Abstract
Artemisinin constitutes the frontline treatment to aid rapid clearance of parasitaemia and quick resolution of malarial symptoms. However, the widespread promiscuity about its mechanism of action is baffling. There is no consensus about the biochemical target of artemisinin but recent studies implicate haem and PfATP6 (a calcium pump). We investigated the role of iron and artemisinin on PfATP6, in search of a plausible mechanism of action, via density functional theory calculations, docking and molecular dynamics simulations. Results suggest that artemisinin gets activated by iron which in turn inhibits PfATP6 by closing the phosphorylation, nucleotide binding and actuator domains leading to loss of function of PfATP6 of the parasite and its death. The mechanism elucidated here should help in the design of novel antimalarials.
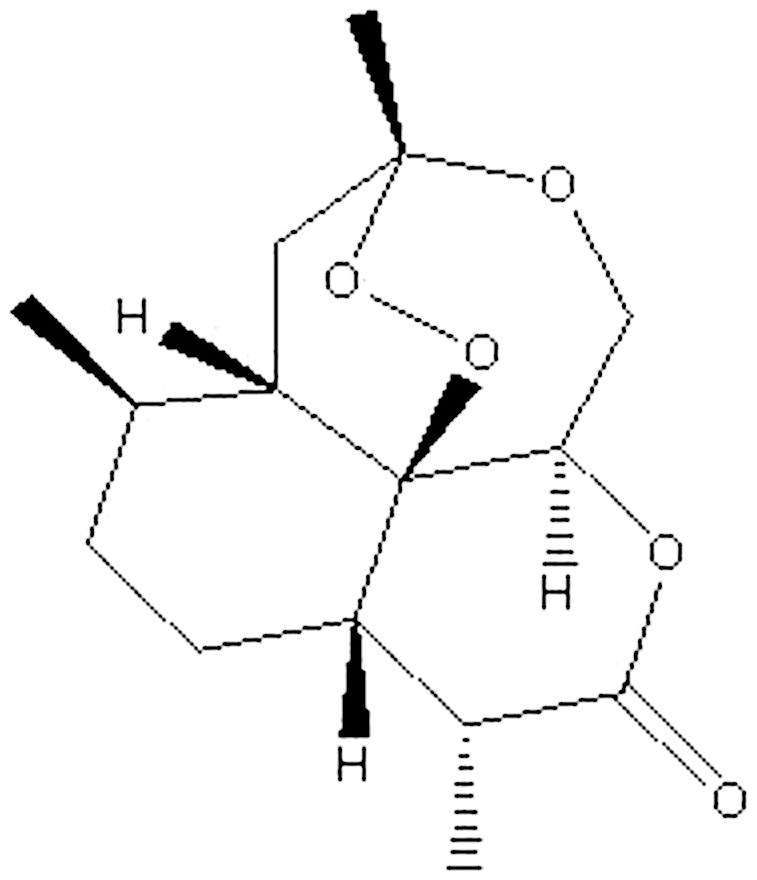
Plasmodium falciparum is among the most prevalent species infecting humans killing more than two million people every year1,2. Several reports have appeared in the recent past highlighting the complexity of its genomic organization, the metabolic pathways involved in infection and the potential for cure3,4,5,6. Resistance of these parasites to the traditional treatments has led to extensive work in discovering newer artemisinin analogs and derivatives7,8. Artemisinin (Fig. 1) is a sesquiterpene lactone endoperoxide containing a structural feature called peroxide bridge which is believed to be the key to its mode of action9. In spite of increasing popularity in the use of artemisinin based therapies, the mechanism of action of these sesquiterpene lactone endoperoxides has eluded researchers due to its controversial nature10.
Figure 1. Molecular structural formula of artemisinin.
According to one school of thought, the cleavage of the peroxide bridge in presence of ferrous ion (Fe2+) from haem forms highly reactive free radicals which rapidly rearrange to more stable carbon-centered radicals11,12. It has been suggested that these artemisinin-derived free radicals chemically modify and inhibit a variety of parasite molecules, resulting in parasite's death13. A rich source of intracellular Fe2+ is haem an essential component of hemoglobin, the malarial parasite is rich in haem iron derived from a breakdown of the host cell hemoglobin. It has long been suspected that Fe2+-haem is responsible for activating artemisinin inside the parasite14. Note that endoperoxides are known to be unstable, especially in the presence of iron. Theoretical studies on the cleavage of the peroxide bond by Taranto et al.15 and Araujo et al.16 suggest a thermodynamically favorable interaction between artemisinin and haem.
Alternative views propose a direct interaction between artemisinin and PfATP6, a calcium pump. PfATP6 is the only SERCA-type Ca2+-ATPase present in the malarial parasite17. Some studies suggested that mutations on PfATP6 of the malarial parasite were able to modulate the affinity of artemisinin for the protein18,19. The malarial enzyme was introduced into frog eggs and its activity was assessed, in the presence of artemisinin as well as thapsigargin, a known inhibitor for mammalian SERCA. It was found that both artemisinin and thapsigargin indeed inhibited SERCA. Furthermore, thapsigargin was found to interfere with the action of artemisinin indicating that they operated on malarial parasite through a similar mechanism18. In the parasite, the endoplasmic reticulum is situated outside the food vacuole throughout the cytoplasm, the same location as the proposed site of artemisinin action14. The parasite ingests and degrades up to 80% of the host-cell hemoglobin during its growth and replication process, in a compartment called food vacuole. Inside the malarial parasite, artemisinin, is activated by free iron neighboring PfATP6 in the endoplasmic reticulum. Abundance of ferrous ions within this vacuole, catalyze the cleavage of the peroxide bridge forming a highly reactive free radical14.
