Abstract
ABCB5 is a multidrug resistance (MDR) member of the ATP-binding cassette (ABC) superfamily of active transporters and represents a marker for chemoresistant malignant melanoma-initiating cells. ABCB5 expression is closely linked to tumorigenicity and progression of diverse human malignancies, including melanoma, and is functionally required for tumor growth. Here, we genotyped 585 melanoma cases and 605 age-matched controls for 44 ABCB5 tagging single nucleotide polymorphisms (SNPs) to span a region covering 108.2kb of the gene on the 7p21.1 locus. We identified three SNPs that were associated with decreased melanoma risk in additive models: rs10231520 (OR: 0.83, 95% CI: 0.70–0.98), rs17817117 (OR: 0.82, 95% CI: 0.68–0.98), and rs2301641 (OR: 0.83, 95% CI: 0.69–0.98). Additionally, the rs2301641 SNP was associated with non-red compared to red hair color (OR: 0.38, 95% CI: 0.14–1.03) in controls. Twelve human melanoma cell lines were genotyped for the rs2301641 SNP, which encodes a non-synonymous ABCB5 amino acid change (K115E). Functional studies revealed that the E form associated with lower melanoma risk correlated significantly with decreased ABCB5 transport capacity (P<0.01) and increased melanin production (P<0.05). Our results identify novel associations of the ABCB5 K115E polymorphism with human pigmentation phenotype and melanoma risk and point to potential functional roles of ABCB5 in melanomagenesis. Moreover, they provide a first example that functional variation in a prospective cancer stem cell marker can be associated with disease risk for the corresponding malignancy.
Keywords: ABCB5, single nucleotide polymorphism, genotype, melanoma, cancer, cancer stem cells, pigmentation, case-controlled study, humans
Introduction
The lifetime risk of developing malignant melanoma, a highly aggressive and therapy-resistant skin cancer, escalates yearly and increases more rapidly than in most adult onset cancers [1]. The search for melanoma susceptibility genes originated with linkage studies in familial melanoma cases that revealed primarily high penetrance, low frequency genetic variants [2]. Genome-wide studies on large cohorts of melanoma patients have also identified several genes and polymorphisms of more common, low penetrance associated with an increased risk of developing melanoma. While the majority of these identified polymorphisms are related to pigmentation, reflecting the importance of gene/environment interactions such as that of ultraviolet radiation, functionally relevant proteins involved in melanoma progression have also served as candidate genes for genomic analyses [3].
ABCB5, a multidrug resistance (MDR) member of the ATP-binding cassette (ABC) superfamily of active transporters [4], is highly expressed in clinical human melanomas, with low expression in normal skin [5; 6]. In melanoma, ABCB5 marks malignant melanoma-initiating cells (MMIC), in which clinical virulence resides as a consequence of unlimited self-renewal capacity, resulting in inexorable tumor progression and metastasis [5; 7; 8; 9; 10; 11]. Furthermore, ABCB5 is associated with clinical drug resistance, tumor progression and disease recurrence in malignant melanoma [5; 6; 8; 10; 12] and serves as an independent biomarker of disease recurrence in this malignancy [13]. ABCB5(+) melanoma subpopulations trigger tumorigenesis and promote neoplastic progression through enhanced self-renewal and proliferative capacity [5]. Preferential evasion of host antitumor immunity and vasculogenic mimicry represent further mechanisms responsible for the enhanced tumorigenicity of ABCB5(+) melanoma subpopulations [7; 11]. Functionally, ABCB5 has been shown to serve as a drug efflux transporter and drug resistance mediator in melanoma for multiple chemotherapeutic compounds [14; 15; 16]. Additionally, ABCB5 regulates tumor growth in a drug-transport independent manner [16]. Consistent with the close association of ABCB5 with the melanoma-initiating cell phenotype and its functional role in regulating tumor growth, a germline MITF-activating mutation, which increases MITF binding to the ABCB5 promoter and increases ABCB5 expression, predisposes to melanoma, and is functionally associated with increased tumor clonogenicity, migration and invasiveness, as shown by Bertolotto and colleagues in 2011. Based on the close association of ABCB5 with melanoma initiation, progression and recurrence, and the functional role of ABCB5 in regulating tumor growth, we hypothesized that genetically determined ABCB5 functionality correlates with melanoma risk.
