Abstract
Microsomal monoxygenase enzymes of the cytochrome-P450 family are found in all biological kingdoms, and play a central role in the breakdown of metabolic as well as xenobiotic, toxic and 70% of the drugs in clinical use. Full-length cytochrome-b5 has been shown to be important for the catalytic activity of cytochrome-P450. Despite the significance in understanding the interactions between these two membrane-associated proteins, only limited high-resolution structural information on the full-length cytochrome-P450 and the cytochromes-b5-P450 complex is available. Here, we report a structural study on a functional ~72-kDa cytochromes-b5-P450 complex embedded in magnetically-aligned bicelles without having to freeze the sample. Functional and solid-state NMR (Nuclear Magnetic Resonance) data reveal interactions between the proteins in fluid lamellar phase bilayers. In addition, our data infer that the backbone structure and geometry of the transmembrane domain of cytochrome-b5 is not significantly altered due to its interaction with cytochrome-P450, whereas the mobility of cytochrome-b5 is considerably reduced.
Protein science in the post-genomic era is beginning to direct its course towards the elucidation of protein-protein interactions. Many biologically important protein-protein interactions, such as signal transduction, electron transport chain and photosynthesis, take place on the surface or across the lipid membrane of living cells. Despite their importance and ongoing investigative efforts1,2,3,4,5,6,7,8,9, there are very few reports on the protein structure determination of the combinatorial complexes composed of protein-protein interactions and lipid bilayers since practicable methods were deemed necessary to characterize the structure of those complex systems10,11,12,13. In this study, we show that solid-state NMR (Nuclear Magnetic Resonance) is capable of characterizing protein-protein interactions in lipid bilayers, and report structural and dynamic interactions in an intact mammalian membrane-bound complex constituting rabbit cytochrome b5 (cyt b5) and cytochrome P450 2B4 (cyt P450) in a fluid lamellar phase lipid bilayer.
Degradation of a wide variety of substances in the liver is catalyzed by microsomal monoxygenase enzymes of the cytochrome P450 family14. The cytochrome P450 super family is found in all biological kingdoms, and plays a central role in the breakdown of metabolic as well as xenobiotic, toxic and pharmaceutical compounds. The key step in oxygenating substrates is the reduction of diatomic atmospheric oxygen by supplying two electrons (Fig. S2)15,16,17,18. The reducing electrons can be delivered by two different electron-carrier proteins, cyt P450 reductase and cyt b5. The interaction between membrane-bound cyt P40 and cyt b5 is the subject of the investigation presented here.
Rabbit cyt b5 is a 16.7 kDa protein composed of three distinct domains; amino acid sequence is given in the supporting information16,17,19. We recently reported the first high-resolution NMR structure of the full-length membrane-bound rabbit cyt b520. The soluble domain of cyt b5 delivers an electron to cyt P450 by means of a prosthetic heme group. Cyt b5 has different effects on different members of the cyt P450 family. In some cases, catalytic action is enhanced, in others it is inhibited. In case of cyt P450 2B4, a 55.8 kDa protein from rabbit, studies of the stoichiometry of the metabolism have shown that cyt b5 increases the efficiency of enzymatic activity by cyt P450 2B4 primarily by decreasing the formation of the side-product superoxide21. A linker region of 15 amino acids length connects the soluble domain to a helical hydrophobic transmembrane domain in cyt b520. It has been shown that the linker and the transmembrane domains are essential for cyt b5 to bind to cyt P450 and to modify enzymatic turnover21. In order to elucidate the mechanism in which cyt b5 increases the efficiency of cyt P450 activity as an oxidation agent, the structure of the cyt b5-cyt P450 complex is important. However, only few structural details of the molecular basis of this interaction are currently known. A recent solution NMR study revealed the structural interactions between the soluble domains of the two full-length proteins embedded in isotropic bicelles and proposed a mechanism for the electron transfer from cyt b5 to cyt P45020. In this study, we report the solid-state NMR investigation of membrane topology and dynamic protein-protein interactions of the cyt b5-cyt P450 complex embedded in fluid lamellar phase lipid bilayers.