Modeling of PfATP6 enzyme based on templates 1IWO20,21 and 2DQS22 and docking of artemisinin derivative on to it was reported earlier. Jung et al.20 and Naik et al.21 obtained good correlation between their computed docking scores and in vitro antimalarial activities. However, no such correlations for several other antimalarial compounds with PfATP6 were observed22. Both 1IWO and 2DQS templates are from the same organism Oryctolagus Cunniculus bound with thapsigargin but having closed conformation of calcium pump. Reason for this contradictory results could be docking of artemisinin derivative to a static model of the enzyme or the starting structure which is in a closed conformation.
In a nutshell, two types of mechanism of action for artemisinin have been proposed, one involving iron (or haem)12,13 and the other independent of iron but involving PfATP618. There have been several supportive19,23,24 and dismissive reports for these mechanisms25,26 which makes the present study challenging27. It seems that a singular role of hemoglobin or PfATP6 may be insufficient to kill the parasite by artemisinin. We propose a mechanism of action of artemisinin which involves both iron and the binding of artemisinin to PfATP6 enzyme. Understanding the issues that govern PfATP6 domain mobility, studied by molecular dynamics simulation and docking in the present study, may have profound implications for elucidating the detailed mechanism of binding of drug to this enzyme and in the design of new therapeutic agents, such as allosteric inhibitors, intended to interfere with topological changes in the protein domains and thereby enzyme function.
Amino acid sequence alignment clearly shows that PfATP6 conserves all of the motifs and residues that are important for the structure or function of a mammalian SERCA pump28. Both PfATP6 and mammalian SERCA shares 51% identity. It is also reported that PfATP6 displays structure and function similar to mammalian SERCA present in skeletal muscle29. Krishna et al.30 and Salas-Burgos et al.31 modeled the structure of PfATP6, (PDB ID: 1U5N) for which 1SU4 was used as a template which is an open conformation crystal structure of calcium pump from the same organism Oryctolagus Cuniculus as studied earlier20,21,22 except that previous studies used a closed conformation. In order to get accurate binding affinities upon docking, the enzyme has to undergo significant conformational changes which no docking procedure allows. Closed conformation of PfATP6 models were built using 1IWO as template. Binding affinity computed using several docking and scoring methods could not differentiate between inhibitor and non-inhibitor (such as deoxy-artemisinin) in closed model of PfATP6. However, our goal is to seek a plausible mechanism action of artemisinin binding the enzyme whether that leads to similar change in conformation of PfATP6 as thapsigargin binding to mammalian SERCA (Open to closed conformations transformation leading to calcium pumping). Hence we studied from the model which is built using open conformation template (1SU4) of the enzyme to see the dynamic effect of enzyme ligand complex along with the solvent. Alignment and superposition of the structures of the two proteins (1SU4 mammalian SERCA and 1U5N (model of PfATP6) is shown in supplementary Fig. S1. In summary, PfATP6 retains all the characteristic features of the P-type ATPase family of SERCA pumps and can occur in broadly two conformations closed or open. There are 41 reported mammalian SERCA crystal structures in the RCSB32 till date, most recent ones being 3W5A33 and 4H1W34. All the structures except 1SU4 and 2C9M are bound with different ligands and have closed or semi closed (E1 and E2)35 conformations. A superposition of all these structures is shown in supplementary Fig. S2. The calculated average RMSD between the open conformation structure 1SU4 and the 39 closed conformations is ~13Å Individual RMSDs of all these above mentioned proteins with respect to 1SU4 are compiled in supplementary table T2.
Structure of PfATP6 is divided into three major parts: the cytoplasmic headpiece, the stalk domain and the transmembrane domain (supplementary Fig. S3). The headpiece is composed of mainly the phosphorylation (P) domain, the nucleotide binding (N) domain, the hinge domain, and the actuator (A) domain or C-terminal region. The ligand binding site of PfATP6 was identified with thapsigargin as a reference ligand. The binding site is near the center of four transmembrane helices. Small changes in these helices could lead to disruption of the Ca2+ binding sites and to alterations in the movement of ions to the cytosol or to luminal spaces36,37,38.
Thapsigargin is known to induce structural change i.e. convert the mammalian SERCA from and open conformation to closed form. It is thus likely that the previous studies on artemisinin which used closed conformation were indecisive with respect to the activity as well as mechanism of action. We thus hypothesize that artemisinin or its Fe-adduct may act in manner similar to thapsigargin on mammalian SERCA. It is thus of interest to study the open conformation of the enzyme and the influence of artemisinin and Fe-artemisinin adduct on the open conformation. Hence we adopted extensive molecular dynamics simulations of enzyme to analyze all the possible dynamic structures and intra domain motions in PfATP6.
