Abstract
Improvement of leaf photosynthesis is an important strategy for greater crop productivity. Here we show that the quantitative trait locus GPS (GREEN FOR PHOTOSYNTHESIS) in rice (Oryza sativa L.) controls photosynthesis rate by regulating carboxylation efficiency. Map-based cloning revealed that GPS is identical to NAL1 (NARROW LEAF1), a gene previously reported to control lateral leaf growth. The high-photosynthesis allele of GPS was found to be a partial loss-of-function allele of NAL1. This allele increased mesophyll cell number between vascular bundles, which led to thickened leaves, and it pleiotropically enhanced photosynthesis rate without the detrimental side effects observed in previously identified nal1 mutants, such as dwarf plant stature. Furthermore, pedigree analysis suggested that rice breeders have repeatedly selected the high-photosynthesis allele in high-yield breeding programs. The identification and utilization of NAL1 (GPS) can enhance future high-yield breeding and provides a new strategy for increasing rice productivity.
Rice (Oryza sativa L.) is a staple food that feeds nearly half of the world's population. Over the past half century, rice production has more than doubled owing to the advent of high-yielding semi-dwarf cultivars, including IR8, sometimes called “miracle rice”. IR8 has a short-stature phenotype, caused by the sd1 (semi-dwarf1) gene, which enables the plant to resist lodging and thus allows treatment with higher levels of fertilizer, resulting in high yields. This surprising success contributed to the “Green Revolution”1,2. However, the current world population of over 7 billion and the predicted growth to 9 billion by 2050 will require a 60% to 70% further increase in rice production without a corresponding increase in the amount of agricultural land3,4.
Rice yield is determined by the balance of a number of factors, including sink size, source strength, and carbohydrate translocation. Several genes underlying quantitative trait loci (QTLs) associated with sink size have been identified, including several for grain number regulation (GN1a5, APO16, and WFP7) and grain size regulation (GW28, GS39, qSW510, and GS511). However, near-isogenic lines (NILs) having favourable GN1a and APO1 alleles have not markedly improved yield in the japonica genetic background, illustrating the importance of the balance among sink size, source strength, and carbohydrate translocation for attaining high yield12.
Leaf photosynthesis is one of the main aspects of source strength, and rice simulation models indicate that leaf photosynthesis, particularly in flag leaves, plays an important role in determining crop yield13,14. Wide natural variation in light-saturated photosynthesis rate under ambient CO2 concentration has been observed among rice cultivars15,16. The photosynthesis rate is generally determined by both CO2 supply to chloroplasts and demand for CO2 in the chloroplasts17,18. The CO2 supply is governed by diffusion of CO2 from the atmosphere through stomata to the sites of carboxylation in the chloroplasts. One of the factors involved in the CO2 supply is stomatal conductance, an indication of stomatal aperture17. The demand for CO2 is governed by the rate of CO2 processing in the chloroplast. One of the factors involved in the demand for CO2 is the amount of active ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco)18. Because large amounts of nitrogen (N) are invested in Rubisco, leaf N content is also recognized as an important factor for photosynthesis19,20. The natural variation in photosynthesis rate is well explained by variation in both stomatal conductance and leaf N content21,22. While these eco-physiological characteristics of rice photosynthesis have been elucidated, genes associated with the variation in photosynthesis rate are yet to be identified, owing to the complexity of this trait23,24.
In this study, QTL analysis combined with map-based cloning using Takanari, a high-yielding, high-photosynthesis rice cultivar, led to the identification and isolation of a major QTL controlling the photosynthesis rate in flag leaves of rice. An NIL carrying the favourable allele for this QTL had an enhanced photosynthesis rate caused by an increase in carboxylation efficiency, which originated from pleiotropic effects of leaf anatomical modifications. A subsequent pedigree analysis revealed that rice breeders have repeatedly selected this allele in high-yield breeding programs. These results suggest that the identification and utilization of photosynthesis-related QTLs can enhance future high-yield breeding and provide a new strategy for achieving increased rice productivity.
Results
QTL analysis
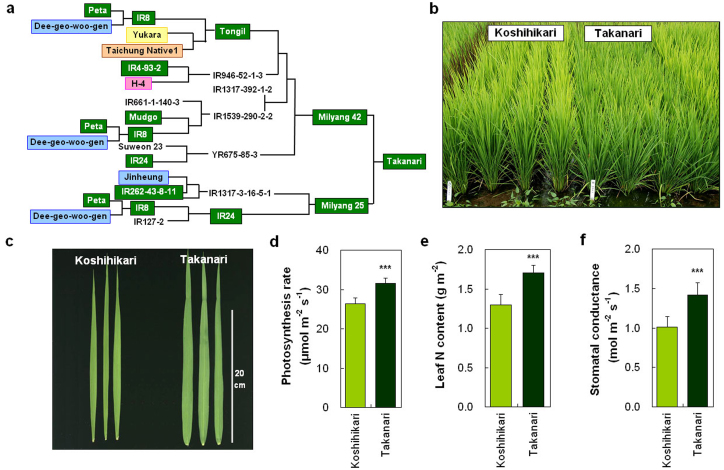
To identify genes controlling photosynthesis rate, we chose a high-yielding indica rice cultivar, Takanari, and a leading japonica cultivar, Koshihikari, both grown in Japan. Takanari has one of the highest photosynthesis rates among cultivars in global rice core collections21, and the greatest difference in photosynthesis rate between Koshihikari and Takanari at any time during the growth period was in flag leaves at the full heading stage25. Takanari, which is descended from high-yielding cultivars including IR8 (Fig. 1a), possesses the sd1 gene26 and has shorter plant stature but darker green and wider flag leaves than Koshihikari (Fig. 1b, c). As expected, the photosynthesis rate of flag leaves at the full heading stage was higher in Takanari than in Koshihikari (Fig. 1d). Leaf N content and stomatal conductance were also higher in Takanari than in Koshihikari (Fig. 1e, f).
Figure 1. Characteristics of high-yielding rice cultivar Takanari.
(a) Pedigree of Takanari. Cultivars marked with the same colour box have the same GPS haplotype. (b, c) Plant morphology (b) and flag leaves (c) of field-grown Koshihikari and Takanari. (d–f) Comparisons of photosynthesis rate (d), N content (e), and stomatal conductance (f) of flag leaves at full heading stage between Koshihikari and Takanari. Each column represents mean ± s.d. (n = 18); ***P < 0.001 versus Koshihikari (Student's t-test).
We developed primary mapping populations of reciprocal backcross inbred lines (BILs) derived from a cross between Koshihikari and Takanari. Both sets of BILs, consisting of 82 lines in the Koshihikari genetic background and 86 in the Takanari background, were used to make detection and mapping of QTLs more precise. We measured the leaf photosynthesis rate in flag leaves of the reciprocal BILs at the full heading stage with a portable photosynthesis system (LI-6400; Li-Cor) under an optimal and constant leaf chamber conditions in the morning on clear days. Photosynthesis rate showed continuous variation in both mapping populations (Supplementary Fig. S1). QTL analysis with 140 molecular markers detected two QTLs in the Koshihikari background and four in the Takanari background (Supplementary Fig. S1, Supplementary Table S1). Among them, a QTL on the long arm of chromosome 4 was commonly detected in both mapping populations, with the allele from Takanari contributing to an increase in photosynthesis rate. Our earlier studies also found QTLs associated with photosynthesis-related traits such as chlorophyll content on the long arm of chromosome 427,28. Thus, we selected this QTL, here named GREEN FOR PHOTOSYNTHESIS (GPS), for further analysis.