Results
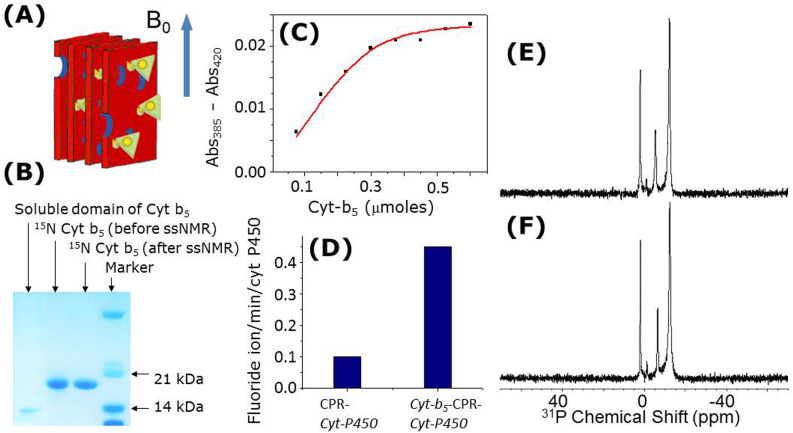
Cyt b5 and cyt P450 were expressed and purified from E.Coli, and were first assembled into a complex, then incorporated into 1,2-Dimyristoyl-sn-Glycero-3-Phosphocholine (DMPC)/1,2-Dihexanoyl-sn-Glycero-3-Phosphocholine (DHPC) bicelles with [DMPC]/[DHPC] = 3.5 that were used for all experimental measurements in this study (Fig. 1A; more details are given in the Supporting Information). Our results show the presence of interactions between cyt b5 and cyt P450 (Fig. 1C), the role of cyt b5 in the enzymatic activity of cyt P450 (Fig. 1D), and the magnetic-alignment of bicelles (Figs. 1E & 1F).
Figure 1.
(A) Representation of magnetically-aligned bicelles containing cyt b5-cyt P450 complex. (B) Gel confirming the intact nature of cyt b5 after solid-state NMR experiments; the transmembrane domain of cyt b5 can be cleaved off by sample mishandling, heat or an enzyme. (C) Complex formation between cyt P450 and cyt b5 is confirmed by the alternation of spin state as measured by type I spectral changes. (D) Activities of cyt P450 embedded in [DMPC]/[DHPC] = 3.5 bicelles with and without cyt b5 and cytochrome P450 reductase (CPR) measured at 37°C, using a substrate of cyt P450, methoxyflurane. (E & F) Proton-decoupled 31P-chemical shift spectra of bicelles containing a complex of 1:1 cyt b5 and cyt P450 demonstrating a high degree of alignment as indicated by the narrow lines for DMPC (in the high-field region) and DHPC (in the low-field region); phosphate buffer peak is observed ~0 ppm.
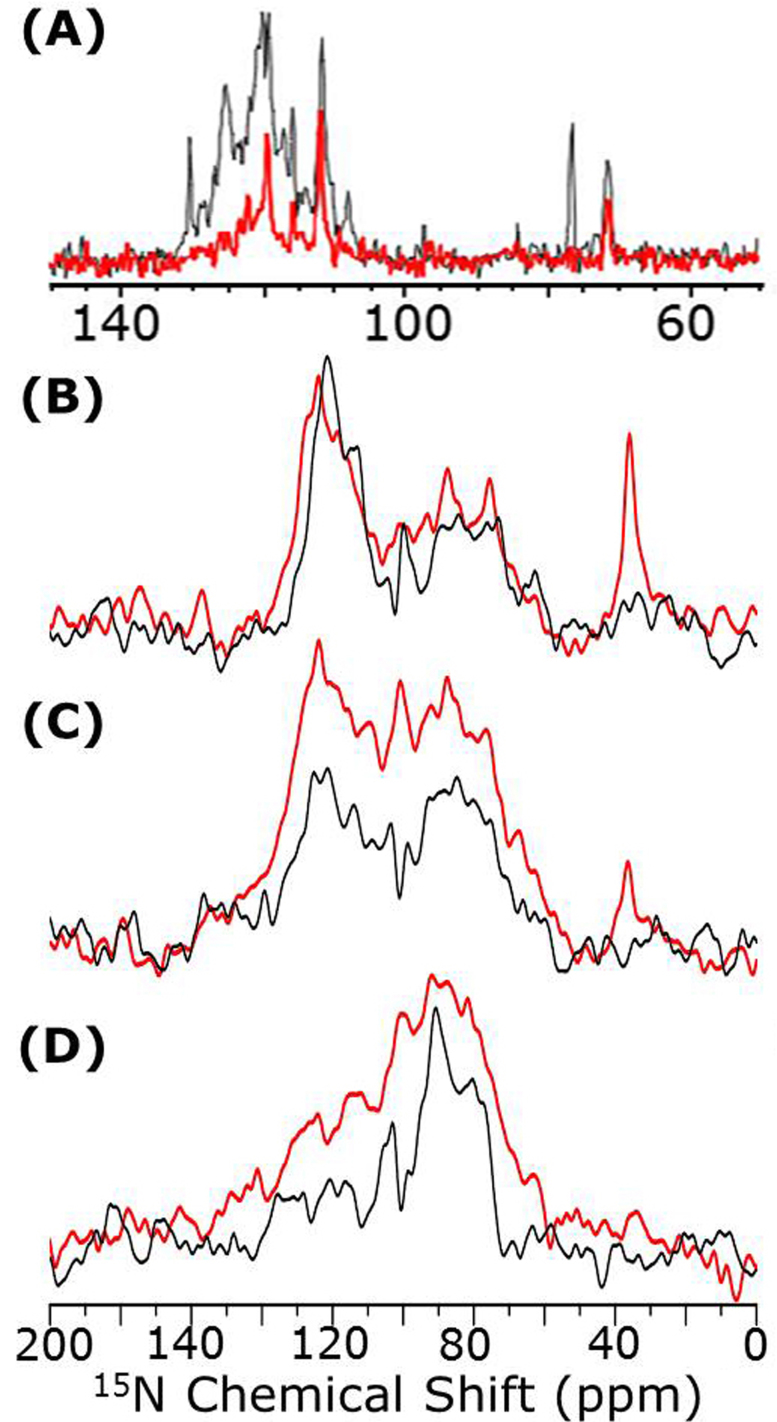