Among single nucleotide polymorphisms (SNPs) of the ABCB5 gene, several, including the rs2301641 SNP, which encodes a non-synonymous ABCB5 amino acid change (K115E), have been predicted to be relevant to ABCB5 function based on bioinformatic analyses [17]. However, experimental evidence for their role in ABCB5 functionality and their potential association with melanoma risk has not been presented to date. Here, we have examined in a systematic manner whether rs2301641 and/or additional SNPs of ABCB5 are associated with melanoma risk, by interrogating a large cohort study of melanoma patients with matched controls. Three SNPs were significantly associated with a decreased melanoma risk. We confirmed the direct association of one of these SNPs, rs2301641 (K115E), with ABCB5 function and, furthermore, uncovered a novel association of ABCB5 with pigmentation.
Material and Methods
Study population
The nested case-control study consisted of participants from the Nurses’ Health Study (NHS) and the Health Professional Follow-up Study (HPFS). The NHS was established in 1976, when 121,700 female registered nurses between the ages of 30 and 55, residing in 11 larger US states, completed and returned the initial self-administered questionnaire on their medical histories and baseline health related exposures. Updated information was obtained by questionnaires every two years, and blood samples were collected from 32,826 participants in the NHS cohort between May 1989 and September 1990. For the HPFS, in 1986, 51,529 men from all 50 US states in health professions (dentists, pharmacists, optometrists, osteopath physicians, podiatrists, and veterinarians) aged 40–75 years answered a detailed mailed questionnaire, forming the basis of the HPFS, and during 1993–1994, 18,159 study participants provided blood samples by overnight courier. All the cases and controls in our study were from the sub-cohorts of NHS and HPFS who had given a blood specimen. Eligible cases consisted of pathologically confirmed melanoma cases diagnosed after the baseline up to 2006 follow-up cycle for both cohorts, who had no previously diagnosed cancer. Controls were randomly selected from participants who were free of diagnosed melanoma up to and including the questionnaire cycle in which the case was diagnosed. One or two controls were matched to each case by age (± 1 year). Cases and their matched controls were selected in the same cohort. All subjects in our study were United States non-Hispanic Caucasians. Finally, we recruited 585 melanoma cases and 605 matched controls. The study protocol was approved by the Committee on Use of Human Subjects of the Brigham and Women’s Hospital, Boston, MA.
Risk factor data
We obtained information regarding melanoma risk factors from prospective biennial questionnaires. Information on natural hair color at age 20, mole count on the left arm, and childhood and adolescent tanning tendency were collected in both the NHS and HPFS prospective questionnaires.
SNP identification
Polymorphisms evaluated in this study were selected for genotyping by an approach of tagging SNPs screening for ABCB5. The ABCB5 gene locates on chromosome 7p21.1, spanning approximately 108.2kb, and all polymorphisms were looked for between nucleotide 35kb upstream from the start codon and 5kb downstream of the 3’-untranslated region. Based on the HapMap phase II SNP genotype data, we chose 44 tag-SNPs as surrogates for untyped polymorphisms in this gene region using the HapMap Project 90 (30 trios) Caucasian samples from a US Utah population with Northern and Western European ancestry collected in 1980 by the Centre d'Etude du Polymorphisme Humain (CEPH) [18]. Briefly, the tag-SNPs (minor allele frequency > 0.05) were selected using the Tagger program of (r2>0.8), which combines the simplicity of pair-wise r2 methods [19] with the potential efficiency of multimarker haplotype approaches [20].