In the present work, we have carried out density functional theory calculations on artemisinin, haem and their complexes and conducted molecular docking experiments of these compounds with PfATP6. We have further carried out all atom molecular dynamics simulations for 100 ns on (i) Ca2+ bound PfATP6, (ii) artemisinin bound PfATP6, and (iii) Fe-artemisinin adduct bound PfATP6 here after referred to as serca, arte-serca and Fe-arte-serca respectively. As a further control, we have also carried out molecular dynamics simulations on (iv) thapsigargin and mammalian SERCA complex.
Results
Density functional theory calculations on the complex between the haem group and artemisinin provided interaction energies, electronic states, partial charges on each atom and geometry of the complex. Resultant structure of the complex was used for docking with PfATP6. The docked conformations of artemisinin and Fe-artemisinin adduct in the active site of PfATP6 are depicted in supplementary Fig S4. The binding pocket of PfATP6 consists of LEU 263, PHE 264, GLN 267, ILE 977, ILE 981, ALA 985, ASN 1039, LEU 1040, ILE 1041 and ASN 1042 residues. Interaction of artemisinin with the residue in the active site in the docked structure are consistent with the results reported earlier20,22.
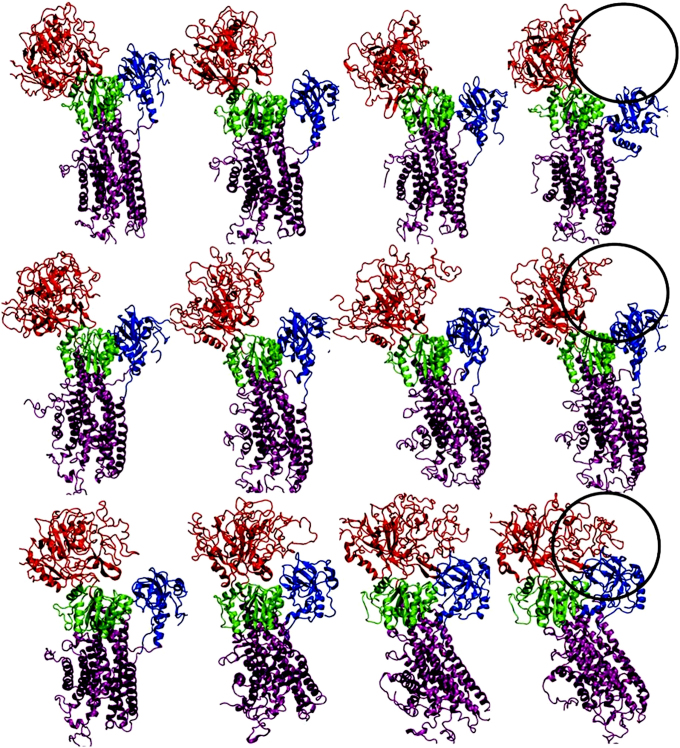
The essential massage of molecular dynamics (MD) simulations are captured in Fig. 2 in which systems (i) and (ii) viz. PfATP6 and artemisinin-PfATP6 complex show no major conformational changes. The two domains N & A, remain (~25Å) away from each other as in the native structure. However, they exhibit significant local fluctuations and some rearrangements. System (iii) namely, Fe-artemisinin adduct bound to PfATP6 shows a dramatic large conformational change resulting in a closure of N- and A- domains (see Fig. 2 and supplementary Fig. S5). Systerm (iv) too, namely thapsigargin bound to mammalian SERCA shows a similar large conformational change consistent with the experiment37 (also see supplementary Fig. S6 and S7).
Figure 2. MD snapshots generated from individual trajectories after every 25 ns for PfATP6 enzyme (top row), artemisinin bound PfATP6 (second row) and Fe-artemisinin adduct bound PfATP6 (third row).
Nucleotide binding (N) domain and the actuator (A) domains are seen to be closing in the case of Fe-artemisinin adduct bound PfATP6 system.
Binding energies of the enzyme and ligand complex were calculated33 as average over 50 structures extracted after every two ns from the 100 ns simulation trajectories for the artemisinin-PfATP6, Fe-artemisinin-PfATP6 and thapsigargin-PfATP6 complexes and reported in Table 1. Fe-Artemisinin binds relatively strongly to PfATP6 but has lesser affinity towards mammalian SERCA (Table 1). However thapsigargin binds strongly with mammalian SERCA and shows reasonable affinity towards PfATP6. Experiments have also shown that thapsigargin interfered with the action of artemisinin18. It is clear from Table 1 that artemisinin and Fe activated artemisinin bind preferentially to PfATP6. thapsigargin which binds strongly in the active site of mammalian SERCA36 has a larger volume 598 Å3 compared to artemisinin (258 Å3) as the active site of mammalian SERCA is large to fit thapsigargin tightly.
Table 1. Average binding free energies computed from the MD trajectories.
| Molecule | Target/Enzyme | Binding Free Energy (kcal/mol) |
|---|---|---|
| Artemisinin | PfATP6* | −6.5 |
| Fe-artemisinin | PfATP6* | −8.3 |
| Thapsigargin | PfATP6* | −6.7 |
| Artemisinin | Mammalian SERCA (1SU4) | −4.2 |
| Fe-artemisinin | Mammalian SERCA (1SU4) | −5.1 |
| Thapsigargin | Mammalian SERCA (1SU4) | −9.1 |
*Modelled using 1SU4 as described in text.
