Abstract
microRNAs regulate the expression of over 60% of protein coding genes by targeting their mRNAs to AGO2-containing complexes in the cytoplasm and promoting their translational inhibition and/or degradation. There is little evidence so far for microRNA-mediated regulation of other classes of non-coding RNAs. Here we report that microRNA-9 (miR-9) regulates the expression of the Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT-1), one of the most abundant and conserved long non-coding RNAs. Intriguingly, we find that miR-9 targets AGO2-mediated regulation of MALAT1 in the nucleus. Our findings reveal a novel direct regulatory link between two important classes of non-coding RNAs, miRs and lncRNAs, and advance our understanding of microRNA functions.
High-throughput technologies have revealed that the vast majority of our genome is transcribed; however only a very small percentage (around 2%) encodes for proteins1,2. So far most of the efforts have been focused on characterizations of short non-coding RNA. Among them, microRNAs (miRNAs) are estimated to regulate the expression of 60% of protein-coding genes by targeting mRNAs in the cytoplasm and leading to translational inhibition or RNA degradation3,4.
Aside from small RNAs, the non-coding human genome contains thousands of different long noncoding transcripts (lncRNAs) that are dynamically expressed in tissue-, differentiation stage-, and cell type-specific patterns5,2, and deregulation of their expression has been reported in various diseases including cancer6.
The Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) is a 7 kb long nuclear non-coding RNA transcribed from the NEAT2 locus7. MALAT1 is one of the most abundant and well-conserved lncRNAs indicating that it is likely to play an important biological function. Its expression is significantly associated with metastasis in non-small-cell lung carcinoma (NSCLC) and it was shown to be an independent prognostic marker for survival in lung adenocarcinoma8. Furthermore, several reports have indicated that MALAT1 could regulate splicing9,10. Surprisingly, in spite of its high degree of conservation, its specific localization at nuclear speckles and its developmentally regulated profile of expression, recent reports indicated that MALAT1 is dispensable for mouse development as well as for survival to adulthood. Surprisingly, deletion of MALAT1 does not significantly influence splicing patterns in various mouse tissues11,12,13. The observation that MALAT1 activity is stimulated by LPS (in HUVEC cells)10 raised, however, the possibility that it plays an important role in stress responses.
Similarly, microRNA-9 (miR-9) is also induced by several stress signals, including LPS treatment14. In addition, we have recently shown that miR-9 deregulation contributes to Hodgkin lymphoma pathogenesis and that its systemic inhibition in a xenograft model impairs tumor growth15.
Here we identify MALAT1 as a bona fide target of miR-9. Importantly, we find that miRNA-mediated regulation of this lncRNA occurs in the nucleus through direct binding to MRE (miRNA recognition elements) and in an AGO2-dependent manner. This highlights the existence of miRNA-mediated regulation of lncRNAs and extends the functions of miRNAs as post-transcriptional regulators to the nuclear compartment.
Results
Direct binding between miR-9 and MALAT1 in vivo
Growing evidence supports an important role for miR-9 in tumorigenesis16,17. In order to gain insights into miR-9 mechanisms of action we performed RNA-seq transcriptome analyses in the L428 Hodgkin Lymphoma cell line transfected with an LNA-modified inhibitor of miR-9 or a scrambled control (supplementary figure 1a).
Among the upregulated transcripts, MALAT1 was found to be >16 fold upregulated (Supplementary Table 1).