Laboratory assays
We genotyped the 44 tagging SNPs in ABCB5 using the OpenArray™ SNP Genotyping System (BioTrove, Woburn, MA). Laboratory personnel were blinded to the case-control status, and blinded quality control samples were inserted to validate genotyping procedures; concordance for the blinded samples was 100%. Primers, probes and conditions for genotyping assays are available upon request.
Statistical methods
We used the Chi-square test to assess whether the genotypes for the 44 SNPs were in Hardy–Weinberg equilibrium among the controls. The association between each genotype and melanoma risk was evaluated by determining the P value in unconditional logistic regression in an additive model, which was according to an ordinal coding for genotype (0, 1 or 2 copies of SNP minor allele). For the SNPs with nominal significant association, odds ratios (ORs) and 95% confidence intervals (95% CIs) were calculated in both the additive model and the co-dominant model using unconditional logistic regression, to assess the risk of melanoma for these genotypes. We evaluated the associations between the variants and pigmentary phenotypes among controls. We regressed an ordinal coding for tanning ability (1 = practically none; 2 = light tan; 3 = average tan; 4 = deep tan) and mole count (1 = none; 2 = 1–2; 3 = 3–5; 4 = 6–9; 5 = 10–14; 6 = 15+) on an ordinal coding for genotype (0, 1 or 2 copies of the SNP minor allele). For hair color, we used two different statistical models: (i) We tested the association between the ordinal genotype coding and an ordinal coding of hair color excluding the women with red hair (1 = black; 2 = dark brown; 3 = light brown; 4 = blonde) using linear regression; and (ii) we used logistic regression to test the association between the ordinal genotype coding and a binary red hair phenotype (red hair vs. non-red hair color). In addition, associations between the SNPs and survival time in melanoma cases were estimated by using the Kaplan-Meier method and the log-rank test, and survival time was calculated from the date of melanoma diagnosis to the date of death or to last follow-up. All statistical analyses were two-sided and carried out using SAS V9.1 (SAS Institute, Cary, NC).
Cell Culture
The G3361 human malignant melanoma cell line was provided by Dr. Emil Frei III (Dana-Farber Cancer Institute, Boston, MA), C8161 melanoma cells by Dr. Mary Hendrix (Children's Memorial Research Center, Chicago, IL), LOX and FEMX1 melanoma cells by Dr. Udo Schumacher (University Hospital Hamburg- Eppendorf, Germany) and the MeWo, SKMel2, SKMel30, Malme3M, M14, UACC257 melanoma cells by Dr. Hans Widlund (Brigham and Women’s Hospital, Boston, MA). Clinical melanoma cells were derived from a surgical patient specimen provided by Dr. Martin Gasser (University of Würzburg, Germany) according to an Institutional Review Board–approved research protocol. Melanoma cells were cultured as described previously [5].
Sequencing and primers
The ABCB5 gene-specific oligonucleotide primer pair 5’- TGGGATTGTCATTTCCTGTTCTAACC-3’ (forward primer) and 5’- ACAGCATCTCCTTCTGTCCTCTAAACC-3’ (reverse primer) was used for PCR sequencing analyses performed with 100 ng of genomic DNA (QIAGEN DNAeasy kit).
Rhodamine-123 efflux transport assay
Efflux transport capacity for the dye rhodamine-123 (Rh-123), a hallmark function of ABCB5 [4], was assessed for all of 12 ABCB5-genotyped melanoma cell lines by flow cytometry as described previously [4]. ABCB5 positivity among melanoma cells was determined after Rh-123 incubation by counterstaining with an APC conjugated-ABCB5 monoclonal antibody (mAb) (clone 3C2-1D12) [4; 5]. Rh-123 efflux capacity by ABCB5 in each melanoma cell line was established by calculating the quotient of [(mean Rh-123 fluorescence in ABCB5(+) cells at T120)/(mean Rh-123 fluorescence in all cells at T120)] and [(mean ABCB5 fluorescence at T120)/(mean autofluorescence in all cells)]. The relative ABCB5-dependent Rh-123 transport capacities of the various melanoma cell lines were then established by dividing individual values to the mean of all values. For assessment of the effects of ABCB5 blockade on Rh-123 efflux, efflux studies were performed in additional experiments for G3361 melanoma cells in the presence of ABCB5-blocking mAb (clone 3C2-1D12 [4]) or isotype control mAb (50 g/ml, respectively).
