Abstract
Objective
An accurate computable representation of food and drug allergy is essential for safe healthcare. Our goal was to develop a high-performance, easily maintained algorithm to identify medication and food allergies and sensitivities from unstructured allergy entries in electronic health record (EHR) systems.
Materials and methods
An algorithm was developed in Transact-SQL to identify ingredients to which patients had allergies in a perioperative information management system. The algorithm used RxNorm and natural language processing techniques developed on a training set of 24 599 entries from 9445 records. Accuracy, specificity, precision, recall, and F-measure were determined for the training dataset and repeated for the testing dataset (24 857 entries from 9430 records).
Results
Accuracy, precision, recall, and F-measure for medication allergy matches were all above 98% in the training dataset and above 97% in the testing dataset for all allergy entries. Corresponding values for food allergy matches were above 97% and above 93%, respectively. Specificities of the algorithm were 90.3% and 85.0% for drug matches and 100% and 88.9% for food matches in the training and testing datasets, respectively.
Discussion
The algorithm had high performance for identification of medication and food allergies. Maintenance is practical, as updates are managed through upload of new RxNorm versions and additions to companion database tables. However, direct entry of codified allergy information by providers (through autocompleters or drop lists) is still preferred to post-hoc encoding of the data. Data tables used in the algorithm are available for download.
Conclusions
A high performing, easily maintained algorithm can successfully identify medication and food allergies from free text entries in EHR systems.
Keywords: Natural language processing, Hypersensitivity, Electronic health records, Electronic medical records, Allergies, RxNorm
Introduction
Maintenance of an active medication allergy list is a meaningful-use core measure requirement for electronic health record (EHR) systems that relates to patient safety.1 When cross-referenced against prescribed medications, providers can be warned if they attempt to prescribe a drug or class of drugs to which the patient has a known or possible allergy, sensitivity, or other adverse reaction (collectively referred to as ‘allergies’ in this paper).2 3 More than 10% of medication prescribing errors may be due to failure to recognize a patient's previously known drug allergies,4 5 and computerized decision support systems have been shown to decrease these errors.6 The efficacy of such decision support systems, however, is dependent on the order entry system being able to identify accurately the listed drugs that should be avoided.7 Modern EHRs typically provide selection lists of common allergies and medication classes, along with lists of specific reactions and sensitivities. However, free text entry boxes are also present to allow the entry of non-listed items, which can confound the matching process. For example, if ‘penicillins’ is selected from the allergy drop-down list, a warning will be provided if the provider attempts to prescribe amoxicillin, an aminopenicillin. However, if the provider instead enters ‘PCN–? anaphylaxis as a child’ or misspells ‘penicilin’ in a free text entry, the system may ignore a potential drug reaction.
The primary objective of this study was to develop a high-performance, easily maintained algorithm that could be applied to free text allergy entries documented in patient allergy lists (eg, as part of semistructured patient summaries or problem lists) from any EHR. Our algorithm used text processing and natural language processing (NLP) techniques to codify free text entries of allergies and sensitivities entered via an anesthesia information management system that receives such information from a variety of clinical applications within a large, academic institution. Although the specific allergy requirement for meaningful use relates to medications, we also formally evaluated the identification of food allergies, because these can be life threatening, and inadvertent exposure can occur in the context of a hospitalization.
Background
NLP methods have previously been applied to the problem of recognizing medication names and signature information from clinical narrative documentation. The third annual Informatics for Integrating Biology and the Bedside (i2b2) NLP challenge centered on this task8 and involved 23 teams. The best-performing systems used machine learning9 or rule-based10 techniques to identify medication names and signature information from discharge summaries. The i2b2 NLP evaluated only medications actually taken by the patients; thus medications to which a patient was allergic were to be skipped by the algorithms. In addition, Levin et al11 studied automated mapping of free text preoperative medication entries in an anesthesia information management system using RxNorm. Fewer studies have specifically investigated the use of NLP for detecting allergies.