Physiological characterization of GPS
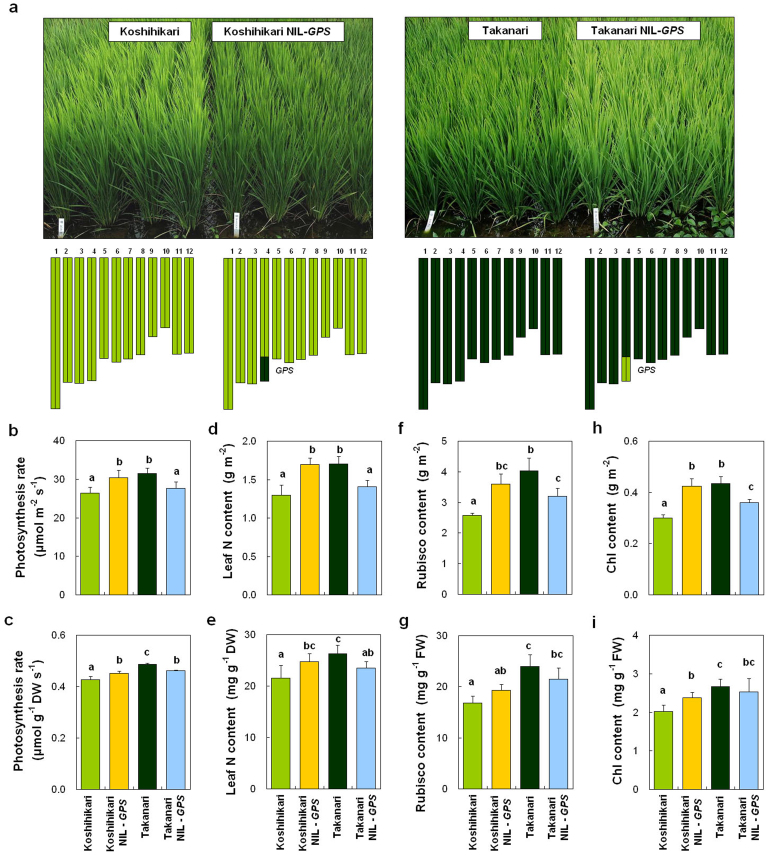
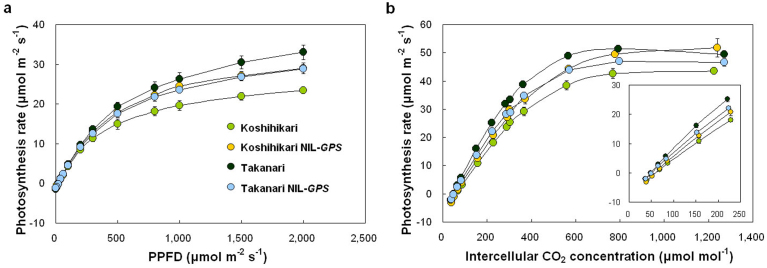
For map-based cloning and characterization of QTLs, we developed reciprocal NILs-GPS, each carrying the GPS region from either Koshihikari or Takanari in the genetic background of the other (Fig. 2a). Koshihikari NIL-GPS (i.e., Koshihikari background containing GPS from Takanari) had darker green leaves than Koshihikari, and Takanari NIL-GPS had lighter green leaves than Takanari (Fig. 2a). Comparison of flag leaf photosynthesis rates confirmed the effect of GPS: Koshihikari NIL-GPS had a higher photosynthesis rate per unit leaf area than Koshihikari, and Takanari NIL-GPS had a lower photosynthesis rate than Takanari (Fig. 2b). These differences in photosynthesis rate were observed even when expressed per unit dry weight (Fig. 2c). Higher photosynthesis rates were associated with enhanced leaf N, Rubisco, and chlorophyll contents per unit leaf area, and in some cases per unit fresh or dry weight (Fig. 2d–i). Koshihikari NIL-GPS also exhibited greater responses to light and CO2 than did Koshihikari, most notably a higher initial slope of the CO2 response curve, reflecting higher carboxylation efficiency (Fig. 3a, b). According to a biochemical model of C3 photosynthesis (the “Farquhar model”), photosynthesis rate is limited either by ribulose-1,5-bisphosphate (RuBP) carboxylation by Rubisco (carboxylation efficiency) or by RuBP regeneration in response to CO2 concentration29. To examine which step limited the photosynthesis rate in the parental cultivars and the reciprocal NILs-GPS under our experimental conditions (light saturation and ambient CO2), we fitted the model to the CO2 response data. Analysis revealed that the photosynthesis rate in all cultivars and NILs-GPS was limited by RuBP carboxylation by Rubisco (Supplementary Fig. S2). These results indicate that the primary role of GPS is related to carboxylation efficiency during photosynthesis. We further examined the relationship between Rubisco content and Vc,max, the maximum rate of RuBP carboxylation by Rubisco, obtained from the model-fitting analysis. Vc,max was closely related to Rubisco content (Supplementary Fig. S3). These results suggest that the increase in Rubisco content seen in lines containing the Takanari allele of GPS led to an increase in Vc,max, in turn increasing carboxylation efficiency and consequently photosynthesis rate. On the other hand, no significant difference was observed in stomatal conductance between each parental cultivar and the corresponding NIL-GPS (Supplementary Fig. S4). Neither stomatal density nor stomatal length differed between each parental cultivar and the corresponding NIL-GPS (Supplementary Fig. S4).
Figure 2. Physiological characterization of GPS alleles.
(a) Plant morphology and graphical genotypes of Koshihikari, Koshihikari NIL-GPS, Takanari, and Takanari NIL-GPS. Yellow-green bars, chromosome regions from Koshihikari; dark green, Takanari. (b–i) Comparisons of photosynthesis rate (b, c), leaf N content (d, e), Rubisco content (f, g), and chlorophyll (Chl) content (h, i) of flag leaves at full heading stage. Each column represents mean ± s.d. (n = 9). Different letters indicate significant difference at the 5% level (Tukey–Kramer HSD test).
Figure 3. Responses of GPS to light and CO2.
(a) Effect of light intensity on photosynthesis in flag leaves at full heading stage in Koshihikari, Koshihikari NIL-GPS, Takanari, and Takanari NIL-GPS (n = 3). PPFD, photosynthetic photon flux density. (b) Effect of intercellular CO2 concentration on photosynthesis in flag leaves at full heading stage in the four genotypes (n = 3).