To compare the flexibility of the structures, root mean square fluctuations were calculated (supplementary Fig. S8). Unbound protein (SERCA) flexibility (supplementary Fig. S8a) was mostly localized in the loop regions of cytoplasmic domains. Differential fluctuation map of artemisinin-PfATP6 (Fig. S8b) Fe-artemisinin bound system (supplementary Fig. S8c) indicates suppressed and aggravated flexibility in specific regions. Fluctuations in the actuator domain, barring the residues which are connected to M1 helix, most of the residues display comparable fluctuations with respect to the native PfATP6 system. The nucleotide binding domain residues from 700 to 750 exhibit large fluctuations (supplementary Fig. S9). A comparison of arte-serca and Fe-arte-serca reveals the requirement for activation of artemisinin by Fe as essential to bringing about the large conformational changes in PfATP6 leading to its inactivation.
It is to be noted that (Fig. S3) the ATP-binding site in the N-domain is more than 25Å away from the critical phosphorylation site SER 357, and about 80Å away from the Ca2+-binding site in both PfATP6 and mammalian SERCA. On the other hand, binding of Ca2+ opens up the cleft in those domains. Thapsigargin, an inhibitor of mammalian SERCA, closes the N- and A- domain cleft, thus affecting the functioning of the enzyme37,39. Interestingly, mammalian SERCA, when inhibited by thapsigargin, leads to substantial conformational changes in the cytoplasmic headpiece. Three cytoplasmic domains that were widely separated in the open structure of the Ca2+-bound structure were found to come close to each other in the Ca2+-free enzyme upon inhibition by thapsigargin29,38,39. A similar conformational change is observed with Fe-artemisinin PfATP6 complex but not with artemisinin PfATP6 or unbound PfATP6, Analogous spontaneous opening and reclosing of flaps of HIV-1 protease with molecular dynamics simulations has been reported earlier40.
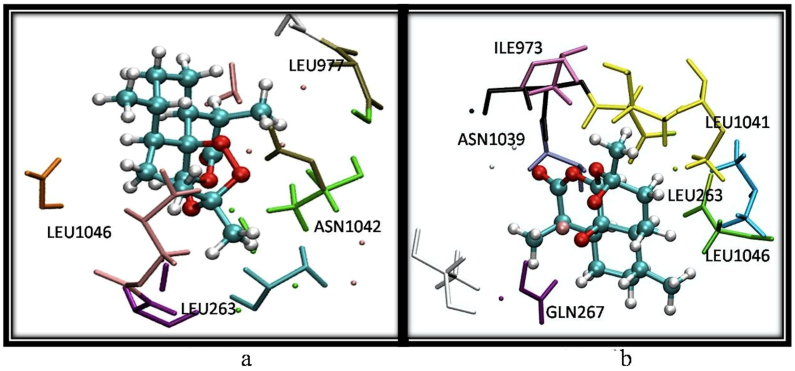
An analysis of the MD trajectories shows that Fe-arte-serca during simulation moves inside the transmembrane region (supplementary Figs. S10 and S11) forming hydrogen bonds with ASN 1040, ILE 1039 which are a part of the M5 helix. Additionally, bending of M5 helix is seen to occur accompanying a large conformational change in the open structure formed by the A-, P- and N-domains (Fig. 2). In Fe-arte-serca, changes in the topology of the enzyme structure are accompanied by a new interacting residue environment (Fig. 3a and 3b). Fe-artemisinin adduct moves towards the helical region forming hydrogen bond interactions with ASN 1039 and GLN 267 allowing the ring part of artemisinin to interact favorably with LEU 263, ILE 1041, ILE 1046, ILE 977 (Fig. 3a and 3b).
Figure 3. Residues of PfATP6 in the vicinity of 5Å after 40 ns from simulation trajectories of (a) artemisinin (system (ii)) and (b) Fe-artemisinin adduct (system (iii)).
Images were generated using VMD56.
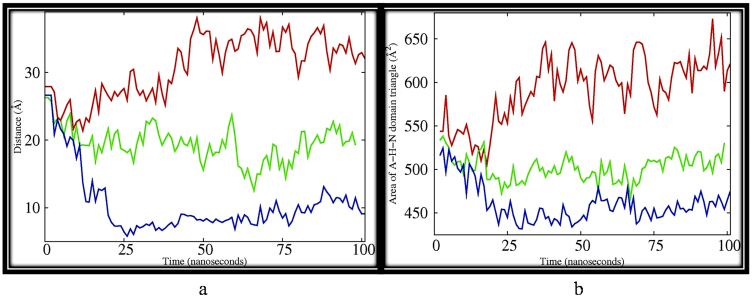
The cleft formed by the P- and N-domains closes which could facilitate phosphorylation of SER 35737. Inter-atomic distances of residues from the nucleotide binding domain and the actuator domain decrease during the simulation of Fe-arte-serca (Fig. 4a). To obtain a quantitative view, we plotted the areas (Fig. 4b) of A-, H- and N-domains which is indicative of the movement of cytoplasmic domains towards each other.
Figure 4.