Available medication identification systems such as MedEx12 use the RxNorm terms to derive a list of medication names to be searched, and can do so with high performance for medication name recognition. RxNorm is an extensive compendium of normalized names of generic and branded drugs, ingredients, relationships, and attributes, and is available for free download from the National Library of Medicine. There are multiple tables in RxNorm that are linked together using unique concept identifiers (called RXCUI) that permit one to perform actions such as finding generic names or ingredients of branded drugs, alternative formulations of the same drug, and drugs in a specified class of medications. There are often multiple representations of the same drug in the database, derived from different reference sources, linked together via RxNorm relationships. Fields used included the medication database source (SAB), concept identifiers (RXCUI, RXCUI1, RXCUI2), the drug formulation string (STR), the term type (TTY), the designated preferred name (PT), and the relationship (RELA).13
For example, the RXNCONSO table contains three entries in the STR field for the branded non-steroidal anti-inflammatory drug ‘Vioxx’ (derived from different reference sources), all under the same concept identifier RXCUI=‘262149’. A look-up in the RXNREL table for the concept identifier RXCUI1=‘262149’ returns 16 rows, one of which has concept identifier RXCUI2=‘232158’. Searching RXNCONSO for RXCUI=‘232158’ returns 16 rows, 14 of which indicate the generic name ‘rofecoxib’ and two that return the chemical name for the drug: ‘4-(4-methylsulfonylphenyl)-3-phenyl-5H-furan-2-one’. Matches can be narrowed by specifying criteria for additional fields, described further in the Algorithm development section.
Zhou et al14 found that RxNorm covered 75% of all drug terms in the partners drug dictionary and 83% of the top 99 most frequently used terms. In addition, RxNorm contains an extensive vocabulary covering food substances, medical supplies, diseases, symptoms and other factors related to healthcare. RxNorm is updated weekly, with data contained in a standardized text file format. Goss et al15 compared five vocabularies in their ability to represent drug allergies in medical records. They found that RxNorm provided the greatest concept coverage for allergens. By mapping medication allergies to RxNorm, the Veterans Affairs and Department of Defense found that the interoperability of allergies between the two systems increased dramatically, with 74% of the Veterans Affairs’ allergies understandable by the Department of Defense.16
Other resources used in our study included the structured query language (SQL) dialect Transact-SQL (Microsoft, Redmond, Washington, USA) and two phonetic matching algorithms, Soundex17 and Double Metaphone.18 Transact-SQL was used for processing of the Microsoft SQL Server database. The phonetic algorithms create approximate spoken English language representations of words and then use these derivations to identify possible misspellings and duplicates. For example, ‘atenalol’ and ‘atenolol’ each match to A354 and ATNL in Soundex and Double Metaphone, respectively.
Methods
Data sources
The Vanderbilt University Institutional Review Board reviewed the study and approved it as non-human subject research. Allergy and sensitivity entries were extracted from the Vanderbilt perioperative information management system for all anesthetics performed between 1 January 2012 and 30 September 2012. No protected health information was retrieved. These data were a combination of free text entry and items selected from drop-down lists provided in the various systems that feed the Vanderbilt perioperative information management system. The dataset was divided into training and testing sets. The training dataset consisted of 9445 cases performed from 1 January to 31 March. The testing dataset consisted of 9430 cases performed from 1 July to 30 September.
After obtaining a license from the US National Library of Medicine, all RxNorm rich release format files dated 5 November 201219 were downloaded. These data were then imported into a SQL Server database created by modifying the supplied Oracle database format (Oracle, Redwood Shores, California, USA). For the study, only the RXNCONSO table (containing the concept names, found in the STR field, and concept numbers, represented as RXCUI) and the RXNREL table (containing relationships between RXCUI) were used. The allergy-matching algorithm was written in Transact-SQL (Microsoft). An additional entry concept with RXCUI=‘NKDA’ corresponding to the RxNorm string ‘No known allergies’ was added to the local RXNCONSO table to allow matching of entries indicating the absence of an allergy in the allergy field. All SQL matches were case insensitive.
Algorithm development
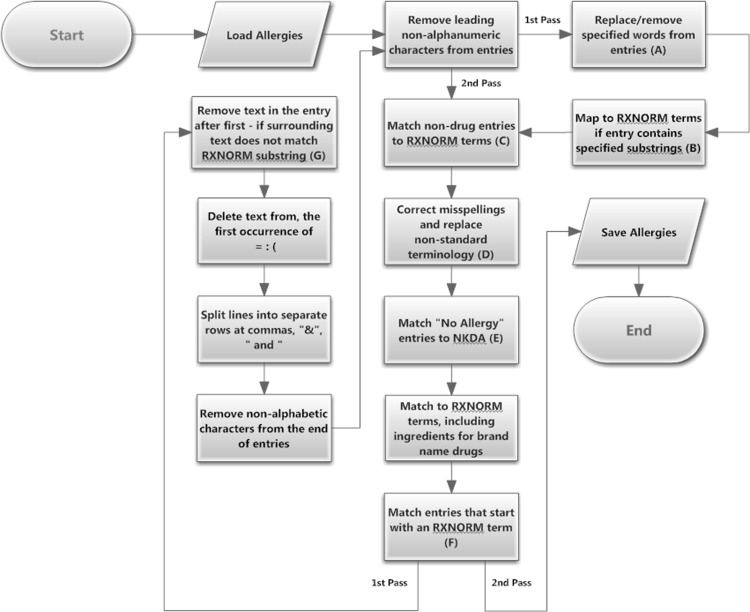
Using the training dataset, the algorithm was iteratively developed (figure 1, appendix 1) by matching the processed entries against the STR field in RXCONSO, removing matched records, and then examining the residual entries to develop additional criteria.