15N-NMR spectra of uniformly 15N-labeled cyt b5, in the absence and presence of unlabeled cyt P450, obtained using RINEPT22 (refocused insensitive nuclei enhanced by polarization transfer) and CP (cross polarization) sequences are shown in Fig. 2. Because of the uniaxial alignment of the complex in bicelles (as shown by 31P spectra in Fig. 1F), amide-15N nuclei in residues from transmembrane (70–100 ppm) and soluble (100–135 ppm) domains of cyt b5 resonate in different chemical shift regions (Fig. 2)23. The appearance of peaks in the 70–100 ppm region even for a short contact period (0.1 ms) in a CP experiment suggest that they arise from the immobile (or undergoing a slow motion in the millisecond time scale) transmembrane domain of the protein (Fig. 2D). Signal from other parts of the protein appear when the contact time is increased as shown in Fig. 2(B, C). On the other hand, the appearance of peaks in the isotropic chemical shift region (100–135 ppm) of the RINEPT spectra (Fig. 2A) reveals the fast dynamics of the soluble domain23.
Figure 2. 15N NMR spectra of uniformly aligned DMPC/DHPC bicelles containing U-15N-labeled cyt b5 without (black) and with cyt P450 (red).
Spectra were obtained using the RINEPT pulse sequence (A) and ramped cross-polarization with a contact time of 3 ms (B), 0.8 ms (C), and 0.1 ms (D). The differences seen in the spectra not only report on the variation in the mobilities of free cyt b5 (black) but also on the cyt b5 bound to P450 (red). The15N RINEPT spectra (A) show spectral intensity only in the 100–135 ppm region, consistent with a high mobility of the soluble domain of free cyt b5 (black trace) and significant reduction in its mobility due to b5-P450 interaction (red trace). In the RINEPT sequence, 2.6 and 1.3 ms were used in the first (before the pair of 90° pulses) and second (after the pair of 90° pulses) delays, respectively.