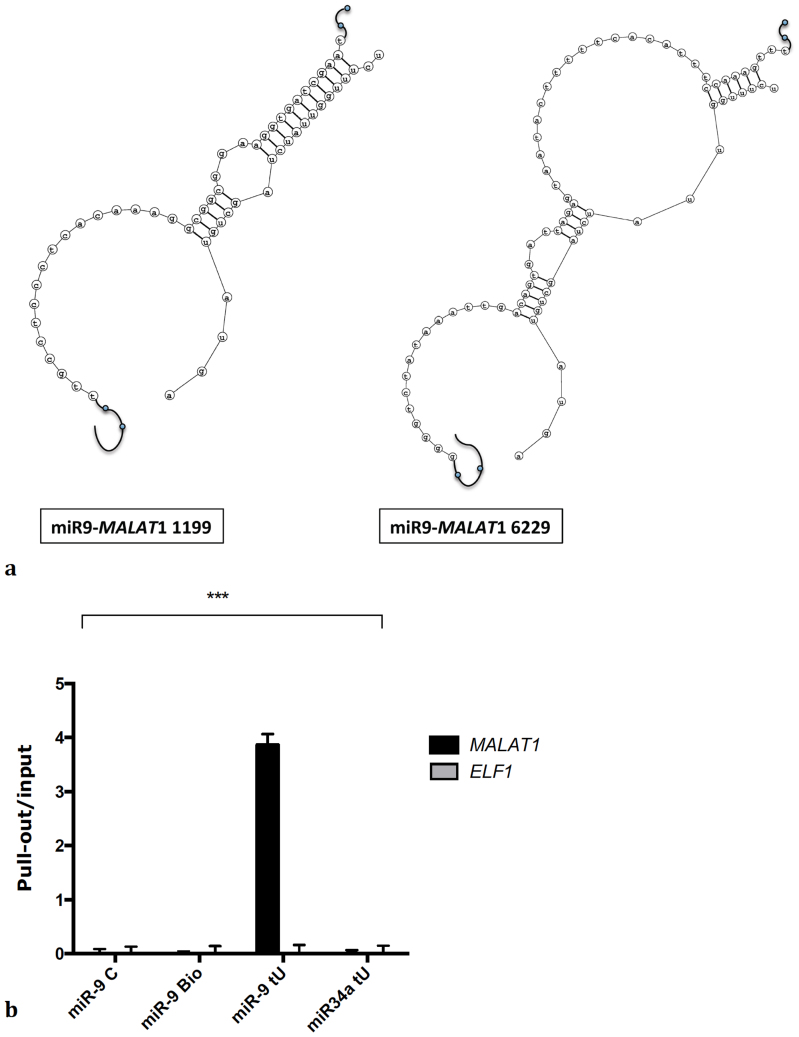
An in silico analysis of the MALAT1 sequence identified two putative binding sites for miR-9 in the MALAT1 sequence (NR_002819.2) at position 1199 and 6229 respectively (Fig. 1a), further supporting a potential interaction between miR-9 and this lncRNA.
Figure 1. miR-9 binds to MALAT1.
(a) Predicted binding sites for miR-9 in MALAT1. (b) L428 were transfected with a miR-9 mimic, a miR-9 mimic labeled with biotin, a miR-9 mimic labeled with biotin and containing 4-thiouridine or miR-34a labeled with biotin and containing 4-thiouridine. qPCR was used to measure MALAT1 enrichment in the pull-out samples over the inputs. Bars show mean +/− SEM (n = 3). P-value (p<0.001) was calculated by two-ways ANOVA.
Consistent with these findings, MALAT1 was readily detected by affinity purifications of miR-9 interacting transcripts from L428 cells transfected with 4-thiouridine and biotin-labelled miR-9, but not in samples transfected with non-biotinylated miR-9 or with a biotinylated and 4-thiouridine-labeled miR-34a control. Moreover, neither miR-9 nor the control miR-34a associated with the ELF1 mRNA, which was used as negative control (Fig. 1b). A similar result was obtained in U87MG cells (supplementary figure 1b), which also express high levels of miR-9. Together, these data indicate that miR-9 directly binds MALAT1 RNA in vivo.