Figure 1.
Algorithm flowchart. This flow diagram outlines the steps in the processing of entries by the Transact-SQL code in an attempt to match to an RxNorm term. The sequential steps were developed preferentially to match the most specific terms, then to match remaining entries to less specific terms. Two passes were made through the algorithm, the second bypassing steps that could not logically result in any additional matches. The second pass was applied to process entries created by splitting lines where multiple potential allergies had been entered in the allergy field (eg, several words separated by commas). The letters in parentheses in the boxes of the flowchart reference the corresponding step in table 1, which provide a description of the processing that is being done, along with examples. Pseudocode for the algorithm is provided in appendix 1.
Examination of non-matching entries revealed a variety of reasons, including misspellings, the presence of conditional text, reaction information appended to the allergen, use of abbreviations and non-standard drug terminology (table 1). Database look-up tables were created to handle these errors, as well as for the identification and mapping of food, environmental, insect bites and stings, and substring matches (table 1). Superfluous punctuation marks (eg, ‘?’, ‘!’) at the beginning or end of entries were also removed, and spaces were trimmed from the start and end of processed strings.
Table 1.
Algorithm look-up tables
| Figure 1 reference* | Table name | Function | Examples |
|---|---|---|---|
| A | NLP_Allergy_Substitutions | Remove descriptors or replace text, subject to exclusions | ▸ Remove ‘possibly allergic to’, ‘intolerant of’ (no exclusions) ▸ Remove ‘oral’ (exclude if ‘Chloral Hydrate’ or ‘Oral Contraceptive’) |
| B | NLP_Like_Substitutions | Replaced entry with RxNorm term if the substring(s) are contained in the entry | ▸ If ‘ASA ’ is contained in the entry, replace with aspirin ▸ If ‘x-ray’ and ‘contrast’ are contained in the entry, replace with ‘Radiographic Contrast Agent’ |
| C | Non_Drugs | Maps foods, environmental sensitivities, and insect bites and stings | ▸ Peanuts mapped to the RxNorm term ‘Food Allergy’ ▸ ‘Cat’ mapped to the RxNorm term ‘Environmental Hypersensitivity’ ▸ ‘Yellow Jackets’ mapped to the RxNorm term ‘Insect Bites and Stings’ |
| D | Mappings | Replace misspelled allergies and non-standard entries | ▸ ‘AMOLODIPINE’ replaced with ‘Amlodipine’ ▸ ‘c-sporins’ replaced with ‘Cephalosporins’ |
| E | NKDA_Abbreviations | Abbreviations and misspellings for ‘no known drug allergies’ mapped | ▸ NKA mapped to NKDA ▸ KNDA mapped to NKDA |
| F | Substring_Exclusions | Prevent matching RXNCONSO STR terms as a substring of the allergy (starting with the first character) if the STR term is in the exclusion list | ▸ ‘Morphine makes me sick’ matches to ‘Morphine’ (‘morphine’ is not in the exclusion list) ▸ ‘Influenza Vaccine’ is prevented from matching to the RxNorm disease term ‘Influenza’ because ‘Influenza’ is in the list of excluded matches |
| G | Hyphenation_Exclusions | Prevent deletion of text to the right of a hyphen if there is a drug in RxNorm with 3 characters of matching text around the hyphen | ▸ ‘Codeine-rash’ is replaced by ‘Codeine’ (ine-ras is not in the exclusion table) ▸ ‘Darvocet-N 100’ is not replaced (‘cet-N 1’ is in the exclusion table) |
*The letters corresponding to each row are identified in the algorithm flow chart in figure 1.
Each pass through the RxNorm section of the algorithm consisted of several steps, executed sequentially, with each step only attempting to match items to the STR field (containing the drug name, and sometimes additional information such as the dose and formulation) in RXCONSO not previously matched. The order of matching was designed to prioritize the most specific match, as follows: 1. Source (Field name: SAB)=Veterans Health Administration National Drug File—Reference Table (NDFRT) and Term Type (Field name: TTY)=Designated Preferred Name (Field name: PT); 2. SAB=RXNORM, any TTY; 3. SAB=NDFRT, any TTY; and 4. Any SAB, any TTY. Following these steps, the generic component(s) of brand names represented in RXNREL were identified by using RXNREL relationships (ie, to map to RXCUI with TTY=‘IN’ (ingredient) and Relationship (Field name: RELA)=‘has_brandname’). For example, using this method, ‘Bactrim’ was mapped to ‘sulfamethoxazole/trimethoprim’, and ‘Altace’ to ‘ramipril’. For medications without definite mappings to ingredients (typically some multi-ingredient medications), the medication (eg, ‘Lortab’) was left in its brand-name form.