Similar experiments were also performed for bicelles containing 1:1 mole ratio of cytochromes-b5:P450 that showed protein-protein interaction (Fig. 1C) and activity (Fig. 1D). The presence of cyt P450 significantly changed the appearance of15N NMR spectra observed from cyt b5 mainly due to changes in the dynamics of cyt b5 (Fig. 2), indicating that protein-protein interaction is actually taking place in bicelles in agreement with the experimental data given in Fig. 1(C & D). In particular, as seen in the RINEPT spectra (Fig. 2A), the signal observed from the soluble domain (100–135 ppm) of cyt b5 in the cytochromes-b5-P450 complex is significantly weaker than that from the free cyt b5 (Fig. 2A). These results indicate the presence of interaction between the soluble domains of the proteins in the ~70-kDa complex that significantly reduces the mobility of the soluble domain of cyt b5. On the other hand, signal observed from the CP experiments are much stronger for the complex even for a shorter contact time (Fig. 2D) as the rate of magnetization transfer from1H to15N is faster for the complex than for the free cyt b5 (Fig. 2(B,C,D)). Interestingly, peaks from the side chain of Lys residues (~35 ppm) appear only from the complex but not from free cyt b5 (Fig. 2(B,C,D)). This indicates that the highly mobility of Lys side chains in free cyt b5 make CP inefficient, whereas this motion is considerably slowed down by the interactions between cyt b5 and cyt P450 so that CP becomes efficient in the complex. These results indicate that the mobility of cyt b5 is significantly slowed down upon binding to cyt P450. These observations are in agreement with a previous study that demonstrated that cyt b5 effectively binds to cyt P450 in a membrane environment21.
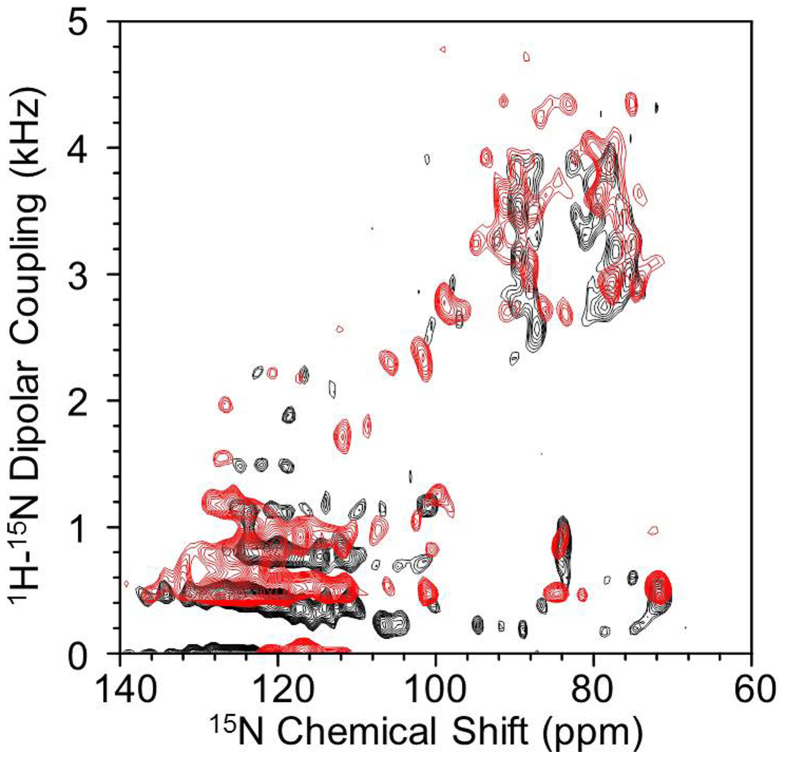
To better understand the interaction between the proteins, 2D HIMSELF24,25,26 (Heteronuclear Isotropic Mixing leading to Spin Exchange via the Local Field) or HERSELF (HEteronuclear Rotating-frame Spin Exchange via the Local Field) spectra, which correlate15N chemical shift with 1H-15N dipolar coupling, were obtained from magnetically-aligned bicelles containing the b5-P450 complex (Figure 3). The distinct geometry of the transmembrane α-helix of cyt b5 gives rise to a circular pattern of resonances seen in the upper right region of the 2D spectrum27,28,29,30. Figure 3 also shows the resolved signals exhibiting residual1H-15N dipolar couplings associated with the soluble domain resonances in the 100–135 ppm range, which could be due to the weak alignment of this domain. These residual dipolar couplings were found to be larger in the complex than in free cyt-b5. This further confirms the interaction between the soluble domains of cyt b5 and cyt P450.