MALAT1 stability is AGO2-dependent and regulated by miR-9
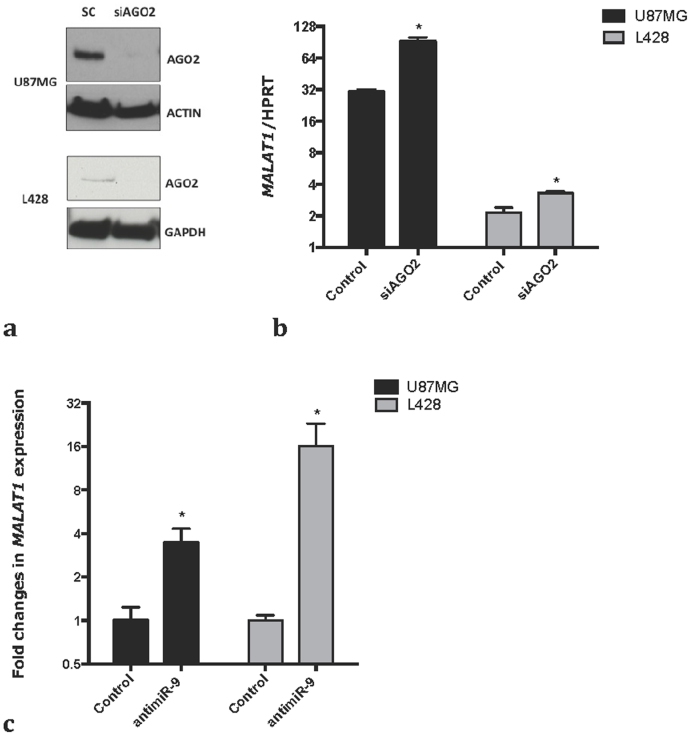
We next investigated whether MALAT1 expression is under the control of microRNAs by knocking down AGO2 in the L428 and U87MG cell lines (Fig. 2a). We observed a significant increase in MALAT1 expression in AGO2 KD cells (Fig. 2b) whereas, as expected, miR-9 stability is impaired by AGO2 knockdown (supplementary figure 2a). Importantly, miR-9 inhibition also resulted in a significant increase in MALAT1 levels (Fig. 2c, supplementary figure 2b) in both cell lines. Moreover, miR-9 overexpression resulted in a significant down-regulation of MALAT1 (supplementary figure 2c). Moreover, miR-9 overexpression resulted in a modest, but significant down-regulation of MALAT1 (supplementary figure 2b), most likely because both cell lines already express high levels of miR-915. Together these data indicate that miR-9 directly regulates the level of MALAT1.
Figure 2.
MALAT1 expression is regulated by miRNA (a) Cells were transfected with a scramble control or a pool of siRNA against AGO2. Western blot for AGO2 shows the efficiency of AGO2 knock down in U87MG and L428 cells. The lower and the upper panel (for both cell lines) derive from the same gel, only the membrane was split in two to perform the incubation with the antibody. A full picture of the gel can be found in the supplementary informations. (b) qPCR shows MALAT1 stabilization upon knock-down of AGO2 in U87MG and L428. p-values were calculated using a t-test. Bars show mean +/− SEM (n = 3). (c) Cells were transfected with an 8-mer against miR-9 or a scrambled 8-mer. qPCR shows MALAT1 stabilization upon inhibition of miR-9 in U87MG and L428. p-values were calculated using a t-test. Bars show mean +/− SEM (n = 3).
MALAT1 expression is regulated through direct miR-9 binding
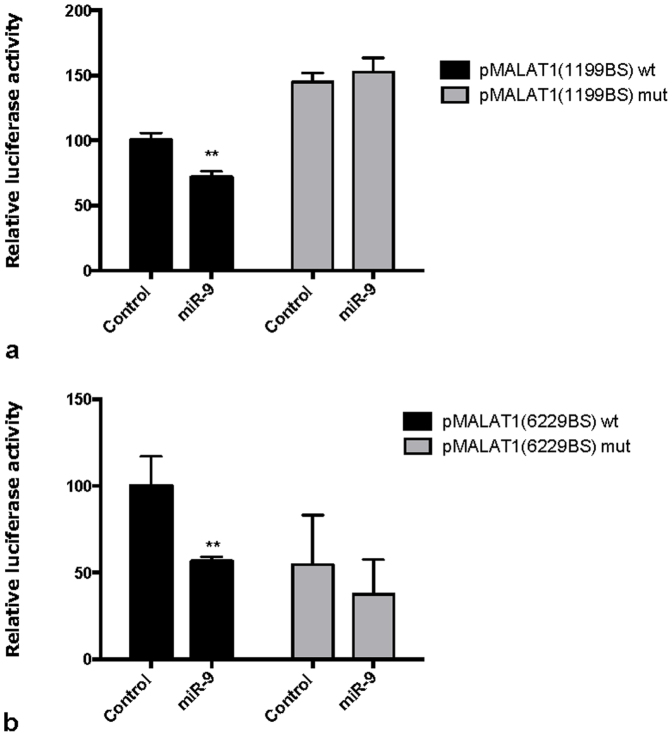
To further confirm that the miR-9:MALAT1 interaction occurs through the predicted miR-9 binding sites (supplementary Figure 1a), we cloned two fragments of MALAT1 encompassing the two putative binding sites downstream the luciferase gene and performed luciferase assays in U87MG cells transfected with reporter plasmids containing the two putative miR-9 binding sites. In both cases, miR-9 over-expression resulted in a significant decrease in luciferase activity (Fig. 3a–b), while an opposite effect was observed upon miR-9 inhibition (supplementary Figure 3). Moreover, directed mutagenesis of the predicted miR-9 binding sites abolished this effect (Fig. 3a–b).