Cloning of GPS
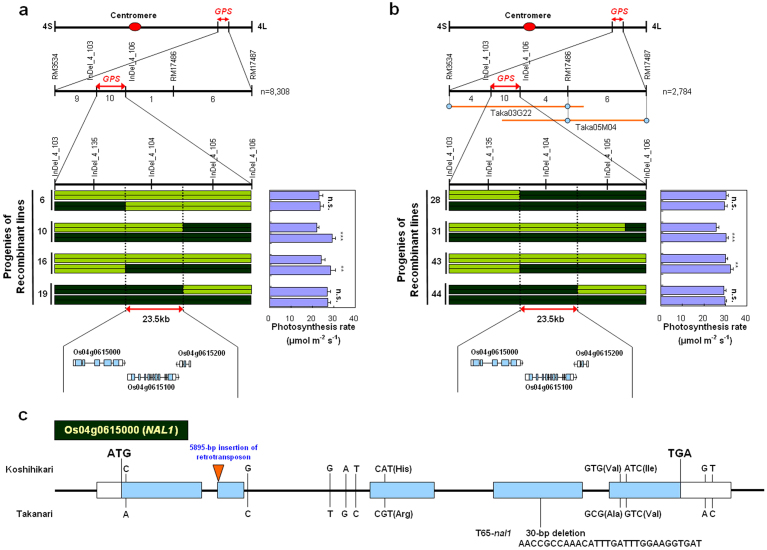
For map-based cloning of GPS, we first used 142 F2 plants derived from Koshihikari × Koshihikari NIL-GPS and 143 from Takanari × Takanari NIL-GPS. GPS was coarsely mapped near the simple sequence repeat (SSR) molecular marker RM3534 in both populations (Fig. 4a, b). For further high-resolution mapping, another 8308 and 2784 F2 plants were used to select plants with recombination near RM3534 through marker-assisted selection in the Koshihikari and Takanari genetic backgrounds, respectively. From this selection, we obtained 26 and 24 plants, respectively. Analysis of recombinant homozygous F3 lines narrowed the candidate region down to the 23.5-kb region between the markers InDel_4_135 and InDel_4_105 in both genetic backgrounds (Fig. 4a, b). The photosynthesis rates in lines carrying the Takanari allele in this region were higher than in those carrying the Koshihikari allele (Fig. 4a, b). The Rice Annotation Project Database (RAP-DB)30 predicts three genes in this region. A bacterial artificial chromosome (BAC) clone containing the candidate genomic region (Taka03G22; Fig. 4b) was obtained by screening a genomic library of Takanari. We determined the sequence of the candidate region in Takanari by using this BAC clone and compared it with the corresponding sequence in Koshihikari, for which the whole genome sequence is now available31. Among the three genes annotated in RAP-DB, we found polymorphisms in the coding region for only one gene, at Os04g0615000. This gene was previously reported as NARROW LEAF1 (NAL1), encoding a plant-specific protein which may be involved in polar auxin transport32. There were ten single-nucleotide polymorphisms (SNPs), three of which encoded amino acid substitutions, in NAL1 between Koshihikari and Takanari (Fig. 4c). A 5895-bp retrotransposon insertion was also found in the second exon of NAL1 in the Koshihikari genome (Fig. 4c).
Figure 4. Molecular cloning of GPS.
(a, b) High-resolution linkage map of GPS region produced with 8308 F2 plants in the Koshihikari background (a) and 2784 F2 plants in the Takanari background (b). Populations were produced by crossing each cultivar with the corresponding NIL-GPS. The number of recombinants between molecular markers is indicated below the second line in each figure part. Yellow-green shading, regions homozygous for alleles from Koshihikari; dark green, Takanari. Each purple bar in the photosynthesis rate graphs represents the mean ± s.d. (n = 6) of the adjacent genotype. ***P < 0.001; **P < 0.01; n.s., not significant within pairs of recombinant lines (Student's t-test). (c) Gene structure and mutation sites of NAL1 in Koshihikari, Takanari, and nal1 mutant line in Taichung 65 genetic background (T65-nal1). Light blue bars represent exons; white bars represent 5′ and 3′ untranslated regions.
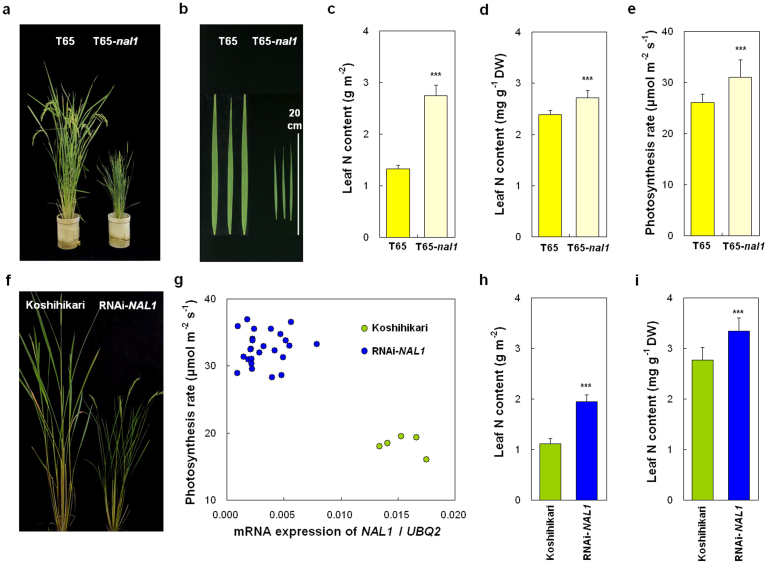
The previously identified nal1 mutation affected plant height as well as lateral leaf growth32. However, the relationship between NAL1 and leaf photosynthesis was not examined in that study. To clarify the effect of NAL1 on leaf photosynthesis, we analysed a nal1 mutant in the Taichung 65 genetic background (T65-nal1), which has the same deletion of 30-bp in the fourth exon as previously reported32 (Fig. 4c). T65-nal1 showed severe dwarf plant stature and remarkably smaller and narrower flag leaves than T65 (Fig. 5a, b, Supplementary Fig. S5a–c). However, T65-nal1 had higher flag leaf N content both per unit leaf area and per unit dry weight at full heading than T65 (Fig. 5c, d) and a higher photosynthesis rate than T65 (Fig. 5e). These results suggest that reduction or loss-of-function of NAL1 increased photosynthesis rate.
Figure 5. Plant morphology and photosynthesis rate in mutant and transgenic plants with differing expression of NAL1.
(a, b) Plant morphology (a) and flag leaves (b) of Taichung 65 (T65) and nal1 mutant line in T65 genetic background (T65-nal1). (c–e) Comparison of N content per unit leaf area (c), N content per unit dry mass (d), and photosynthesis rate (e) of flag leaves at full heading stage between T65 and T65-nal1. Each column represents mean ± s.d. (n = 10); ***P < 0.001 versus T65 (Student's t-test). (f) Plant morphology of Koshihikari and transgenic plants with RNAi-induced suppression of NAL1 in the Koshihikari genetic background (RNAi-NAL1). (g) Relationship between expression level of NAL1 and photosynthesis rate of flag leaves at full heading stage in Koshihikari and RNAi-NAL1 T0 plants. Each circle represents an individual Koshihikari or T0 plant. (h, i) Comparison of N content per unit leaf area (h) and per unit dry mass (i) between Koshihikari and RNAi-NAL1. Each column represents mean ± s.d. (n = 25 for RNAi-NAL1, n = 5 for Koshihikari); ***P < 0.001 versus Koshihikari (Student's t-test).
To confirm that NAL1 controls photosynthesis rate, we generated RNA interference (RNAi) transgenic plants to decrease the expression level of NAL1 in the Koshihikari genetic background (RNAi-NAL1). The RNAi-NAL1 plants (T0) showed dwarf plant stature and smaller and narrower flag leaves than Koshihikari (Fig. 5f, Supplementary Fig. S5e–g), and the reduced levels of NAL1 expression induced a higher photosynthesis rate in the flag leaves of the RNAi-NAL1 plants at full heading (Fig. 5g). The enhanced photosynthesis rate was associated with higher flag leaf N content (Fig. 5h, i). Thus, we conclude that NAL1 pleiotropically controls both plant stature and photosynthesis rate and corresponds to GPS.