Figure 3. Two-dimensional15N-HIMSELF spectrum of U-15N-cytb5 (black) and the complex of U-15N-cyt b5 with cyt P450 (red).
Spectra were obtained using a Low-E probe30 at 900 MHz. The use of Low-E probe and higher magnetic field is advantageous for this study because the intact cyt P450 is sensitive to heat induced by the radiofrequency pulses. Other experimental parameters include 0.5 ms cross polarization contact time, 64 t1 experiments, 1024 scans, 10.24 ms acquisition time, 25 kHz SPINAL16 decoupling, and 2 s recycle delay. Total measurement time was 18.2 hours at 30°C.
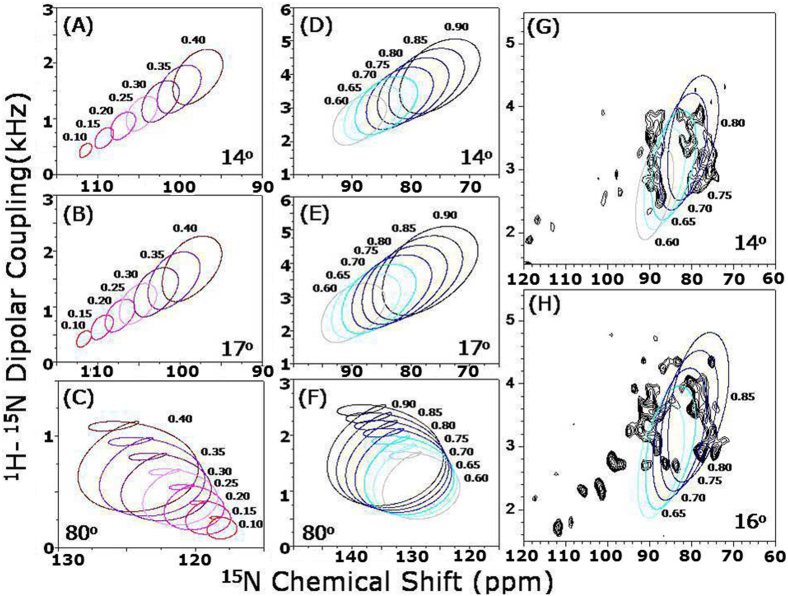
Simulations were performed to determine the tilt angle of the transmembrane helix in cyt b5 (Figure 4; more details are given in the Supporting Information). The resonance pattern analysis revealed that the free cyt b5 helix is tilted by 14 ± 3° away from the lipid bilayer normal at a molecular order parameter of Smol = 0.70, whereas it is found to be 16 ± 3° in the cyt b5-cyt P450 complex at a molecular order parameter of Smol = 0.76. These observations suggest that the transmembrane helix of cyt b5 is immobilized and could be involved in a direct interaction with the transmembrane domain of cyt P450. This finding could be useful to explain why cyt b5 lacking the transmembrane region does not influence the function of cyt P4506.
Figure 4. Simulation showing the sensitivity of the helical wheel pattern observed in 2D HIMSELF spectra (in Figure 3) to the tilt angle.
14° (A & D), 17° (B & E) and 80° (C & F) for indicated order parameters. The transmembrane region of 2D15N-HIMSELF spectra of U-15N-cyt b5 in the absence (G) and presence of (H) cyt P450 along with best-fitting simulated results.
Discussion
Our experimental results reported in this study demonstrate that not only cyt b5 but also a complex of cyt b5 and cyt P450 can be prepared in bicelles, aligned in a magnetic field for NMR experiments, and high-resolution spectra and meaningful structural and dynamic information can be obtained. While it is known that cyt P450 is unstable at ambient temperatures and both proteins are sensitive to impurities and heat, it is remarkable that the bicelles containing the complex was quite stable under our experimental conditions even up to 30°C for more than two weeks. The differences in the spectra obtained using CP and RINEPT pulse sequences, and with and without cyt P450, and the resonance pattern observed in 2D HIMSELF spectra provide a wealth of dynamic information about cyt b5 and also about the interaction between cyt b5 and cyt P450. Interestingly, the interaction between the proteins neither significantly altered the helical structure of the transmembrane domain of cyt b5 nor does it alter its geometry. This observation considerably simplifies our understanding on the interaction between these two proteins. Since the soluble domain of cyt-b5 is highly mobile in the NMR time scale20,23, the interaction between the soluble domains of cyt b5 and cyt P450 reduced the mobility of cyt b5. On the other hand, the interaction between the transmembrane helices could be essential in restricting the motion of cyt b5 soluble domain to those orientations that will result in a productive complex with cyt P450. Cyt P450 is believed to be rigidly anchored to the membrane and may, therefore, be limited in its ability to form a productive complex unless its binding partner is appropriately orientated. More experiments are needed to completely characterize the structural changes and the changes in the dynamics of the full-length membrane-bound proteins in the complex form. We expect the results reported in this study to be valuable for structural and dynamic studies of membrane protein complexes using solid-state NMR techniques.