Figure 3.
miR-9 targets MALAT1 through canonical miRNA binding sites (a) Cells were transfected with constructs encoding luciferase activity under control of miR-9 binding site 1199 wt (wild-type) or mut (mutated) in the MALAT1 sequence, in presence of a miR-9 mimic or a control oligonucleotide. miR-9 mimic induces a significant decrease in luciferase activity only in wt construct. p-values were calculated by multiple t-tests. Bars show mean +/− SEM (n = 3). (b) Cells were transfected with constructs encoding luciferase activity under control of miR-9 binding site 6229 wt (wild-type) or mut (mutated) in MALAT1 sequence, in presence of a miR-9 mimic or a control oligonucleotide. miR-9 mimic induces a significant decrease in luciferase activity only in wt construct. p-values were calculated by multiple t-tests. Bars show mean +/− SEM (n = 3).
miR-9 targets MALAT1 in the nucleus
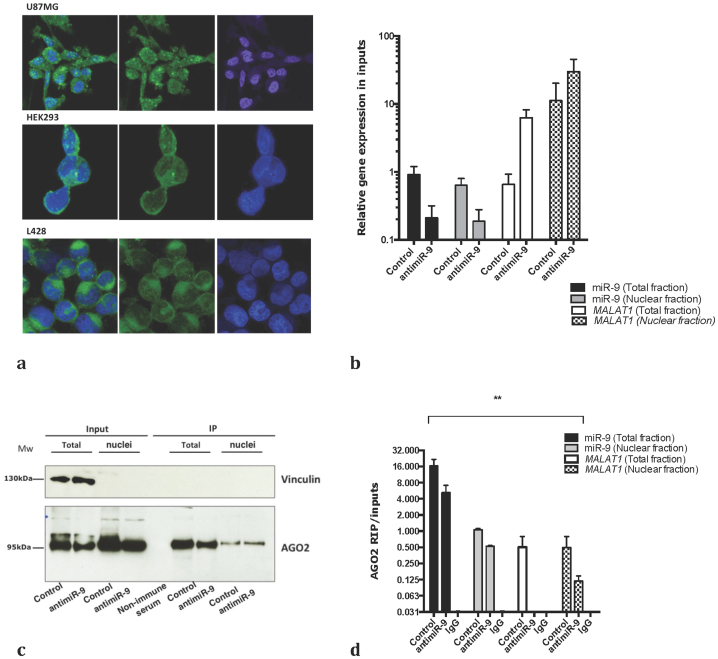
MicroRNAs are known to exert their functions mainly, if not exclusively, in the cytoplasm18. However, MALAT1 is a nuclear long ncRNA that localizes to nuclear speckles in association with splicing regulators19 raising the possibility that miR-9 targets MALAT1 in the nucleus. Using in situ hybridization and confocal microscopy, we detected miR-9 both in the cytoplasm and the nucleus of L428 Hodgkin lymphoma cells, HEK293 and U87MG glioblastoma cells (Fig. 4). Since previous studies have already demonstrated the presence of AGO2 in the nucleus20,21,22, we next investigated whether miR-9 regulates MALAT1 in the nucleus in an AGO2-dependent manner. To this end, we transfected U87MG cells and L428 with the antimiR-9 oligonucleotide or a scrambled control (Fig. 4b), isolated the nuclei and immunoprecipitated AGO2 and the associated RNAs. AGO2 was efficiently immunoprecipitated from both total cell extracts and nuclear extracts (Fig. 4c–d). Notably, whereas MALAT1 was readily detected in AGO2 immunoprecipitates from scrambled control treated cells, its levels were significantly reduced in AGO2 complexes purified from cells treated with antimiR-9, similar results were obtained in L428 (supplementary figure 4a–b). These results are consistent with the notion that MALAT1 is targeted by miR-9 in the nucleus in an AGO2-dependent manner (Fig. 2c).