We explored the potential of adding Transact-SQL implementations of the phonetic Soundex and Double Metaphone algorithm to identify misspelled drugs not matched by dictionary look-ups.11 However, we found that there was considerable ambiguity due to the presence of many ‘sound-alike’ drugs and vowel misrepresentations in the unmatched drugs. For example, the Double Metaphone algorithm matched ‘Fiorcet’ (which should be spelled ‘Fioricet’) to ‘Versed’, and ‘paroxatine’ (which should be spelled ‘paroxetine’) to ‘Percocet’. Soundex matched ‘Acupril’ (which should be spelled ‘Accupril’) to both ‘aspirin’ and ‘aspariginase’.
The algorithm tuning process was repeated until the raw match rate for both drugs and food allergies was above 98%, as compared to manual review (see below).
Evaluation of algorithm performance
Following refinement of the algorithm using the training dataset, the code was executed against the testing dataset and performance manually determined. For the purpose of assessing performance of the RxNorm matching algorithm, all provider entries were considered to be valid allergies or sensitivities, even if implausible (eg, an ‘allergy’ to epinephrine). Each entry was classified as a drug, food, environmental exposure, insect bite/sting, or as ‘other’. All potential matches in the testing dataset were reviewed by the third author (MS), a fourth-year medical student, and checked for accuracy by the first author (RHE), a board-certified anesthesiologist. Matches were considered to be true positives if the algorithm identified a match within the RXNCONSO table that corresponded to the entry. Matches were classified as false positives if the entry was not an allergy but a match was found (eg, ‘Can take Morphine’ matching to a morphine allergy). Matches were considered to be false negatives if no match was found but the entry was an allergy. Matches were also considered to be false negatives if the entry was an allergy and was matched incorrectly (eg, ‘glaucoma gtts?’ matching to ‘glaucoma’) or the match was incomplete (eg, ‘pentazocine-naloxone’ to ‘pentazocine’). Matches were considered to be true negatives if the entry was not an allergy and no match was found. This final category usually resulted from information inappropriately placed in the allergy text field (eg, ‘No IV-Venipunctures right side’).
Allergy and sensitivity entries in the training and testing datasets were manually inspected to determine performance of the model using all entries. Accuracy 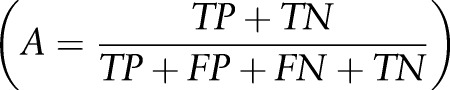 , specificity
, specificity 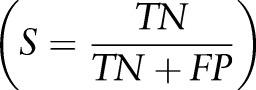 precision
precision 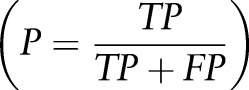 recall
recall 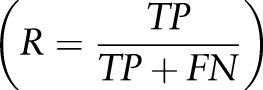 , and F-measure
, and F-measure 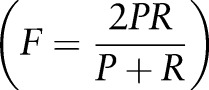 were calculated. Performance was also calculated separately for drug and food allergies.
were calculated. Performance was also calculated separately for drug and food allergies.
To avoid inflating the performance metrics, the analysis was repeated including only free text entries and non-standard entries from drop lists (ie, without an exact match in RxNorm).
Results
The training dataset consisted of 24 599 entries from 9445 cases performed from 1 January to 31 March 2012. The testing dataset consisted of 24 857 entries from 9430 cases performed from 1 July to 30 September 2012. After splitting entries containing a comma, the word ‘and’, or the symbol ‘&’ into multiple lines for processing, there were 25 158 rows in the training dataset and 25 164 rows in the testing dataset. The algorithm processed approximately 1000 entries per second for both datasets on an HP Proliant DL380 G6 server (Hewlett-Packard Co., Palo Alto, California, USA) with dual hex core 2.6 GHz CPU processors, 128 GB of RAM, running SQL Server 2008 R2 Enterprise Edition (SP2) on a Windows Server 2008 R2 Enterprise Edition 64 bit operating system (Microsoft).
The overall raw match rate (ie, before algorithmic processing) in the training dataset between the allergy and sensitivity entries was 68.2% (72.3% for drugs and 38.3% for foods). Similar percentages for raw matches were found in the testing dataset (66.7% overall, 73.5% for drugs, 39.5% for foods). These baseline match rates represent selection by providers of items from drop-down lists or free text entries that exactly matched an existing RxNorm term.