Methods
Expression, purification and functional assay of proteins
All proteins were expressed and purified as detailed in the Supplementary Information. The experimental details on the CO assay are also given in the Supplementary Information.
Preparation of bicelles is given in the Supporting Information.
NMR experiments
One-dimensional 31P and 15N experiments were carried out on a Agilent/Varian 400 MHz solid-state NMR spectrometer using a home-built 5mm double-resonance static probe. 31P-chemical shift spectra were obtained using a spin echo sequence (90°-τ-180°-τ-acquire, τ = 100 μs) and15N chemical shift spectra were obtained using RINEPT and CP pulse sequences as mentioned in Fig. 2 caption. A 50 kHz proton decoupling during 31P signal acquisition and a recycle delay of 4 s were used. 2D HIMSELF experiments were initially optimized using the 400 MHz Agilent/Varian solid-state NMR spectrometer using a home-built 5mm double-resonance static probe, and the final spectra were obtained from the 900 MHz Bruker NMR spectrometer. All other experimental parameters are given in the figure captions.
Author Contributions
K.Y. and A.R. planned the experiments, K.Y., U.H.N.D., J.X., S.I., L.W. and A.R. performed the experiments and analyzed the results, K.Y., U.H.N.D. and A.R. wrote the paper, and A.R. designed and directed the research. All authors reviewed the manuscript.
Supplementary Material
Supporting Information
Acknowledgments
This research is supported by funds from NIH (GM084018 and GM095640 to A.R. and GM35533 to L.W.). Preliminary 2D HIMSELF/HERSELF spectra was obtained using the 900 MHz NMR spectrometer at the National High Magnetic Field Laboratory, which is supported by National Science Foundation Cooperative Agreement No. DMR-1157490, the State of Florida, and the U.S. Department of Energy. We thank Peter L. Gor'kov, Dr. Riqiang Fu, and Dr. William W. Brey for the help at NHMFL.
References
- Renault M. et al. Cellular solid-state nuclear magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. USA 109, 4863–4868 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bgate M. P. & McDermott A. E. Protonation state of E71 in KcsA and its role for channel collapse and inactivation. Proc. Natl. Acad. Sci. USA. 109, 15265–15270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison E. A. et al. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature 481, 45–50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy M. T. et al. Lipid dynamics and protein-lipid interactions in 2D crystals formed with the β-barrel integral membrane protein VDAC1. J. Am. Chem. Soc. 134, 6375–6387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupasinghe S. G. et al. High-yield expression and purification of isotopically labeled cytochrome P450 monooxygenases for solid-state NMR spectroscopy. Biochim. Biophis. Acta. 1768, 3061–3070 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijac A. Z., Li Y., Sligar S. G. & Rienstra C. M. Magic-angle spinning solid-state NMR spectroscopy of nanodisc-embedded human CYP3A4. Biochemistry 46, 13696–13703 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cady S. D. et al. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 463, 689–692 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verardi R., Shi L., Traaseth N. J., Walsh N. & Veglia G. Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc. Natl. Acad. Sci. USA 108, 9101–9106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M. et al. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science 330, 509–512 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca S. et al. The conformation of neurotensin bound to its G protein-coupled receptor. Proc. Natl. Acad. Sci. USA. 100, 10706–10711 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Pallavi P., Golebiewska U., McLaughlin S. & Smith S. O. Structure of the membrane reconstituted transmembrane-juxtamembrane peptide EGFR(622-660) and its interaction with Ca2+/calmodulin. Biochemistry 45, 12704–12714 (2006). [DOI] [PubMed] [Google Scholar]