Figure 4.
miR-9 targets MALAT1 to AGO2 containing complexes in the nuclei (a) RNA fluorescent in situ hybridization for miR-9 in different cell lines. Magnification 63X. DAPI is in blue, miR-9 in green. (b) Expression of miR-9 and MALAT1 in nuclear and cytoplasmic fractions used as inputs for AGO2 IP was measured by RT-qPCR. (c) Nuclear and total fractions were isolated from U87MG cells transfected with a scramble control or an antimiR-9, and immunoprecipitated using an AGO2 antibody. The western blot shows the purity of the extracts and the efficiency of AGO2 immunoprecipitation. The lower and the upper panel derive from the same gel, only the membrane was split in two to perform the incubation with the antibody. A full picture of the gel can be found in the supplementary informations. (d) The amount of miR-9 and MALAT1 was measured in inputs RNA used for the RIP by qPCR. d) The amount of miR-9 and MALAT1 bound to AGO2 was measured by qPCR in nuclear and cytoplasmic fractions in presence of a scramble control or an antimiR-9. P-value (p<0.0001) was calculated by two-ways ANOVA.
Discussion
The human genome contains thousands of lncRNA2 that are expressed at low levels and in a tissue specific manner. LncRNA are in general poorly conserved and about one-third of them are primate specific2. Unlike these lncRNA, MALAT1 is extremely abundant, ubiquitously expressed and highly conserved among mammals, reaching up to 90% identity between human and mouse23. In addition, the transcript appears to be quite stable due to a triple helix in the 3′end, which prevents its degradation24,25. Even though the physiological role of MALAT-1 is currently unclear, data suggest that deregulation of its expression contributes to cancer development and/or progression8,26,27. The absence of obvious phenotypes in mice lacking Malat1 raises the possibility that its activities may be cell type or context dependent. Moreover, very little is known about the mechanisms responsible for its biogenesis and stability that might prime its deregulation in cancer.
microRNAs are frequently deregulated in cancer and a large body of evidence indicates that they play key roles in mediating stress responses28. miR-9 in particular, was shown to be induced by several stress signals including LPS treatment15 and it is frequently deregulated in cancer16,17.
Here we showed that depletion of AGO2 stabilize MALAT-1 suggesting that it is regulated by miRNA. In addition, we showed that miR-9 targets MALAT-1 for degradation in the nucleus by directly binding to two miRNA binding sites in the MALAT-1 sequence.
Our data support a role for miRNAs in the regulation of expression of not only protein-coding genes but also other ncRNAs. This observation is consistent with a recent report by Hansen and colleagues showing that miR-671 can target the circular ncRNA NAT (natural antisense transcript) for AGO2- mediated degradation29. Recently, a number of ncRNAs have been suggested to act as sponges for miRNA competing for their binding to bone fide mRNA targets30,31,32. Whereas the high abundance and stability of MALAT1 could suggest a ceRNA function titrating the amount of miR-9, we do not detect any significant chances in miR-9 levels or in the expression in validated miR-9 targets following MALAT1 knockdown.
Concluding, our data demonstrates that canonical miRNA targeting is not merely confined to the cytoplasm, thus enlarging the complexity of these small molecules functions.
Methods
Cell transfection
The Hodgkin Lymphoma cell line L428 was grown at standard conditions in 5% CO2 at 37°C in RPMI 1640-glutamax (Gibco, Invitrogen) supplemented with 10% FBS (Hyclone, Thermo Scientific). Cells were transfected by nucleofection using solution L and program X-001 (Amaxa) and Nucleofector II (Amaxa). For siRNA experiments the cells were transfected with 80 nM siAGO2 and harvested 19 hours after transfection. For miR-9 overexpression cells were transfected with 50 nM miR-9 (Biosynthesis) and harvested 9 hours after transfection.