Performance of the algorithm for all allergy entries is listed in table 2. Overall accuracy, precision, recall, and F-measure were all above 98% in the training dataset and above 95% in the testing dataset. Accuracy, precision, recall, and F-measure for drug matches were all above 98% in the training and above 97% in the testing dataset. Corresponding values for food matches were above 97% and above 93%, respectively. Overall specificity (true negative rate) was above 95% in the training and testing datasets. Specificity for drug matches was 90.3% and 85.0% for drug matches and 100% and 88.9% for food matches in the training and testing datasets, respectively.
Table 2.
Algorithm performance: all allergy entries
| All allergies | Drug allergies | Food allergies | All unique strings only | |||||
|---|---|---|---|---|---|---|---|---|
| Dataset | Dataset | Dataset | Dataset | |||||
| Measure | Training | Testing | Training | Testing | Training | Testing | Training | Testing |
| # evaluated | 22 995 | 23 910 | 21 019 | 20 737 | 1755 | 1828 | 3275 | 3662 |
| Accuracy (%) | 98.22 | 95.98 | 98.63 | 96.69 | 98.38 | 93.63 | 99.60 | 81.13 |
| Specificity (%) | 95.97 | 97.79 | 90.32 | 85.00 | 100.00 | 88.89 | 75.00 | 97.60 |
| Precision (%) | 99.98 | 99.98 | 99.99 | 99.99 | 100.00 | 99.95 | 99.97 | 99.89 |
| Recall (%) | 98.23 | 95.97 | 98.83 | 96.70 | 98.38 | 93.65 | 99.63 | 80.55 |
| F-measure (%) | 99.10 | 97.93 | 99.30 | 98.30 | 99.18 | 96.69 | 99.80 | 89.18 |
F-measure=harmonic mean of precision and recall.
There were 3275 and 3662 unique allergy rows in the training and testing dataset, respectively. In the training and testing dataset, 13.3% and 14.6% of these items accounted for 80% of the total number of rows, respectively. Overall accuracy, precision, recall, and F-measure were virtually identical to the performance using all rows in the training dataset, but specificity was reduced from 90% to 75% (table 2). This resulted from the equal weighting of high frequency allergies (eg, ‘penicillin’) to rare singleton entries that could not be matched (eg, ‘unknown anti-emetic’). Because of the additional number of rare entries that did not match in the testing dataset, performance was reduced when calculated according to unique entries. This was mostly due to spelling errors not identified in the training dataset.
There were 723 total drug entries (477 unique strings) in the testing dataset that did not match an RxNorm entry. Of these, 310 (65%) were correctable by simple additions to the look-up tables. The majority of these errors were due to spelling mistakes.
Including only allergy entries entered via free text and non-standard entries selected from drop lists, performance of the algorithm was less than when all entries were considered, as would be predicted (table 3). However, performance was still quite good, with overall accuracy, specificity, precision, recall, and F-measure all above 94% in the training dataset, and all above 88% in the testing dataset.
Table 3.
Algorithm performance: free text and non-standard drop list allergy entries only
| All allergies | Drug allergies | Food allergies | All unique strings only | |||||
|---|---|---|---|---|---|---|---|---|
| Dataset | Dataset | Dataset | Dataset | |||||
| Measure | Training | Testing | Training | Testing | Training | Testing | Training | Testing |
| # evaluated | 8053 | 8958 | 6121 | 6044 | 1120 | 1888 | 2447 | 2570 |
| Accuracy (%) | 94.26 | 88.84 | 96.03 | 91.62 | 97.40 | 93.38 | 87.32 | 74.05 |
| Specificity (%) | 95.93 | 97.78 | 90.32 | 85.00 | 95.93 | 97.78 | 96.43 | 97.64 |
| Precision (%) | 99.93 | 99.94 | 99.96 | 99.96 | 99.93 | 99.94 | 99.85 | 99.83 |
| Recall (%) | 94.23 | 88.61 | 96.05 | 91.64 | 94.23 | 88.61 | 86.98 | 72.82 |
| F-measure (%) | 97.00 | 93.93 | 97.94 | 95.60 | 97.00 | 93.93 | 96.43 | 97.64 |
F-measure=harmonic mean of precision and recall.