- Linden A. H. et al. Neurotoxin II bound to acetylcholine receptors in native membranes studied by dynamic nuclear polarization NMR. J. Am. Chem. Soc. 133, 19266–19269 (2011). [DOI] [PubMed] [Google Scholar]
- Arakawa T. et al. Dynamic structure of pharaonis phoborhodopsin (sensory rhodopsin II) and complex with a cognate truncated transducer as revealed by site-directed 13C solid-state NMR. FEBS Lett. 536, 237–240 (2003). [DOI] [PubMed] [Google Scholar]
- Dürr U. H., Waskell L. & Ramamoorthy A. The cytochromes P450 and b5 and their reductases—promising targets for structural studies by advanced solid-state NMR spectroscopy. Biochim. Biophys. Acta. 1768, 3235–3259 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang H., Myshkin E. & Waskell L. Role of cytochrome b5 in catalysis by cytochrome P450 2B4. Biochem. Biophys. Res. Commun. 338, 499–506 (2005). [DOI] [PubMed] [Google Scholar]
- Vergéres G. & Waskell L. Cytochrome b5, its functions, structure and membrane topology. Biochimie. 77, 604–620 (1995). [DOI] [PubMed] [Google Scholar]
- Schenkman J. B. & Jansson I. The many roles of cytochrome b5. Pharmacol Ther. 97, 139–152 (2003). [DOI] [PubMed] [Google Scholar]
- Cheng Q., Sohl C. D. & Guengerich F. P. High-throughput fluorescence assay of cytochrome P450 3A4. Nat. Protoc. 4, 1258–1261 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrooney S. B. & Waskell L. High-level expression in Escherichia coli and purification of the membrane-bound form of cytochrome b5. Protein Expr. Purif. 19, 173–178 (2000). [DOI] [PubMed] [Google Scholar]
- Ahuja S. et al. A model of the membrane-bound cytochrome b5-cytochrome P450 complex from NMR and mutagenesis data. J. Biol. Chem. (2013) in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T. A., Im S. C., Bidwai A. & Waskell L. The role of the length and sequence of the linker domain of cytochrome b5 in stimulating cytochrome P450 2B4 catalysis. J. Biol. Chem. 279, 36809–36818 (2004). [DOI] [PubMed] [Google Scholar]
- Burum D. P. & Ernst R. R. Net polarization transfer via a J-ordered state for signal enhancement of low-sensitivity nuclei. J. Magn, Reson. 39, 163–168 (1980). [Google Scholar]
- Dürr U. H., Yamamoto K., Im S. C., Waskell L. & Ramamoorthy A. Solid-state NMR reveals structural and dynamical properties of a membrane-anchored electron-carrier protein, cytochrome b5. J. Am. Chem. Soc. 129, 6670–6671 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinskikh S. V., Yamamoto K. & Ramamoorthy A. Heteronuclear isotropic mixing separated local field NMR spectroscopy. J. Chem. Phys. 125, 034507 (2006). [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Dvinskikh S. V. & Ramamoorthy A. Measurement of heteronuclear dipolar couplings using a rotating frame solid-state NMR experiment. Chem. Phys. Lett. 419, 533–536 (2006). [Google Scholar]
- Divinskikh S. V., Yamamoto K. & Ramamoorthy A. Separated local field NMR spectroscopy by windowless isotropic mixing. Chem. Phys. Lett. 419, 168–173 (2006). [DOI] [PubMed] [Google Scholar]
- Ramamoorthy A., Wei Y. F. & Lee D.-K. PISEMA solid-state NMR spectroscopy. Ann. Rep. NMR Spectrosc. 52, 1–52 (2004). [Google Scholar]
- Gor'kov P. L. et al. Using low-E resonators to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz. J. Magn. Reson. 185, 77–93 (2007). [DOI] [PubMed] [Google Scholar]
- Soong R. et al. Proton-evolved local-field solid-state NMR studies of cytochrome b5 embedded in bicelles, revealing both structural and dynamical information. J. Am. Chem. Soc. 132, 5779–5788 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. et al. INEPT-based separated-local-field NMR spectroscopy: a unique approach to elucidate side-chain dynamics of membrane-associated proteins. J. Am. Chem. Soc. 132, 9944–9947 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information