Melanin assay
Cellular melanin content was determined spectrophotometrically as described previously [21]. Melanin contents were normalized to total protein concentrations using the Pierce microBCA Protein Assay Kit (ThermoScientific, Rockford, IL).
Results
Descriptive characteristics of cases and controls
Basic characteristics of cases and controls in our study are presented in Table 1. Age and gender were two matched variables. Melanoma cases were more likely to possess red hair color and more moles on the arms, while the childhood tanning ability of cases was less than that of controls, similar to the results of our previous study among women only [22].
Table 1.
Characteristics of cases and controls in the nested case-control study
| Characteristic | Controls | Melanoma |
|---|---|---|
| (n=609) | (n=585) | |
| Age at diagnosis (mean, years) | 61.8 | 61.5 |
| Gender, male (%) | 30.2 | 31.5 |
| Natural hair color at age 20, red (%) | 3.0 | 6.2 |
| Tanning ability, tan or deep tan (%) | 69.8 | 54.0 |
| Moles count on the left arm, 3+ (%) | 8.9 | 21.7 |
Association between the 44 tag-SNPs in the ABCB5 gene and melanoma risk
Information on the 44 tag-SNPs in the ABCB5 gene region is presented in Supplementary Table 1. Among them, there are two non-synonymous SNPs (rs2301641 and rs6401515), which may have putative function, and another SNP (rs10254317) which is a synonymous SNP. Forty-one out of the 44 SNPs were successfully genotyped with call rate > 95%, with the other three in a range of 90–95%. The distributions of genotypes among controls were in Hardy-Weinberg equilibrium (P>0.001, 0.05/44), except for rs10216013. We evaluated the associations of the genotypes with the risk of melanoma in an additive model. We observed nominal significant associations for three SNPs. In the analyses controlling for age and gender, we observed that the variant genotypes of rs10231520, rs17817117 and rs2301641 were associated with decreased melanoma risks (OR: 0.83, 95% CI: 0.70–0.98, P=0.026 for rs10231520; OR: 0.82, 95% CI: 0.68–0.98, P=0.028 for rs17817117; and OR: 0.83; 95% CI: 0.69–0.98, P=0.032 for rs2301641; assuming an additive model) (Table 2). These associations remained similar after further adjusting for risk factors including hair color, childhood tanning ability and moles on the arms. We analyzed the age at diagnosis for the top three SNPs and found no difference among genotypes (data not shown).
Table 2.
Association between genotypes and melanoma risk
| SNP | Genotype | Controls | Melanoma | ORa) | P value |
|---|---|---|---|---|---|
| (n=609) | (n=585) | (95% CI) | |||
| rs10231520 | |||||
| CC | 232(39.5) | 260(45.8) | 1.00 | ||
| CT | 268(45.6) | 238(41.9) | 0.79 (0.62–1.02) | ||
| TT | 88(15.0) | 70(12.3) | 0.71 (0.49–1.01) | ||
| Additive modelb | 0.83 (0.70–0.98) | 0.026 | |||
| rs17817117 | |||||
| GG | 248(41.7) | 273(47.8) | 1.00 | ||
| GC | 284(47.7) | 251(44.0) | 0.81 (0.63–1.03) | ||
| CC | 63(10.6) | 47(8.23) | 0.68 (0.45–1.03) | ||
| Additive modelb | 0.82 (0.68–0.98) | 0.028 | |||
| rs2301641 | |||||
| K155E | AA | 260(43.6) | 284(49.6) | 1.00 | |
| AG | 268(44.9) | 237(41.4) | 0.81 (0.64–1.04) | ||
| GG | 69(11.6) | 52(9.08) | 0.70 (0.47–1.04) | ||
| Additive modelb | 0.83 (0.69–0.98) | 0.032 |
Adjusted by age and gender
Assuming an additive effect of the variant alleles
Association between the three significant SNPs and pigmentary phenotypes
We evaluated the associations between the three SNPs and pigmentary phenotypes including hair color, tanning ability and moles on the arms among controls. We observed that the carriers of the SNP rs2301641 were more likely to have non-red hair color compared to red hair color (OR: 0.38, 95% CI: 0.14–1.03). However, in those with non-red hair color, no significant association was found with hair color (from black to blond). This SNP was not associated with tanning ability or mole counts. Besides that, we did not find any association of the other two SNPs with pigmentary phenotypes.