The glioblastoma cell line U87MG was grown in DMEM-glutamax (Gibco, Invitrogen), supplemented with 10% FBS (Hyclone, Thermo Scientific). Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions and harvested 48 h after transfection with 80 nM siAGO2. For miR-9 overexpression cells were transfected with 10 nM miR-9 mimic and harvested 6 hours after transfection.
AntimiR-9 and negative control have been described elsewhere15.
Paired-end sequencing
Isolation of mRNA from total RNA was achieved by two tandem purifications using a Dynabead mRNA purification kit (Invitrogen). The RNA samples for library preparation were obtained by mixing equal amounts of RNA from three different biological replicates. The RNA (2 μg) was reverse transcribed using Superscript double-stranded cDNA synthesis kit (Invitrogen). After adapter ligation, 75 bp paired-end RNA-seq reads were produced on the Illumina Genome Analyzer IIX and Hiseq 2000. Sequences were mapped to the GRCh37/hg19 genome and analyzed using the Bowtie v. 0.12.8, Tophat v. 2.0.4 and Cufflinks v. 2.0.233, with standard parameters. Sequencing data will be submitted to the NCBI Short Read Archive with accession number upon manuscript acceptance.
Luciferase reporters cloning and MALAT1 binding sites identification
The miR-9 binding sites on MALAT1 were identified by RNAhybrid 2.234. Please notice that, the actual binding site sequence starts at position 1192. pMALAT1(6229) was obtained by cloning MALAT1 (NR_002819.2) nucleotides 3171–7161 in pmiRreport (Ambion, Applied Biosystems) between SacI and MluI. pMALAT1(1199) was obtained by cloning MALAT1 (NR_002819.2) nucleotides 360–1620 in pmiRreport (Ambion, Applied Biosystems) between SpeI and HindIII All the constructs were checked by sequencing.
Mutants of the two plasmids were obtained mutating 2 bases in the miR-9 binding motif, using QuickChange Multi-site directed mutagenesis kit (Agilent technologies) and the following primers (mutated nucleotides are in bold):
5′-atactttttcacatttccagggtttgcatgttaactttaaatg-3′ for pMALAT1(6229)
5′-ggcggcggaaggtccgcgaattccggtg-3′ for pMALAT1(1199).
Pull-out assay and qPCRs
For affinity purification of miRNA targets, L428 cells were transfected with 2 nM miR-9 duplex, 3′-biotin-tagged miR-9 duplex or a mix of three 3′-biotin-tagged miR-9 duplexes containing a UV-reactive 4-thiouridine nucleotide at positions 4- and 17- and 21- respectively or a 4-tU mix for miR-34a as a control.
MicroRNA pull-down and luciferase reporter assays were performed as previously described15. The washes were performed in RNase free Water.
For detection of MALAT1 and ELF1 a TaqMan (Applied Biosystems) assays were used. Similarly, a TaqMan (Applied Biosystems) assay for HPRT was used as reference for relative quantification.
For detection of miR-9 TaqMan (Applied Biosystems) assays was used using as a reference RNU6B.
Fluorescent in situ hybridization for detection of miR-9
Fluorescent in situ Hybridization (FISH) for miR-9, was conducted as previously described35 on 4% paraformaldehyde (PFA) fixed slides with monolayers of U87MG, HEK293 and L428 cell lines, using a 5′-DIG-labelled probe (Exiqon) at 37.5 nM concentration. Hybridization was performed at 55°C for 1 hour, followed by visualization using anti-DIG-POD (Roche Applied Science) and FITC-labeled TSA (Perkin Elmer). Slides were subsequently counterstained using DAPI.
RNA immunoprecipitation
RNA immunoprecipitation was performed as previously described36. Briefly, nuclei were isolated from U87MG or L428 cells 24 hours after transfection using Nuclei EZ prep (Sigma) according to the manufacturer's instructions. Western blotting for vinculin was used to assess the purity of the separation. AGO2 was immunoprecipitated by using 500 μl of a monoclonal anti-Ago2 11A9 hybridoma supernatant (Kind gift from Gunter Meister) coupled to 25 μl of protein G Dynabeads (Invitrogen).