(a) Inter-atomic distances of backbone atom of VAL 221 of actuator domain and PHE 535 of nucleotide domain for (i) PfATP6 (red), (ii) artemisinin bound PfATP6 (green) and (iii) Fe-artemisinin adduct bound PfATP6 (blue) depicting the two residues coming closer from 25Å to almost 8 Å for most part of the simulation (iii). However smaller variation in the distances were noticed with PfATP6 (red), (26 to 32 Å). For artemisinin bound PfATP6 system (green) (26- 20 Å). (b) Area between the geometric centers of N-, H- and A- domains of PfATP6 system (red), artemisinin–PfATP6 system (green) and Fe-artemisinin adduct PfATP6 system (blue). Areas are calculated from triangles formed by the geometric centers of the three corresponding domains.
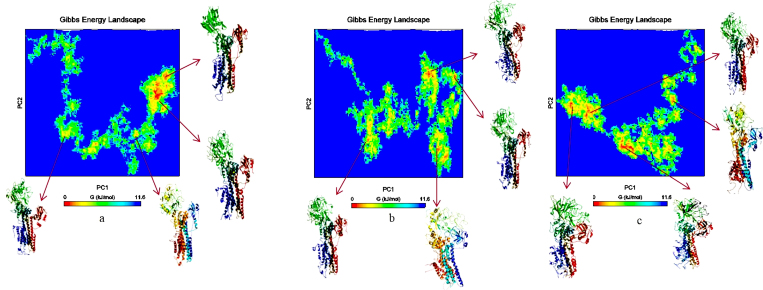
To further understand the structural information with thermodynamics of the open to closed transition, we constructed free energy landscapes from the MD trajectories performing principal component analysis (PCA). PCA41,42 uses the actual dynamics of the protein to generate the motions of the native state of the protein conformation. In Fig. 5 the energy landscape in Fe-arte-serca system reveals characteristics that are not observed either with systems serca or arte-serca. There is a well-defined minimum structurally representing the closed conformation state in Fe-arte-serca. The red region in all the plots has the least Gibbs free energy and their corresponding structures, which are most probable, are pointed with arrows.
Figure 5. PCA based free energy landscape of open to closed conformational transition of (i) SERCA; (i) Ca2+ bound SERCA, (ii) artemisinin bound SERCA, (iii) Fe-artemisinin adduct bound SERCA.
The abscissa and ordinates correspond to the first and second principal components respectively.
A closer look (supplementary Fig. S4) at the ligand binding region shows that, Fe-artemisinin adduct forming directional hydrogen bonds with the backbone atoms of the functionally important residues LEU 26319, ILE 97720 and ASN 103920. These H-bonds may guide Fe-artemisinin adduct deep into the membrane disrupting the helical region leading to strong and stable electrostatic interaction between iron and side chain of ASN 980 from ~10 Å to ~3Å (supplementary Fig. S12). ASN 1039 (equivalent to mammalian PRO 827) and LEU263 (equivalent to mammalian GLU 255) seem to be giving selectivity to the artemisinin binding to PfATP6.
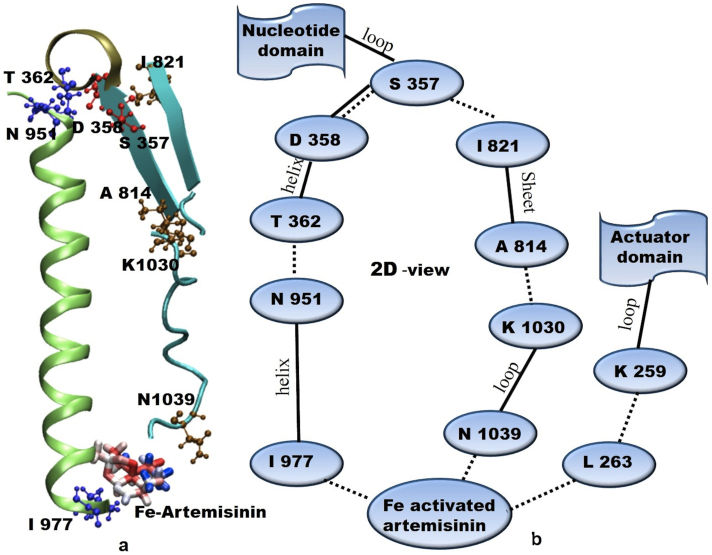
A key question is how the ligand binding site and the phosphorylation site, 25Å apart, communicate with each other. In this regard, it is important to note that the cytoplasmic end of M5 is integrated into the P-domain near the phosphorylation site and hydrogen bonded to the M4 helix. We analyzed the hydrogen bond linkage from the simulation trajectory of Fe-arte-serca system. There is an apparent linkage of network between Fe-artemisinin adduct and SER 357 (phosphorylation site) which plays a pivotal role in the functioning of SERCA. Also, SER 357 is directly connected to the nucleotide domain through a loop. Fe-artemisinin adduct makes hydrogen bond with ASN 1039 which is connected to SER 357 (phosphorylation site) through hydrogen bonds, loop region and sheets. (Fig. 6). A tilt and lateral shift of M5 which leads to the large movement of nucleotide domain can also be attributed to the analogous linkage of Fe-artemisinin adduct with ILE 977 mentioned earlier. Main chain and H-bond connectivity are shown in Fig. 6a. Residue ASN 951, at one terminal of the M5 helix shows strong interaction with THR 362 of phosphorylation domain, thus directly modulating the motion of the nucleotide domain. In addition to this, interaction of Fe-artemisinin adduct and residues of M4 (ASN 1039 and loop) helix also strongly influence the movement of the nucleotide domain. A network of hydrogen bonds between Fe-artemisinin adduct, LEU 263 and LYS 259, attached to the actuator domain through a loop, regulates the motion of the actuator domain as shown in Fig. 6b. There are other networks of ASP 358 alongside M5 and M4 where Fe-artemisinin adduct is located. However, the interactions mentioned above were found to be stronger indicated by a high survival of hydrogen bonds throughout the trajectory (almost 70% of the simulation run time taking hydrogen bond distance cut off as 3.2 Å). These hydrogen bond linkages were not found with artemisinin bound complex during the simulation (supplementary Fig. S13). It is quite probable that many H-bond networks amongst P domain and the M5 and M4 helices in association with other sites (Fig. 6) are responsible for the allosteric effect between the ligand binding site and the cytosolic regions, causing the closure of two domains leading to inaccessibility of calcium ion at the calcium binding site. Thus, the events that occur at either site can be mechanically transmitted to the other site.