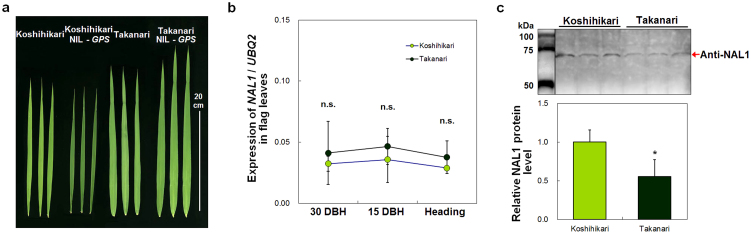
When we compared the flag leaf shapes and culm lengths of Koshihikari, Takanari, the reciprocal NILs-GPS, T65-nal1, and RNAi-NAL1, flag leaf width and length in Koshihikari NIL-GPS were decreased compared with Koshihikari (Fig. 6a, Supplementary Fig. S5i, j), but the reduction was less severe than in T65-nal1 and RNAi-NAL1 (Supplementary Fig. S5a, b, e, f, i, j). No difference was observed in culm length between each parental cultivar and the corresponding NIL-GPS (Supplementary Fig. S5k). On the basis of these results, we considered that the Takanari GPS allele might be functional but weaker than the Koshihikari allele. On the other hand, the presence of a retrotransposon insertion in the coding region of Koshihikari (Fig. 4c) might reduce the function relative to the Takanari allele. Therefore, we compared the expression level of NAL1 in the flag leaves of Koshihikari and Takanari at several developmental stages. Quantitative real-time PCR (qRT-PCR) detected the expression of NAL1 in both Koshihikari and Takanari, with no significant difference at any flag leaf developmental stage (Fig. 6b). Further investigation of NAL1 transcripts found that about 20% of the Koshihikari transcripts sequenced contained no retrotransposon insertion, whereas many other transcripts sequenced contained insertion of the full, or partial retrotransposon (Supplementary Fig. S6). Takanari also had many splice variants in NAL1 transcripts (Supplementary Fig. S7). Although a recent study reported several kinds of splice variants for NAL1 transcripts only in an indica cultivar33, our results revealed many splice variants for NAL1 transcripts in both a japonica cultivar, Koshihikari, and an indica cultivar, Takanari. These results indicate that NAL1 expression mechanism is too complicated to explain the cause of the two alleles of GPS by the expression or transcription level of NAL1.
Figure 6. Expression and protein differences between GPS alleles in Koshihikari and Takanari.
(a) Flag leaves in Koshihikari, Koshihikari NIL-GPS, Takanari, and Takanari NIL-GPS. (b) Expression analysis by quantitative real-time PCR of NAL1 in flag leaves at three developmental stages. DBH, days before heading. Each symbol represents mean ± s.d. (n = 4); n.s., not significant between Koshihikari and Takanari (Student's t-test). (c) Western blot analysis of NAL1 protein extracted from flag leaves. Molecular weight marker and samples were run on the same gel and were electro-transferred onto the same membranes. Each column in the graph represents mean ± s.d. (n = 3). *P < 0.05 versus Koshihikari (Student's t-test).
Then, we next examined the level of NAL1 protein in the flag leaves of Koshihikari and Takanari. Western blot analysis detected a 64-kDa protein band corresponding to NAL1 in both Koshihikari and Takanari, but the signal was less abundant in Takanari (Fig. 6c). These results suggest that the three amino acid substitutions or the different levels of NAL1 protein might have been the cause of the difference in NAL1 function in the two cultivars, detected in this study as the two alleles of GPS, although the detailed mechanisms underlying the differences between these alleles are still unknown.
Mechanisms underlying control of photosynthesis rate by NAL1
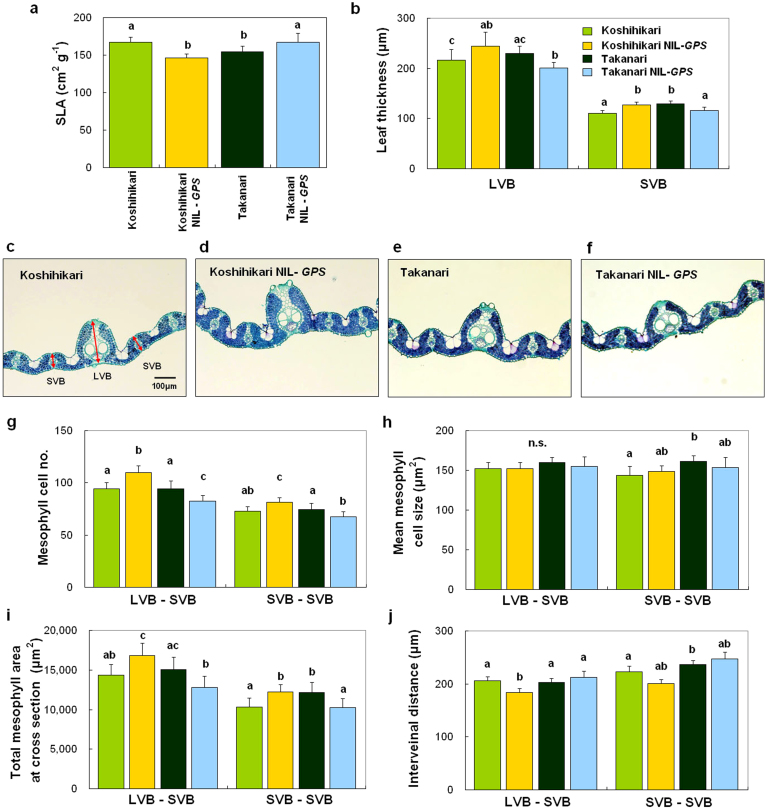
The finding that GPS and NAL1 are the same gene suggests that leaf morphological changes affect the photosynthesis rate. Previous studies have shown that leaf anatomical changes can affect leaf photosynthesis34,35. We examined specific leaf area (SLA), which is inversely related to leaf thickness36, of flag leaves in Koshihikari, Takanari, and the reciprocal NILs-GPS, and found that the Takanari allele of GPS decreased SLA (Fig. 7a). Both T65-nal1 mutant plants and RNAi-NAL1 transgenic plants also showed lower SLA than either wild-type cultivar (Supplementary Fig. S5d, h). We next observed cross-sections of the central parts of flag leaves in Koshihikari, Takanari, and the reciprocal NILs-GPS, and confirmed that the Takanari allele increased leaf thickness at both large and small vascular bundles (LVB and SVB, respectively; Fig. 7b–f). Because the majority of rice leaf tissue consists of mesophyll cells, we examined mesophyll cell number and size in cross-sections of flag leaves. Although mean mesophyll cell size did not differ between each parental cultivar and the corresponding NIL-GPS, the Takanari allele increased mesophyll cell number between the vascular bundles (Fig. 7g, h), and consequently enlarged the total mesophyll area between the vascular bundles in the cross-sections (Fig. 7i). The Takanari allele did not increase the distance between vascular bundles and in some cases decreased it slightly (Fig. 7j). Taken together, these results suggest that increased mesophyll cell number produced thicker flag leaves and ultimately led to higher photosynthesis rate per unit leaf area in plants with the Takanari allele.
Figure 7. Leaf anatomical characteristics controlled by GPS.