Author Contributions
E.L. and A.H.L. designed the overall study. K.H. performed miR-9 ISH; F.P. and E.L. performed all the experiments. J.W. and B.P. analyzed the sequencing data. S.K. and M.L. designed and provided all the antisense oligonucleotides used in this study. E.L. wrote the manuscript.
Supplementary Material
Supplementary informations
References
- Carnici P. et al. The Transcriptional Landscape of the Mammalian Genome. Science 309, 1559–1563 (2005). [DOI] [PubMed] [Google Scholar]
- Derrien T. et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Research 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 (2011). [DOI] [PubMed] [Google Scholar]
- Guo H., Ingolia N. T., Weissman J. S. & Bartel D. P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T. et al. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Research (2012). 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J. E., Freier S. M. & Spector D. L. 3′ End Processing of a Long Nuclear-Retained Noncoding RNA Yields a tRNA-like Cytoplasmic RNA. Cell 135, 919–932 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P. et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 (2003). [DOI] [PubMed] [Google Scholar]
- Tripathi V. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. et al. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 585, 671–676 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. et al. The lncRNA Malat1 Is Dispensable for Mouse Development but Its Transcription Plays a cis-Regulatory Role in the Adult. Cell Reports 2, 111–123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiβmann M. et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA biology 9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S. et al. Malat1 is not an essential component of nuclear speckles in mice. RNA 18, 1487–1499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoni F. et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. PNAS 106, 5282–5287 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucci E. et al. Inhibition of miR-9 de-represses HuR and DICER1 and impairs Hodgkin lymphoma tumour outgrowth in vivo. Oncogene 31, 1–9 (2012). [DOI] [PubMed] [Google Scholar]
- Yuva-Aydemir Y., Simkin A., Gascon E. & Gao F.-B. MicroRNA-9: Functional evolution of a conserved small regulatory RNA. rnabiology 8, 557–564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang G. et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. The EMBO Journal 31, 3513–3523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J., Loedige I. & Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610 (2010). [DOI] [PubMed] [Google Scholar]
- Hutchinson J. N. et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8, 39 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohrt T. et al. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 36, 6439–6449 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernilogar F. M. et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature 480, 391–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameyar-Zazoua M. et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 19, 998–1004 (2012). [DOI] [PubMed] [Google Scholar]
- Bernard D. et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. The EMBO Journal 29, 3082–3093 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. A., Valenstein M. L., Yario T. A., Tycowski K. T. & Steitz J. A. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENβ noncoding RNAs. PNAS. 109, 1–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J. E. et al. A triple helix stabilizes the 3' ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 26, 2392–2407 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Maeda S., Liu C., Karin M. & Edgington T. S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 26, 851–858 (2006). [DOI] [PubMed] [Google Scholar]
- Yamada K. et al. Phenotypic characterization of endometrial stromal sarcoma of the uterus. Cancer Science 97, 106–112 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A. K. L. & Sharp P. A. microRNAs: A Safeguard against Turmoil? Cell 130, 581–585 (2007). [DOI] [PubMed] [Google Scholar]
- Hansen T. B. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. The EMBO Journal 30, 4414–4422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., Kats L. & Pandolfi P. P. A ceRNA Hypothesis: The Rosetta Stone of a Hidden RNA Language? Cell 146, 353–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth F. A. et al. In Vivo Identification of Tumor- Suppressive PTEN ceRNAs in an Oncogenic BRAF-Induced Mouse Model of Melanoma. Cell 147, 382–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M. et al. A Long Noncoding RNA Controls Muscle Differentiation by Functioning as a Competing Endogenous RNA. Cell 147, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 516–520 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M. Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–1517 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soe M. J., Moller T., Dufva M. & Holmstrom K. A Sensitive Alternative for MicroRNA In Situ Hybridizations Using Probes of 2'-O-Methyl RNA + LNA. Journal of Histochemistry & Cytochemistry 59, 661–672 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peritz T. et al. Immunoprecipitation of mRNA-protein complexes. Nat Protoc 1, 577–580 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary informations