Figure 6. Interaction network of Fe-artemisinin adduct and phosphorylation site through bonded and non-bonded network.
(a) A three dimensional view; (b) A two dimensional representation of the same linkage.
Discussion
Present study shows that Fe-artemisinin adduct upon binding to an open jaw like structure of PfATP6 comprising phosphorylation, nucleotide binding and actuator domains, is seen to cause large conformational changes in PfATP6 leading to a closure of these three domains causing inaccessibility of the calcium binding site and hence loss of function of Ca2+ pump of the parasite and its resultant death. The dynamic behavior in terms of flexibility, conformation and the inhibitor-enzyme interaction of artemisinin and Fe-artemisinin adduct in the PfATP6 binding pocket are successfully explained by molecular simulations. The studies lead to the inference that Fe- artemisinin adduct inhibits PfATP6 through an allosteric mechanism. Present studies on the binding mechanism of PfATP6 and Fe-artemisinin adduct could prove helpful in the design of new antimalarials.
Methods
Density functional theory (DFT) calculations
The geometries of both artemisinin and the haem group were fully optimized using density-functional theory as implemented in the ADF package43 and as carried out by Araujo et al. The states with singlet, triplet and quintet multiplicities (S = 0, 1 and 2, respectively) were calculated for haem, while triplet, quintet, and septet states (S = 1, 2, and 3, respectively) were calculated for the complex between haem and artemisinin. Triple zeta with single polarization (TZP) basis set and PBE functional44 was used. The computed charges on each oxygen atoms are provided in Supplementary Table T1. It was found that for interaction at short distances, due to artemisinin reduction by the haem group, the most stable complex has a septet spin state. Further computational details and the more insight into the interaction of haem and artemisinin can be found in that work. The resulting geometry which were of the same order as in the work of Araujo et al. were used for docking.
Docking
Docking of artemisinin and Fe-artemisinin adduct to PfATP6 was carried out using Pardock45,46,47,48,49. Scoring of these docked structures was done by Bappl49. Details of the docking methodology are provided in the supplementary information. These docked structures were taken as input for molecular dynamics simulations.
Molecular dynamics
All MD simulations were carried out using GROMACS50. Amber FF99SB51 force field was used. The complexes obtained from Pardock were solvated in a box of TIP3P52 water molecules (and 30 counter ions to ensure electro-neutrality) and minimized for an additional 2500 steps (1000SD + 1500CG). All these systems sizes comprised around ~275,000 atoms. MD Simulations were started by slowly heating the solvent to 300 K over a period of 100 ps keeping the complex fixed. After this, the entire system was gradually brought to 300 K over a period of 300 ps. The simulation was then carried out under NPT conditions for 100 ns. A 2 femto second time step was used for integrating the equations of motion. Periodic boundary conditions were applied throughout the MD simulations, along with PME summation53 for treating electrostatics. The temperature was kept constant by coupling heat bath through the Berendsen algorithm54 using separate solute and solvent scaling. Pressure was adjusted by isotropic position scaling using a Berendsen like algorithm. Covalent bonds to hydrogen atoms were constrained by the SHAKE algorithm55. Convergence of energy, density were monitored. The initial 10 ns was treated as equilibration and the subsequent 100 ns was treated as production phase for structural and energy analyses.
Author Contributions
B.J. and I.G. designed and conceived the study. A.S. and S.C. carried out the various computational studies and contributed equally. All authors contributed in writing and approved the final manuscript.
Supplementary Material
A plausible mechanism for the antimalarial activity of artemisinin: A computational approach
Acknowledgments
We acknowledge the program support from the Department of Biotechnology (DBT), Government of India, to COE and the Supercomputing Facility for Bioinformatics and Computational Biology (SCFBio), IIT Delhi. Support from the Department of Science and Technology (DST) to the High Performance Computing Facility, Inter University Accelerator Centre and financial assistance to SC are also gratefully acknowledged.