Discussion
The algorithm identified drug and food allergies well. Performance for matching food allergies was slightly less than it was for drug allergies, but results were still excellent. The better performance of drug matches versus food allergies is not unexpected, because nearly all food allergies were entered as free text and identification depended on matching to the manually created look-up table. Because updates to the RxNorm supplementary tables (available online from the RxNorm website19) are provided on a weekly basis, updating the entries in the two tables used for the matching algorithm (RXNCONSO and RXNREL) is easily performed. No alterations are made to these RxNorm tables other than the insertion of a single ‘No drug allergy’ concept (not present in RxNorm), which was assigned a non-colliding RXCUI of ‘NKDA’. Because the algorithm is driven by supplementary tables (available online, see Data sharing statement, below), updating with new foods, additional drug spellings, and other corrections is simple, not requiring modification of the algorithm. Periodic examination of the residual entries not matched by the algorithm would allow identification of required additions to the tables, based on patterns of user entry.
Despite the presence of dedicated fields with drop-down lists for selection of allergies and sensitivities, nearly a third of such entries involved either free text entries or controlled vocabulary terms that did not match a term in RxNorm. Because computerized drug order entry systems require recognition of allergies and sensitivities to generate decision support warnings when providers attempt to prescribe conflicting medications, this method of entry should be discouraged. Because RxNorm is the de facto standard for the identification of drugs,20 ensuring that lists contain terminology that matches entries in RxNorm is recommended. This process will also help facilitate interoperability among systems, because RxNorm comprises concepts from a number of vocabularies (eg, SNOMED Clinical Terms, Veterans Health Administration National Drug File, MicroMedex RED BOOK, FDA National Drug Code Directory). However, as generating an all-inclusive list of potential allergens and sensitivities is neither possible nor desirable (due to performance issues), there remains a need to allow users to enter free text. Nonetheless, experience teaches us that these free text entries are often used even when structured representations would be available due to inadequate synonymy, speed of system implementation, or other user interface issues.21 An allergy-matching algorithm such as the one we developed could easily be applied to such entries and the user prompted to use the discrete lists if items are recognized (even if misspelled).
In the analysis of our datasets, we noted that users occasionally entered important non-allergy information into allergy fields (eg, presence of implants incompatible with MRI). Presumably they were doing this to help ensure that subsequent caregivers would be likely to see the information, because review of patient allergies before prescribing medications is expected. Providing alternative fields within EHRs to enter and display such high acuity information is recommended.
We suggest three potential applications of the approach outlined in this study. First, the allergy-matching algorithm could be used for research purposes related to the identification and classification of patient drug allergies and sensitivities. Second, the SQL process described could easily be integrated within EHR systems to parse free text allergy and sensitivity entries in real time to suggest corrections, potentially with visual feedback for confirmation to the provider. Finally, automated drug dispensing systems used in operating room suites could use the algorithm to parse allergies transmitted from the hospital EHR to provide relevant warnings to anesthesia care providers. This latter use is particularly relevant because the process of drug ordering and administration in the operating room by anesthesia care providers bypasses the decision support provided in the main hospital order entry system, and automated anesthesia drug dispensing systems (eg, Pyxis Anesthesia System 4000; CareFusion, San Diego, California, USA) are unmindful of patient allergies.
A limitation of our study is that we only studied a single hospital, and the look-up tables developed may not be fully portable to other institutions. Although an independent analysis of performance will be required, starting with the already developed tables (available from http://knowledgemap.mc.vanderbilt.edu/research/content/tables-allergy-nlp-matching) should greatly reduce the burden of this process. Additional items can easily be added to the tables, and RxNorm is freely available, simplifying reuse and testing in other environments. To the extent that other hospitals are using standard terminology found in RxNorm in their allergy selection lists, the improvement from our allergy-matching algorithm over raw matching of entered items may differ from that seen in our data.
Another limitation is that periodic analysis of performance of the matching algorithm is necessary, as there are an unlimited number of misspellings and other oddly formatted entries that providers can enter into their systems, and new drugs and terms are added to RxNorm over time. Indeed, spelling errors were the most common cause of failed matches (ie, false negatives). Therefore, development of a more advanced spelling-correction algorithm would improve on our algorithm. Medication-specific spelling correction algorithms have been developed before with high specificity using the open-source Aspell package.10 Other studies of medical spelling correction have seen variable performance,22–24 as noted in this study. However, even given the current method of manual entry of spelling errors, the degradation in accuracy between the training and testing datasets was small (less than 3%), and only a small number of entries was identified that would require updates to the look-up tables. This should thus not be a burdensome process.
Conclusions
We have shown that the identification of allergies via algorithmic processing of free text entry of allergy and sensitivity information was highly successful, practical, and maintainable. However, use of standardized vocabularies in allergy and sensitive lists and encouragement of care providers to use these lists is preferable to free text entry, whenever possible. Interoperability among systems depends on accurate knowledge of allergy information that would be facilitated by using standard terms. Although overall matching performance can be expected to be excellent, there will always be a small percentage of hand-entered allergies that are significant and will be missed.