(a, b) Comparison of specific leaf area (SLA) (a) and thickness (b) of flag leaves in Koshihikari, Koshihikari NIL-GPS, Takanari, and Takanari NIL-GPS. LVB, large vascular bundle, SVB, small vascular bundle. (c–f) Cross-sections of flag leaves stained with toluidine blue in Koshihikari (c), Koshihikari NIL-GPS (d), Takanari (e), and Takanari NIL-GPS (f). (g–j) Comparisons of mesophyll cell number (g), mean mesophyll cell size (h), total mesophyll area of cross-section (i), and interveinal distance (j) between LVB and SVB and between SVBs in the four genotypes. Each column represents mean ± s.d. (n = 9). Different letters indicate significant difference at the 5% level (Tukey-Kramer HSD test).
Selection of GPS in rice breeding history
Takanari is descended from high-yielding rice cultivars, including IR8, IR24, and the Korean high-yielding cultivar Tongil37 (Fig. 1a). To examine whether the Takanari allele of GPS was selected in high-yield breeding programs, we analysed the nucleotide sequences of NAL1 from 14 cultivars in the pedigree of Takanari for which seeds were available. The sequences could be classified into five haplotypes, and high-yielding cultivars such as IR8, IR24, and Tongil were categorized into the same haplotype as Takanari (Supplementary Table S2). Although there is no direct evidence that the Takanari allele originated from IR8, we discovered that IR8 inherited the allele from Peta and that Tongil inherited the allele from IR8 (Fig. 1a). These results suggest that rice breeders have repeatedly selected this favourable allele for photosynthesis in breeding programs for high yield.
Effects of GPS on productivity
To examine whether GPS affects rice productivity, we conducted yield trials with Koshihikari, Takanari, and the reciprocal NILs-GPS. In this experiment, Takanari and Takanari NIL-GPS were compared under high-N conditions, because higher yield can be achieved in Takanari and related materials under high N inputs. On the other hand, Koshihikari and Koshihikari NIL-GPS were compared under standard-N conditions because of their tendency to lodge. Takanari had higher grain yield than Takanari NIL-GPS owing to higher ripening percentage and 1000-grain weight (Table 1). These results indicate the importance of the Takanari allele of GPS for the performance of Takanari. However, grain yield was similar between Koshihikari and Koshihikari NIL-GPS (Table 1). The result suggests that other factors in addition to the Takanari allele of GPS are necessary to enhance grain yield in the Koshihikari genetic background.
Table 1. Yield and yield components in Koshihikari, Koshihikari NIL-GPS, Takanari, and Takanari NIL-GPS.
| Cultivar or NIL | Yield (g m−2) | Spikelets (no. m−2) | Ripening (%) | 1000-grain wt. (g) |
|---|---|---|---|---|
| Koshihikari | 560 | 29,968 | 84.2 | 21.7 |
| Koshihikari NIL-GPS | 559 | 28,029 | 86.1 | 22.8 |
| (n = 3) | n.s. | n.s. | n.s. | ** |
| Takanari | 1,051 | 59,217 | 83.1 | 21.0 |
| Takanari NIL-GPS | 1,002 | 60,797 | 78.4 | 20.5 |
| (n = 5) | * | n.s. | * | ** |
N fertilization was 6 g N m−2 for Koshihikari and its NIL-GPS and 18 g N m−2 for Takanari and its NIL-GPS.
**P < 0.01;
*P < 0.05; n.s., not significant (Student's t-test).
Comparisons were made between each cultivar and the corresponding NIL-GPS.
Discussion
Until now, genetic factors underlying natural variation in photosynthesis rate have not been identified, because photosynthesis is rapidly affected by environmental changes and is hard to measure precisely for a large number of lines in a single experiment. We measured leaf photosynthesis only in the morning on clear days, because leaf photosynthesis did not reach its maximum levels on cloudy days or in the afternoon38,39. The reciprocal materials also facilitated precise mapping of QTLs for photosynthesis rate, because the use of two populations enabled us to compare the positions of QTLs and the effects of QTL alleles between the two populations. By these methods, we identified GPS, a QTL that regulates photosynthesis rate in rice.
We revealed that GPS did not affect stomatal conductance (Supplementary Fig. S4a) but did change carboxylation efficiency by regulating Rubisco content under our experimental conditions (Fig. 2f, g, Fig 3b). Because Rubisco is a key enzyme in CO2 fixation during photosynthesis, many research groups are attempting to improve Rubisco activity by using transgenic technology40,41,42. Although GPS might not be involved in the activity of Rubisco, it is involved in controlling the amount of Rubisco (Fig. 2f, g). Therefore, our results show that manipulation of Rubisco content by using GPS is an effective strategy to enhance carboxylation efficiency and consequently photosynthesis rate.
High-resolution mapping and subsequent mutant and transgenic experiments revealed that GPS was identical to NAL1, which encodes a plant-specific protein involved in controlling lateral leaf growth32. Although the relationship between NAL1 and photosynthesis was not tested at all in the previous study32, the discovery that GPS and NAL1 are the same locus led us to hypothesize that the leaf morphological modifications produced by NAL1 affect leaf photosynthesis properties. Our subsequent investigation of flag leaf anatomy revealed that NAL1 (GPS) controlled not only lateral leaf growth, but also vertical leaf growth, namely leaf thickness, by modulating mesophyll cell divisions between vascular bundles (Fig. 7). Because leaf mesophyll cells are the major site of photosynthesis18 and make up the majority of rice leaf tissue (Fig. 7c–f), an increase in the number of mesophyll cells without size reduction or lateral extension can expand the vertical mesophyll tissue volume. Our results demonstrate the importance of NAL1 (GPS) in the development of flag leaves, where it led to modulation of leaf N, Rubisco, and chlorophyll contents per unit leaf area and thereby contributed to carboxylation efficiency and leaf photosynthesis. Moreover, NAL1 (GPS) also regulated leaf N, Rubisco, and chlorophyll contents per unit fresh or dry mass (Fig. 2e, g, i, Fig. 5d, i), which was reflected in photosynthesis rate per unit dry mass (Fig. 2c). Because these trait values expressed per unit fresh or dry mass reflect concentration, NAL1 (GPS) may also regulate the amount of photosynthesis machinery per mesophyll cell, which may also contribute to carboxylation efficiency and leaf photosynthesis.
The importance of thicker leaves in relation to photosynthesis has been evident for a long time from observations that sun leaves are thicker, have more photosynthesis machinery per cell, and show higher photosynthesis rate than shade leaves34,43. However, little is known about genes controlling leaf thickness and photosynthesis rate, although these two traits show a close relationship in diverse plant species and show varietal differences35,44. In this study, we revealed that NAL1 had pleiotropic effects on leaf anatomy and photosynthesis. Interestingly, a similar function has been reported for the Arabidopsis ERECTA gene45, which encodes a putative leucine-rich-repeat receptor protein kinase46. The functional allele of ERECTA led to a greater number of mesophyll cells, tighter packing of mesophyll cells, and higher carboxylation efficiency than loss-of-function alleles45. However, unlike NAL1, ERECTA was also associated with epidermal morphology. The functional allele of ERECTA reduced stomatal density and thereby lowered stomatal conductance relative to loss-of-function alleles; for this reason, ERECTA is regarded as a transpiration efficiency gene. Recently, rice mutants generated by artificial mutagens were screened for mesophyll cell architecture with the goal of improving photosynthetic capacity47. Several mutant lines showed favourable alterations such as increased mesophyll cell size. However, none of them had an enhanced photosynthesis rate relative to wild-type plants48. Because artificial mutations often cause detrimental side effects, these effects may have cancelled out the effects of the favourable mesophyll alterations necessary for improved photosynthesis rate. In this context, natural variations of NAL1 (GPS) and ERECTA are valuable for obtaining enhanced photosynthesis capacity through leaf anatomical modifications.