References
- Snow R. W., Guerra C. A., Noor A. M., Myint H. Y. & Hay S. I. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434, 214–217 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty P. The numbers game. Nature 484, S14–S15 (2012). [DOI] [PubMed] [Google Scholar]
- Winzeler E. A. Malaria research in the post-genomic era. Nature 455, 751–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily J. et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature 450, 1091–1095 (2007). [DOI] [PubMed] [Google Scholar]
- Wiesner J., Ortmann R., Jomaa H. & Schlitzer M. New Antimalarial Drugs. Angewandte Chemie International Edition 42, 5274–5293 (2003). [DOI] [PubMed] [Google Scholar]
- Li J. & Zhou B. Biological actions of artemisinin: insights from medicinal chemistry studies. Molecules 15, 1378–1397 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennerstrom J. L. et al. Identification of an antimalarial synthetic trioxolane drug development candidate. Nature 430, 900–904 (2004). [DOI] [PubMed] [Google Scholar]
- Kim H.-S. et al. Synthesis and Antimalarial Activity of Cyclic Peroxides, 1,2,4,5,7-Pentoxocanes and 1,2,4,5-Tetroxanes. Journal of Medicinal Chemistry 42, 2604–2609 (1999). [DOI] [PubMed] [Google Scholar]
- Klayman D. L. Qinghaosu (artemisinin): an antimalarial drug from China. Science 228, 1049–1055 (1985). [DOI] [PubMed] [Google Scholar]
- Posner G. H. & O'Neill P. M. Knowledge of the proposed chemical mechanism of action and cytochrome P450 metabolism of antimalarial trioxanes like artemisinin allows rational design of new antimalarial peroxides. Accounts of chemical research 37, 397–404 (2004). [DOI] [PubMed] [Google Scholar]
- Meshnick S. R., Taylor T. & Kamchonwongpaisan S. Artemisinin and the antimalarial endoperoxides: from herbal remedy to targeted chemotherapy. Microbiological reviews 60, 301–315 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner G. H. et al. Further evidence supporting the importance of and the restrictions on a carbon-centered radical for high antimalarial activity of 1, 2, 4-trioxanes like artemisinin. Journal of medicinal chemistry 38, 2273–2275 (1995). [DOI] [PubMed] [Google Scholar]
- Meshnick S. R., Thomas A., Ranz A., Xu C.-M. & Pan H.-Z. Artemisinin (qinghaosu): the role of intracellular hemin in its mechanism of antimalarial action. Molecular and biochemical parasitology 49, 181–189 (1991). [DOI] [PubMed] [Google Scholar]
- Ridley R. G. Malaria: to kill a parasite. Nature 424, 887–889 (2003). [DOI] [PubMed] [Google Scholar]
- Taranto A., Carneiro J. d. M. & Oliveira, F. MNDO/d calculations on the interaction between artemisinin and heme. Journal of Molecular Structure: Theochem 539, 267–272 (2001). [Google Scholar]
- Araujo J. Q., Carneiro J. W., de Araujo M. T., Leite F. H. & Taranto A. G. Interaction between artemisinin and heme. A Density Functional Theory study of structures and interaction energies. Bioorg Med Chem 16, 5021–9 (2008). [DOI] [PubMed] [Google Scholar]
- Gardner M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckstein-Ludwig U. et al. Artemisinins target the SERCA of Plasmodium falciparum. Nature 424, 957–961 (2003). [DOI] [PubMed] [Google Scholar]
- Uhlemann A. C. et al. A single amino acid residue can determine the sensitivity of SERCAs to artemisinins. Nat Struct Mol Biol 12, 628–9 (2005). [DOI] [PubMed] [Google Scholar]
- Jung M., Kim H., Nam K. Y. & No K. T. Three-dimensional structure of Plasmodium falciparum Ca2+ -ATPase(PfATP6) and docking of artemisinin derivatives to PfATP6. Bioorg Med Chem Lett 15, 2994–7 (2005). [DOI] [PubMed] [Google Scholar]
- Naik P. K. et al. The binding modes and binding affinities of artemisinin derivatives with Plasmodium falciparum Ca2+-ATPase (PfATP6). Journal of molecular modeling 17, 333–357 (2011). [DOI] [PubMed] [Google Scholar]
- Garah F. B., Stigliani J. L., Cosledan F., Meunier B. & Robert A. Docking studies of structurally diverse antimalarial drugs targeting PfATP6: no correlation between in silico binding affinity and in vitro antimalarial activity. ChemMedChem 4, 1469–79 (2009). [DOI] [PubMed] [Google Scholar]
- Krishna S., Pulcini S., Fatih F. & Staines H. Artemisinins and the biological basis for the PfATP6/SERCA hypothesis. Trends in parasitology 26, 517–523 (2010). [DOI] [PubMed] [Google Scholar]
- Krishna S., Pulcini S., Fatih F. A. & Staines H. M. More depth of field not wider focus needed. Trends in parasitology 27, 3–4 (2011). [Google Scholar]
- Del Pilar Crespo M. et al. Artemisinin and a series of novel endoperoxide antimalarials exert early effects on digestive vacuole morphology. Antimicrobial agents and chemotherapy 52, 98–109 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodrow C. J. & Bustamante L. Y. Mechanisms of artemisinin action and resistance: wider focus is needed. Trends in parasitology (2010). [DOI] [PubMed] [Google Scholar]
- Padmanaban G., Nagaraj V. A. & Rangarajan P. N. Drugs and drug targets against malaria. Current science 92, 1545–1555 (2007). [Google Scholar]
- Masatsugu K., Yoshiko Y., Suehisa T. & Kazuyuki T. Cloning of a Ca2+-ATPase gene of Plasmodium falciparum and comparison with vertebrate Ca2+ -ATPases. Journal of cell Science 104, 1129–1136 (1993). [DOI] [PubMed] [Google Scholar]
- Møller J. V., Olesen C., Winther A.-M. L. & Nissen P. The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Quarterly reviews of biophysics 43, 501–566 (2010). [DOI] [PubMed] [Google Scholar]
- Krishna S. et al. Homology model of PfATP6. www.rcsb.org. Retrieved on 6 December, 2009.
