOI] [PMC free article] [PubMed]
- 20.Zhou L, Hripcsak G. Temporal reasoning with medical data—a review with emphasis on medical natural language processing. J Biomed Inform 2007; 40:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008;35:128–44 [PubMed] [Google Scholar]
- 22.ISO 8601:2004 Data elements and interchange formats—Information interchange—Representation of dates and times. http://www.iso.org/iso/catalogue_detail?csnumber=40874 (accessed 20 Jan 2012)
- 23.Temporal Histories of Your Medical Events, THYME. https://clear.colorado.edu/TemporalWiki/index.php/Main_Page (accessed 29 Feb 2012)
- 24.UzZaman N, Allen JF. Temporal evaluation. Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies (Short Paper); PA, USA: Association for Computational Linguistics Stroudsburg, 2011;2:351–6 [Google Scholar]
- 25.Lafferty J, McCallum A, Pereira F. Conditional random fields: probabilistic models for segmenting and labeling sequence data. Proceeding of the 18th International Conf. on Machine Learning. San Francisco, CA, US: Morgan Kaufmann Publishers Inc. 2001, 282–9
- 26.Cortes C, Vapnik V. Support-vector networks. Mach Learning 1995;20:273–97. [Google Scholar]
- 27.Tang B, Wu Y, Jiang M, et al. A hybrid system for temporal information extraction from clinical text. J Am Med Inform Assoc 2013;20:828–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Wang Y, Liu T, et al. An end-to-end system to identify temporal relation in discharge summaries: 2012 i2b2 challenge. J Am Med Inform Assoc 2013;20:849–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004;32(Suppl 1):D267–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhagen M, Mani I, Sauri R, et al. Automating temporal annotation with TARSQI. Stroudsburg, PA, USA: Proceedings of the ACL 2005 on Interactive Poster and Demonstration Sessions. Association for Computational Linguistics 2005:81–4
- 31.Brown PF, Desouza PV, Mercer RL, et al. Class-based n-gram models of natural language. Comput Linguistics 1992;18:467–79 [Google Scholar]
- 32.Roberts K, Rink B, Harabagiu S. A flexible framework for recognizing events, temporal expressions, and temporal relations in clinical text. J Am Med Inform Assoc 2013;20:867–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin YK, Chen H, Brown RA. MedTime: A Temporal Information Extraction System for Clinical Narratives. Journal of Biomedical Informatics. 2013 [DOI] [PubMed]
- 34.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings of the AMIA Symposium. 2001:17–21. [PMC free article] [PubMed]
- 35.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovačeviç A, Dehghan A, Filannino M, et al. Combining rules and machine-learning for extraction of temporal expressions and events from clinical narratives. J Am Med Inform Assoc 2013 (in press) [DOI] [PMC free article] [PubMed]
- 37.Sohn S, Wagholikar K, Li D, et al. Comprehensive temporal information discovery from discharge summaries: medical events, time, and TLINK identification. J Am Med Inform Assoc. 2013 (in press) [DOI] [PMC free article] [PubMed]
- 38.D'Souza J, Ng V. Classifying Temporal Relations in Clinical Data: A Hybrid, Knowledge-Rich Approach. Journal of Biomedical Informatics. 2013 [DOI] [PMC free article] [PubMed]
- 39.Guillen R. Rule-Based Methodology to Identify and Classify Temporal Expressions. Journal of Biomedical Informatics 2013 (in press)
- 40.Prateek J, Dan R. Timexes and Events Extraction With Global Inference for Clinical Narratives. J Biomed Inform 2013 (in press)
- 41.Grouin C, Grabar N, Hamon T, et al. Eventual Situations for Timeline Extraction from Clinical Reports. J Am Med Inform Assoc 2013;20:820–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang AX, Manning CD. SUTIME: a library for recognizing and normalizing time expressions. Language resources and evaluation. 2012 [Google Scholar]
- 43.Cherry C, Zhu X, Martin J, et al. A la Recherche du Temps Perdu—extracting temporal relations from medical text in the 2012 i2b2 NLP challenge. J Am Med Inform Assoc 2013;20:843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang Y-C, Wu JC-Y, Chen J-M, et al. TEMPTing System: A Hybrid Method of Rule and Machine Learning for Temporal Relation Extraction in Patient Discharge Summaries. Journal of Biomedical Informatics 2013 (in press) [DOI] [PubMed]
- 45.Nikfarjam A, Emadzadeh E, Gonzalez G, et al. Towards Generating Patient's Timeline: Extracting Temporal Relationships from Clinical Notes. Journal of Biomedical Informatics 2013 (in press) [DOI] [PMC free article] [PubMed]