With regard to detrimental side effects, T65-nal1 and RNAi-NAL1 plants showed severe dwarf plant stature not seen in Koshihikari NIL-GPS (Fig 2a, Fig. 5a, f), although all three of these materials displayed an increase in photosynthesis rate. The differences in phenotype might be attributable to the level of NAL1 function. The 30-bp deletion in the fourth exon in T65-nal1 and the reduction of NAL1 expression induced by RNAi seem to have caused severe loss-of-function of NAL1. On the other hand, the reduced NAL1 protein level in Takanari compared with Koshihikari and the presence of three amino acid substitutions between the two cultivars suggest partial loss-of-function of the NAL1 encoded by the Takanari allele. Other cases have been reported in which a moderate level of gene function produces a preferable plant phenotype49. For example, APO1, which controls panicle structure, increased spikelet number per panicle with no decline in panicle number when expressed to a level twice that of the wild type, but caused a remarkable decline in panicle number when overexpressed to levels much higher than that of the wild type50,51. The partial loss-of-function allele of GN1a also increased spikelet number per panicle, with only a slight decrease in panicle number, compared with the gain-of-function allele5. On the basis of these results, we suggest that a weak NAL1 allele such as the Takanari allele of GPS increases the photosynthesis rate without detrimental side effects and is suitable for breeding. This is supported by the fact that the Takanari allele appears to have been repeatedly selected in high-yield breeding programs (Fig. 1a). While it is well known that the semi-dwarf gene sd1 was selected from Dee-geo-woo-gen when IR8 was developed52, IR8 also contains the Takanari allele of GPS that was inherited from Peta (Fig. 1a). Likewise, the GPS allele from IR8 is present in Tongil, the first high-yielding cultivar in Korea (Fig. 1a). After several more generations of breeding and selection, Takanari inherited this allele, although we could not determine the original cultivar that contributed the allele to Takanari because it is found in several ancestors of Takanari. These facts provide strong evidence that breeders regarded the phenotype conferred by the Takanari allele of GPS as an important factor when developing high-yielding cultivars. The contribution of Dee-geo-woo-gen as the source of sd1 has frequently been highlighted in discussions of the “Green Revolution” in rice. Our results reveal the importance of another gene that the traditional cultivar Peta provided to IR8 and subsequent high-yielding cultivars. But how did breeders select an allele for higher photosynthesis without directly observing photosynthesis? One possibility is that breeders might have used leaf colour as a selection criterion, because the Takanari allele of GPS produces dark green leaves (Fig. 2a, Fig. 6a). Thick, dark green leaves were also targeted in new-plant-type (NPT) breeding in the 1990s53. Because high photosynthesis rates and dark green leaves are common to Takanari, IR8, and Tongil26, the Takanari allele of GPS is likely to have been selected through leaf colour.
The ultimate goal of our studies of GPS is to increase rice yield potential. Thus, we examined grain yield of Koshihikari, Takanari, and the reciprocal NILs-GPS, and discovered that the Takanari allele of GPS was indispensable for Takanari to achieve its normal yield level (Table 1). On the other hand, the Takanari allele did not increase grain yield in Koshihikari (Table 1). When yield components were compared between Koshihikari and Koshihikari NIL-GPS, we found no significant difference in spikelet number per m2, but found a slight increase in ripening percentage and a significant increase in 1000-grain weight for Koshihikari NIL-GPS (Table 1). The comparison indicates that combination of GPS with sink size QTLs such as GN1a and APO1 may be necessary to increase grain yield in the Koshihikari background. An alternative is that GPS may require high N inputs to have maximum effect on yield.
We found that GPS, a newly identified QTL regulating photosynthesis rate, is identical to NAL1, a gene previously identified for its effects on plant morphology. To our knowledge, NAL1 is the first gene shown to account for natural variation in rice photosynthesis rate, although its molecular function is still unknown. Nevertheless, this finding opens up possibilities for further identification of other photosynthesis-related QTLs and further understanding of the genetic mechanisms underlying natural variation in photosynthesis rate. Combining photosynthesis QTLs with sink size QTLs will enhance future high-yield breeding and provide a new and efficient strategy for increased rice productivity.
Methods
Plant growth conditions
Field experiments were conducted in a paddy field at the NARO Institute of Crop Science, Ibaraki, Japan, and at the University Farm, Tokyo University of Agriculture and Technology, Tokyo, Japan. Seeds were sown in nursery boxes, and 21-day-old seedlings were transplanted at a density of one seedling per hill, at a spacing of 15 cm between hills and 30 cm between rows. For the QTL mapping experiments and for yield trials in standard-N plots, basal fertilizer was applied at 60 kg N, 52 kg P, and 75 kg K ha−1. For yield trials in high-N plots, N was applied at 180 kg ha−1 (120 kg as basal fertilizer and 60 kg as topdressing at the panicle initiation stage), with 70 kg P and 100 kg K ha−1 as basal fertilizer. Experimental plots for yield trials were arranged in a randomized complete block design with three or five replications.
Plant materials and QTL mapping
For QTL analysis of leaf photosynthesis, reciprocal BILs (BC1F6) were developed between a high-yielding indica rice cultivar, Takanari, and a leading japonica cultivar, Koshihikari, both grown in Japan. The reciprocal BILs consisted of 87 lines each in the Koshihikari and Takanari genetic backgrounds, but some lines were excluded because of hybrid weakness or extremely late heading; thus, 82 and 86 lines, respectively, were used for QTL analysis. The genome-wide genotype of each line was determined by using 140 PCR-based DNA markers, and linkage maps were constructed with MAPMAKER/EXP 3.0 software54. The chromosomal positions and effects of putative QTLs were determined by composite interval mapping with QTL Cartographer 2.0 software55. The threshold of QTL detection was based on 1000 permutation tests at the 5% level of significance. The reciprocal NILs-GPS were developed through four generations of backcrossing and marker-assisted selection. For map-based cloning, we used F2 progeny derived from crosses of each NIL-GPS to the corresponding recurrent parent cultivar. Consequently, 26 and 24 plants with recombination near the QTL were identified from 8308 and 2784 F2 plants in the Koshihikari and Takanari genetic backgrounds, respectively. The primer sets for the InDel markers used in map-based cloning are listed in Supplementary Table S3.
Leaf photosynthesis measurement
Leaf photosynthesis was measured in flag leaves at the full heading stage with a portable photosynthesis system (LI-6400; Li-Cor). Measurement was conducted on clear days between 0900 and 1300 h under a constant saturated light level of 2000 μmol m−2 s−1 provided by red/blue light-emitting diodes. The leaf chamber temperature was maintained at 30°C, the reference CO2 concentration was 390 μmol mol−1, and the mean leaf-to-air vapour pressure difference was 1.1 kPa. Gas-exchange parameters were recorded once the topmost expanded leaf was enclosed in the chamber and the photosynthesis rate reached steady state. Leaf photosynthesis was also measured at different light levels between 0 and 2000 μmol m−2 s−1 and different reference CO2 concentrations between 0 and 1500 μmol mol−1. To fit the biochemical Farquhar model of C3 photosynthesis to CO2 response data29, we used the Rubisco kinetic parameters determined by the temperature response functions56. Leaf N, Rubisco, and chlorophyll contents were investigated using the same flag leaves for which leaf photosynthesis was measured in the field. Leaf N was quantified with an NC analyser (Sumigraph NCH-22F; Sumik Chemical Analysis Service). Rubisco was quantified by the single radial immunodiffusion method with rabbit polyclonal antibodies raised against purified Rubisco from rice28. Chlorophyll content was determined by extraction with N,N′-dimethylformamide (DMF)57.