- Salas-Burgos A., Iserovich P., Zuniga F., Vera J. C. & Fischbarg J. Predicting the three-dimensional structure of the human facilitative glucose transporter glut1 by a novel evolutionary homology strategy: insights on the molecular mechanism of substrate migration, and binding sites for glucose and inhibitory molecules. Biophysical journal 87, 2990–2999 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M. et al. The protein data bank. Nucleic acids research 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman P. A. et al. Calculating structures and free energies of complex macromolecules: Combining molecular mechanics and continuum models. Accounts of chemical research 33, 889–897 (2000). [DOI] [PubMed] [Google Scholar]
- Winther A. M. L. et al. The sarcolipin-bound calcium pump stabilizes calcium sites exposed to the cytoplasm. Nature 495, 265–269 (2013). [DOI] [PubMed] [Google Scholar]
- Lee A. Ca2+-ATPase structure in the E1 and E2 conformations: mechanism, helix–helix and helix–lipid interactions. Biochimica et Biophysica Acta-Biomembranes 1565, 246–266 (2002). [DOI] [PubMed] [Google Scholar]
- Toyoshima C., Nakasako M., Nomura H. & Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature 405, 647–655 (2000). [DOI] [PubMed] [Google Scholar]
- Toyoshima C. & Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature 418, 605–611 (2002). [DOI] [PubMed] [Google Scholar]
- Ogawa H., Stokes D. L., Sasabe H. & Toyoshima C. Structure of the Ca2+ pump of sarcoplasmic reticulum: a view along the lipid bilayer at 9-A resolution. Biophysical journal 75, 41 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martonosi A. N. & Pikula S. The structure of the Ca 2+-ATPase of sarcoplasmic reticulum. Acta Biochmica Polonica-English Edition 50, 337–366 (2003). [PubMed] [Google Scholar]
- Hornak V., Okur A., Rizzo R. C. & Simmerling C. HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations. Proceedings of the National Academy of Sciences of the United States of America 103, 915–920 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisuradze G. G., Liwo A. & Scheraga H. A. Principal component analysis for protein folding dynamics. Journal of molecular biology 385, 312–329 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsera M. A., Wriggers W., Oono Y. & Schulten K. Principal component analysis and long time protein dynamics. The Journal of Physical Chemistry 100, 2567–2572 (1996). [Google Scholar]
- Te Velde G. et al. Chemistry with ADF. Journal of Computational Chemistry 22, 931–967 (2001). [Google Scholar]
- Ernzerhof M. & Scuseria G. E. Assessment of the Perdew–Burke–Ernzerhof exchange-correlation functional. The Journal of chemical physics 110, 5029 (1999). [DOI] [PubMed] [Google Scholar]
- Shaikh S. A., Jain T., Sandhu G., Latha N. & Jayaram B. From drug target to leads-sketching a physicochemical pathway for lead molecule design in silico. Current pharmaceutical design 13, 3454–3470 (2007). [DOI] [PubMed] [Google Scholar]
- Gupta A., Gandhimathi A., Sharma P. & Jayaram B. ParDOCK: An all atom energy based Monte Carlo docking protocol for protein-ligand complexes. Protein and peptide letters 14, 632–646 (2007). [DOI] [PubMed] [Google Scholar]
- Jayaram B. et al. Sanjeevini: a freely accessible web-server for target directed lead molecule discovery. BMC Bioinformatics 13, S7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T., Biswas D. & Jayaram B. AADS-An Automated Active Site Identification, Docking, and Scoring Protocol for Protein Targets Based on Physicochemical Descriptors. Journal of chemical information and modeling 51, 2515–2527 (2011). [DOI] [PubMed] [Google Scholar]
- Jain T. & Jayaram B. An all atom energy based computational protocol for predicting binding affinities of protein–ligand complexes. FEBS letters 579, 6659–6666 (2005). [DOI] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D. & Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of chemical theory and computation 4, 435–447 (2008). [DOI] [PubMed] [Google Scholar]
- Cornell W. D. et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. Journal of the American Chemical Society 117, 5179–5197 (1995). [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of simple potential functions for simulating liquid water. The Journal of chemical physics 79, 926 (1983). [Google Scholar]
- Essmann U. et al. A smooth particle mesh Ewald method. Journal of Chemical Physics 103, 8577–8593 (1995). [Google Scholar]
- Berendsen H. J., Postma J. P. M., van Gunsteren W. F., DiNola A. & Haak J. Molecular dynamics with coupling to an external bath. The Journal of chemical physics 81, 3684 (1984). [Google Scholar]
- Ryckaert J.-P., Ciccotti G. & Berendsen H. J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics 23, 327–341 (1977). [Google Scholar]
- Humphrey W., Dalke A. & Schulten K. VMD: visual molecular dynamics. Journal of molecular graphics 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A plausible mechanism for the antimalarial activity of artemisinin: A computational approach