Appendix 1: Allergy-matching algorithm
The following is a pseudocode description of the steps in the current version of the SQL allergy identification algorithm. Knowledge of RxNorm and use of its concept identifiers are assumed, as is a general understanding of SQL syntax. Referenced database look-up tables are described in table 1. Note that once an allergy is matched to the RXCUI of an RxNorm STR term, no further processing of that allergy takes place. In other words, the algorithm is designed to provide for a preferential order of matching to possible RxNorm terms.
Once an allergy is matched to an RxNORM RXCUI concept identifier, no further processing takes place for that allergy (ie, it is excluded from processing if RXCUI is not null).
Algorithm pseudocode
Read allergies into a temporary table
Remove non-alphanumeric characters at the start of each allergy until the first alphanumeric character is encountered (ie, [a–z], [A–Z], or [0–9])
Replace substrings in the allergy using the TargetString and ReplacementString fields in the look-up table NLP_Allergy_Substitutions, excepting allergies that contain a listed Exception
Replace the allergy with the value in the field ReplacementString from the look-up table NLP_Like_Substitutions if the allergy contains a substring matching the value of the field TargetString
Identify non-drug allergies by matching the allergy to the value of the field NonDrugName in the look-up table NLP_Allergy_NonDrug and applying the generic RxNorm RXCUI code for ‘Food Allergy’ [1025015], ‘Environmental Hypersensitivity’ [712208], ‘Insect Bites and Stings’ [1029906]
Redo step 5 by creating plurals of the entries in the NonDrugName field
Replace misspelled allergies with the value in the field AllergyName where the complete allergy entry matches the AllergyNameOriginal field NLP_Allergy_Mapping
Repeat step 7 for matches to the plural of the AllergyNameOriginal
Match synonyms to ‘No known documented allergies’ in the complete allergy entry using the NKDA_Synonym field in the NLP_NKDA look-up table
Identify the RXCUI of allergies where the complete allergy entry matches the STR field in the look-up table RXNCONSO where SAB=NDFRT and TTY=PT
Identify the RXCUI of allergies where the complete allergy entry matches the STR field in the look-up table RXNCONSO where SAB=RXNORM
Identify the RXCUI of allergies where the complete allergy entry matches the STR field in the look-up table RXNCONSO where SAB=NDFRT
Identify the RXCUI of allergies where the complete allergy entry matches the STR field in the look-up table RXNCONSO
Identify allergies where the STR value after removing substring ‘ (Chemical/Ingredient)’ in the RXCONSO table (where SAB=NDFRT and TTY=PT) is a substring of the allergy entry starting at the first character of the allergy entry and where the allergy string does not contain any ‘,’ characters (these get processed later after the string is split), and the STR value is not included in the list of ExcludeMatch values in the table NLP_ExcludedMatches
Get rid of all characters in the allergy entry from the first ‘-’ in the string and to the right except if the seven character string including the three characters on either side of the ‘-’ in the allergy substring match an entry in the ExcludedDrug field of the NLP_DashedDrugExclusionsRxNORM table
Remove all characters in the allergy entry including and to the right of the first ‘:’ character
Remove all characters in the allergy entry including and to the right of the first ‘=’ character
Remove all characters in the allergy entry including and to the right of the first ‘(’ character
Replace all substrings ‘ and ’ or ‘&’ with ‘,’
Split all allergy entries containing a ‘,’ character into new allergy table entries, and remove the original entry (eg, ‘a, b, c’ would become three rows ‘a’, ‘b’, and ‘c’)
Remove all non-alphanumeric characters from the end of all allergy entries (as in 2 above)
Remove all non-alphanumeric characters from the start of all allergy entries (as in 2 above)
Repeat steps 5–14 above
Footnotes
Contributors: RHE: substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; drafting the article or revising it critically for important intellectual content; and final approval of the version to be published. This author is the guarantor. PSJ: substantial contributions to conception and design and interpretation of data; drafting the article or revising it critically for important intellectual content; and final approval of the version to be published. MS: substantial contributions to analysis and interpretation of data; drafting the article or revising it critically for important intellectual content; and final approval of the version to be published. BR: substantial contributions to conception and design, analysis and interpretation of data; drafting the article or revising it critically for important intellectual content; and final approval of the version to be published. JME: substantial contributions to conception and design, analysis and interpretation of data; drafting the article or revising it critically for important intellectual content; and final approval of the version to be published. JCD: substantial contributions to conception and design, analysis and interpretation of data; drafting the article or revising it critically for important intellectual content; and final approval of the version to be published.