Observation of the cross-section of flag leaves
After leaf photosynthesis was measured, central sections of flag leaf blades were sampled, fixed in 4% paraformaldehyde/0.25% glutaraldehyde fixative solution, and embedded in paraffin wax. Transverse sections of leaves 7 μm thick were cut on a rotary microtome (HM335E; Microm) and stained with 0.05% toluidine blue. Leaf mesophyll anatomy at the second large vascular bundles from the midrib was observed under a fluorescence microscope (BX51N-34-FL; Olympus) at ×200 magnification and analysed with digital image processing software (Win-ROOF v. 6.1; Mitani). To measure the distance between vascular bundles, stomatal density, and stomatal length, we observed the abaxial epidermis of fixed flag leaves under a scanning electron microscope (SEM) (VE-9800; Keyence) and analysed the measurements with Win-ROOF.
Transformation of rice with RNAi construct
To produce a construct for knockdown of NAL1, we amplified a gene-specific DNA fragment of 308 bp including the fifth exon and 3′ UTR of NAL1 by PCR using specific primers (Supplementary Table S3). The fragments were cloned in head-to-head orientation into the RNA interference vector pZH2Bik (a generous gift from M. Kuroda)58. This plasmid was introduced into callus derived from Koshihikari by a high-efficiency Agrobacterium-mediated transformation system for rice callus59.
RNA isolation and expression and transcript analysis
Total RNA was extracted from flag leaf tissues at each developmental stage by using an RNeasy Mini Kit (Qiagen). First-strand cDNA was synthesized by using SuperScript III reverse transcriptase (Invitrogen). Quantitative real-time PCR (qRT-PCR) was performed by using SYBR Green Realtime PCR Master Mix (Toyobo) and specific primers (Supplementary Table S3) on an ABI7300 system (Applied Biosystems). The results were confirmed by analysing four biological replicates, with three technical repeats for each biological replicate. The UBQ2 gene from rice was used as an internal standard (Supplementary Table S3). For transcript analysis, cDNA was amplified by specific primers of 5′-UTR and 3′-UTR (Supplementary Table S3). The PCR products were cloned by using Zero Blunt TOPO PCR Cloning Kit (Invitrogen) and sequenced with a BigDye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems) on an automated fluorescent laser sequencer (3130xl; Applied Biosystems).
Western blot analysis
Antiserum to NAL1 was generated by injection of rabbits with the synthetic oligopeptide SPVRDDQDAPRSITN, corresponding to the C-terminal region of NAL1. Total protein was extracted from flag leaf tissues at the full heading stage and quantified by the Bradford method60. The protein (14 μg) was subjected to SDS-PAGE in 10% polyacrylamide gel for separation and electro-transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories), which were then incubated with rabbit antiserum to NAL1. The NAL1 oligopeptide antibodies were used at a 1:1000 dilution. Immunodetection was performed with an ECL Plus western blotting kit (GE Healthcare) on a luminescent image analyser (LAS-3000; Fujifilm).
Sequence analysis
For comparison of NAL1 sequences among cultivars in the pedigree of Takanari, the NAL1 genomic regions were amplified by PCR with seven primer sets (Supplementary Table S3). The PCR products were sequenced with a BigDye Terminator v. 3.1 Cycle Sequencing Kit (Applied Biosystems) on an automated fluorescent laser sequencer (3130xl; Applied Biosystems).
Statistical analysis
All statistical analyses were done with JMP v. 8.0 software (SAS Institute).
Author Contributions
T.T. and T.Y. designed the experiments and wrote the manuscript. T.T., T.I., and T.Y. developed the reciprocal BILs. T.T., S.A., T.H., and T.Y. performed phenotyping for cloning GPS. T.T., T.A., I.K., S.I., A.S., and T.Y. performed genotyping for cloning GPS. J.W. and T.M. performed BAC clone analysis. T.T. and F.T.-S. performed sequencing analysis. T.T., U.Y., S.H., Y.T., and T.Y. performed molecular analysis. T.T., Y.S.-A., N.I., S.Y., and M.K. performed yield trials. K.S., K.K., T.O., T.H., M.Y., and M.K. provided advice on the experiments.
Supplementary Material
Supplementary Information
Acknowledgments
The nal1 mutant line was kindly provided by the National Institute of Genetics (Oryzabase, available at http://www.shigen.nig.ac.jp/rice/oryzabaseV4/). Seeds of the reciprocal BILs between Koshihikari and Takanari can be obtained from the Rice Genome Resource Center (http://www.rgrc.dna.affrc.go.jp/index.html). This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, QTL1002 and NVR-0001).
References
- Hargrove T. R. & Cabanilla V. L. The impact of semidwarf varieties on Asian rice-breeding programs. Bioscience 29, 731–735 (1979). [Google Scholar]
- Khush G. S. Green revolution: the way forward. Nat. Rev. Genet. 2, 815–822 (2001). [DOI] [PubMed] [Google Scholar]
- Tester M. & Langridge P. Breeding technologies to increase crop production in a changing world. Science 327, 818–822 (2010). [DOI] [PubMed] [Google Scholar]
- Foley J. A. et al. Solutions for a cultivated planet. Nature 478, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- Ashikari M. et al. Cytokinin oxidase regulates rice grain production. Science 309, 741–745 (2005). [DOI] [PubMed] [Google Scholar]
- Terao T., Nagata K., Morino K. & Hirose T. A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theor. Appl. Genet. 120, 875–893 (2010). [DOI] [PubMed] [Google Scholar]
- Miura K. et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549 (2010). [DOI] [PubMed] [Google Scholar]
- Song X. J., Huang W., Shi M., Zhu M. Z. & Lin H. X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630 (2007). [DOI] [PubMed] [Google Scholar]
- Fan C. et al. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171 (2006). [DOI] [PubMed] [Google Scholar]
- Shomura A. et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028 (2008). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269 (2011). [DOI] [PubMed] [Google Scholar]
- Ohsumi A. et al. Evaluation of yield performance in rice near-isogenic lines with increased spikelet number. Field Crops Res. 120, 68–75 (2011). [Google Scholar]
- Yoshida H., Horie T. & Shiraiwa T. A model for explaining genotypic and environmental variation in vegetative biomass growth in rice based on observed LAI and leaf nitrogen content. Field Crops Res. 108, 222–230 (2008). [Google Scholar]
- Yoshida H. & Horie T. A process model for explaining genotypic and environmental variation in growth and yield of rice based on measured plant N accumulation. Field Crops Res. 113, 227–237 (2009). [Google Scholar]
- Sasaki H. & Ishii R. Cultivar differences in leaf photosynthesis of rice bred in Japan. Photosynthesis Res. 32, 139–146 (1992). [DOI] [PubMed] [Google Scholar]
- Hubbart S., Peng S., Horton P., Chen Y. & Murchie E. H. Trends in leaf photosynthesis in historical rice varieties developed in the Philippines since 1966. J. Exp. Bot. 58, 3429–3438 (2007). [DOI] [PubMed] [Google Scholar]
- Farquhar G. D. & Sharkey T. D. Stomatal conductance and photosynthesis. Annu. Rev. Plant Physiol. 33, 317–345 (1982). [Google Scholar]
- Lambers H., Chapin III F. S. & Pons T. L. Plant Physiological Ecology. (Springer Verlag, 1998). [Google Scholar]
- Evans J. R. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 78, 9–19 (1989). [DOI] [PubMed] [Google Scholar]
- Makino A. Rubisco and nitrogen relationships in rice: Leaf photosynthesis and plant growth. Soil Sci. Plant Nutr. 49, 319–327 (2003). [Google Scholar]
- Kanemura T., Homma K., Ohsumi A., Shiraiwa T. & Horie T. Evaluation of genotypic variation in leaf photosynthetic rate and its associated factors by using rice diversity research set of germplasm. Photosynthesis Res. 94, 23–30 (2007). [DOI] [PubMed] [Google Scholar]
- Ohsumi A. et al. A model explaining genotypic and ontogenetic variation of leaf photosynthetic rate in rice (Oryza sativa) based on leaf nitrogen content and stomatal conductance. Ann. Bot. 99, 265–273 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y. Z. & Zhang Q. F. Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442 (2010). [DOI] [PubMed] [Google Scholar]
- Flood P. J., Harbinson J. & Aarts M. G. Natural genetic variation in plant photosynthesis. Trends Plant Sci. 16, 327–335 (2011). [DOI] [PubMed] [Google Scholar]
- Taylaran R. D., Adachi S., Ookawa T., Usuda H. & Hirasawa T. Hydraulic conductance as well as nitrogen accumulation plays a role in the higher rate of leaf photosynthesis of the most productive variety of rice in Japan. J. Exp. Bot. 62, 4067–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai T. et al. Comparative mapping suggests repeated selection of the same quantitative trait locus for high leaf photosynthesis rate in rice high-yield breeding programs. Crop Sci. 52, 2649–2658 (2012). [Google Scholar]
- Takai T., Kondo M., Yano M. & Yamamoto T. A quantitative trait locus for chlorophyll content and its association with leaf photosynthesis in rice. Rice 3, 172–180 (2010). [Google Scholar]
- Adachi S. et al. Identification and characterization of genomic regions on chromosomes 4 and 8 that control the rate of photosynthesis in rice leaves. J. Exp. Bot. 62, 1927–1938 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar G. D., von Caemmerer S. & Berry J. A. A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149, 78–90 (1980). [DOI] [PubMed] [Google Scholar]
- Rice Annotation Project. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 36, D1028–1033 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T. et al. Fine definition of the pedigree haplotypes of closely related rice cultivars by means of genome-wide discovery of single-nucleotide polymorphisms. BMC Genomics 11, 267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J. et al. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 147, 1947–1959 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. et al. Fine mapping of a major QTL for flag leaf width in rice, qFLW4, which might be caused by alternative splicing of NAL1. Plant Cell Rep. 31, 863–872 (2011). [DOI] [PubMed] [Google Scholar]
- Boardman N. K. Comparative photosynthesis of sun and shade plants. Annu. Rev. Plant Physiol. 28, 355–377 (1977). [Google Scholar]
- Niinemets U. Components of leaf dry mass per area—thickness and density—alter leaf photosynthetic capacity in reverse directions in woody plants. New Phytol. 144, 35–47 (1999). [Google Scholar]
- Vile D. et al. Specific leaf area and dry matter content estimate thickness in laminar leaves. Ann. Bot. 96, 1129–1136 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung G. S. & Heu M. H. Innovative Approaches to Rice Breeding, 135–152 (IRRI, 1980). [Google Scholar]
- Ishihara K. & Saito K. Diurnal courses of photosynthesis, transpiration, and diffusive conductance in the single-leaf of the rice plants grown in the paddy field under submerged condition. Japan. J. Crop Sci. 56, 8–17 (1987). [Google Scholar]
- Takai T., Yano M. & Yamamoto T. Canopy temperature on clear and cloudy days can be used to estimate varietal differences in stomatal conductance in rice. Field Crops Res. 115, 165–170 (2010). [Google Scholar]
- Ishikawa C., Hatanaka T., Misoo S., Miyake C. & Fukayama H. Functional incorporation of sorghum small subunit increases the catalytic turnover rate of Rubisco in transgenic rice. Plant Physiol. 156, 1603–1611 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney S. M. et al. Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) carboxylation rate in Flaveria. Proc. Natl. Acad. Sci. USA 108, 14688–14693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W., Masumoto C., Fukayama H. & Makino A. Rubisco activase is a key regulator of non-steady-state photosynthesis at any leaf temperature and, to a lesser extent, of steady-state photosynthesis at high temperature. Plant J. 71, 871–880 (2012). [DOI] [PubMed] [Google Scholar]
- Terashima I., Hanba Y., Tazoe T., Vyas P. & Yano S. Irradiance and phenotype: comparative eco-development of sun and shade leaves in relation to photosynthetic CO2 diffusion. J. Exp. Bot. 57, 343–354 (2006). [DOI] [PubMed] [Google Scholar]
- Pearce R. B., Carlson G. E., Barnes D. K., Hart R. H. & Hanson C. H. Specific leaf weight and photosynthesis in Alfalfa. Crop Sci. 9, 423–426 (1969). [Google Scholar]
- Masle J., Gilmore S. R. & Farquhar G. D. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436, 866–870 (2005). [DOI] [PubMed] [Google Scholar]
- Torii K. U. et al. The arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie I. R., Pyke K. A. & Murchie E. H. Variation in vein density and mesophyll cell architecture in a rice deletion mutant population. J. Exp. Bot. 63, 4563–4570 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie I. R. A. Analysis of leaf morphology and photosynthesis in deletion mutants of rice (Oryza sativa L.) Ph.D. thesis available at http://etheses.nottingham.ac.uk/2569/ (University of Nottingham, 2012). [Google Scholar]
- Takeda S. & Matsuoka M. Genetic approaches to crop improvement: responding to environmental and population changes. Nat. Rev. Genet. 9, 444–457 (2008). [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K. et al. Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiol. 150, 736–747 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookawa T. et al. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 1, 132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A. et al. A mutant gibberellin-synthesis gene in rice. Nature 416, 701–702 (2002). [DOI] [PubMed] [Google Scholar]
- Peng S., Cassman K. G., Virmani S. S., Sheehy J. & Khush G. S. Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential. Crop Sci. 39, 1552–1559 (1999). [Google Scholar]
- Lander E. S. et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181 (1987). [DOI] [PubMed] [Google Scholar]
- QTL Cartographer. Version 1.16. A Reference Manual and Tutorial for QTL Mapping (North Carolina State University, 2002). [Google Scholar]
- Bernacchi C. J., Singsaas E. L., Pimentel C., Portis J. R. & Long S. P. Improved temperature response functions for models of Rubisco-limited photosynthesis. Plant Cell Environ. 24, 253–259 (2001). [Google Scholar]
- Porra R. J., Thompson W. A. & Kriedemann P. E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 (1989). [Google Scholar]
- Kuroda M., Kimizu M. & Mikami C. A simple set of plasmids for the production of transgenic plants. Biosci. Biotechnol. Biochem. 74, 2348–2351 (2010). [DOI] [PubMed] [Google Scholar]
- Ozawa K. Establishment of a high efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.). Plant Sci. 176, 522–527 (2009). [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information