Funding: This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors. The study was solely funded from departmental resources.
Competing interests: None
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Tables used in the NLP algorithm are available at http://knowledgemap.mc.vanderbilt.edu/research/content/tables-allergy-nlp-matching. There are no restrictions on their use other than they cannot be used for commercial purposes.
References
- 1.HealthIT.gov Step 5: Achieve Meaningful Use. http://www.healthit.gov/providers-professionals/achieve-meaningful-use/core-measures/medication-allergy-list (accessed 16 Feb 2013).
- 2.Abookire SA, Teich JM, Sandige H, et al. Improving allergy alerting in a computerized physician order entry system. Proceedings AMIA Symposium; 2000:2–6 [PMC free article] [PubMed] [Google Scholar]
- 3.Kuperman GJ, Bobb A, Payne TH, et al. Medication-related clinical decision support in computerized provider order entry systems: a review. J Am Med Inform Assoc 2007;14:29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lesar TS, Briceland L, Stein DS. Factors related to errors in medication prescribing. JAMA 1997;277:312–17 [PubMed] [Google Scholar]
- 5.Gandhi TK, Burstin HR, Cook EF, et al. Drug complications in outpatients. J Gen Intern Med 2000;15:149–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bates DW, Leape LL, Cullen DJ, et al. Effect of computerized physician order entry and a team intervention on prevention of serious medication errors. JAMA 1998;280:1311–16 [DOI] [PubMed] [Google Scholar]
- 7.Hsieh TC, Kuperman GJ, Jaggi T, et al. Characteristics and consequences of drug allergy alert overrides in a computerized physician order entry system. J Am Med Inform Assoc 2004;11:482–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patrick J, Li M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J Am Med Inform Assoc 2010;17:524–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doan S, Bastarache L, Klimkowski S, et al. Integrating existing natural language processing tools for medication extraction from discharge summaries. J Am Med Inform Assoc 2010;17:528–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levin MA, Krol M, Doshi AM, et al. Extraction and mapping of drug names from free text to a standardized nomenclature. AMIA Annu Symp Procs 2007;2007:438–42 [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010;17:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.US National Library of medicine 2012AB RxNorm Source Information. http://www.nlm.nih.gov/research/umls/sourcereleasedocs/current/RXNORM/sourcerepresentation.html (accessed 5 May 2013). [DOI] [PubMed]
- 14.Zhou L, Plasek JM, Mahoney LM, et al. Mapping partners master drug dictionary to RxNorm using an NLP-based approach. J Biomed Inform 2012;45:626–33 [DOI] [PubMed] [Google Scholar]
- 15.Goss FR, Zhou L, Plasek JM, et al. Evaluating standard terminologies for encoding allergy information. J Am Med Inform Assoc. 2013;20:969–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warnekar PP, Bouhaddou O, Parrish F, et al. Use of RxNorm to exchange codified drug allergy information between Department of Veterans Affairs (VA) and Department of Defense (DoD). AMIA. Annual Symposium Proceedings/AMIA Symposium.2007 Oct 11:781–5 [PMC free article] [PubMed] [Google Scholar]
- 17.Microsoft Soundex (Transact-SQL). http://msdn.microsoft.com/en-us/library/ms187384.aspx (accessed 16 Feb 2013).
- 18.Philips L. Metaphone algorithm. http://www.sqlteam.com/forums/topic.asp?TOPIC_ID=13574 (accessed 16 Feb 2013).
- 19.US National Library of Medicine RxNorm Files. http://www.nlm.nih.gov/research/umls/rxnorm/docs/rxnormfiles.html (accessed 16 Feb 2013). [DOI] [PubMed]
- 20. Consolidated Health Informatics (CHI). Initiative; health care and vocabulary standards for use in federal health information technology systems. Federal Register, December 2005: 70 No. 246.
- 21.Rosenbloom ST, Miller RA, Johnson KB, et al. Interface terminologies: facilitating direct entry of clinical data into electronic health record systems. J Am Med Inform Assoc 2006;13:277–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tolentino HD, Matters MD, Walop W, et al. A UMLS-based spell checker for natural language processing in vaccine safety. BMC Med Inform Decis Mak 2007;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denny JC, Spickard A, III, Johnson KB, et al. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc 2009;16:806–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruch P, Baud R, Geissbühler A. Using lexical disambiguation and named-entity recognition to improve spelling correction in the electronic patient record. Artif Intell Med 2003;29:169–84 [DOI] [PubMed] [Google Scholar]