Association between the three significant SNPs and melanoma survival
Among the 585 melanoma cases, 46 died of melanoma and 84 died from other causes. For disease-specific survival analysis, the latter were considered as censored data in the analyses. However, none of the three SNPs showed a significant association with melanoma survival.
Association between rs2301641 genotype and ABCB5-mediated Rh-123 efflux capacity
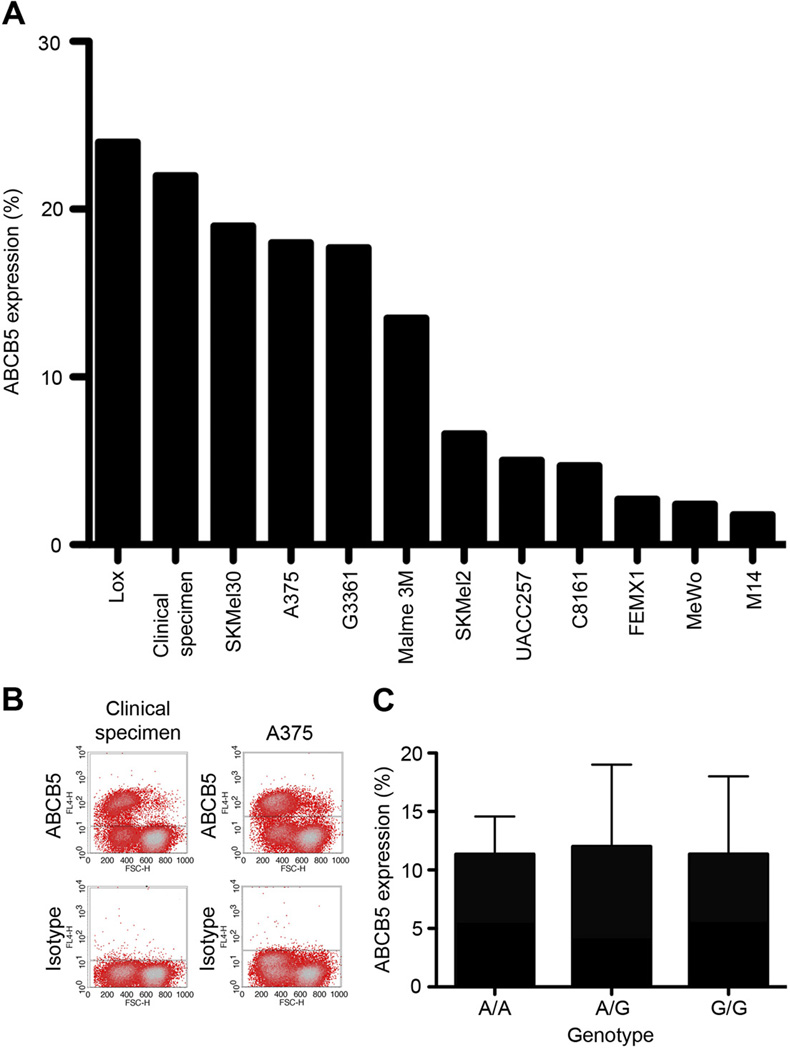
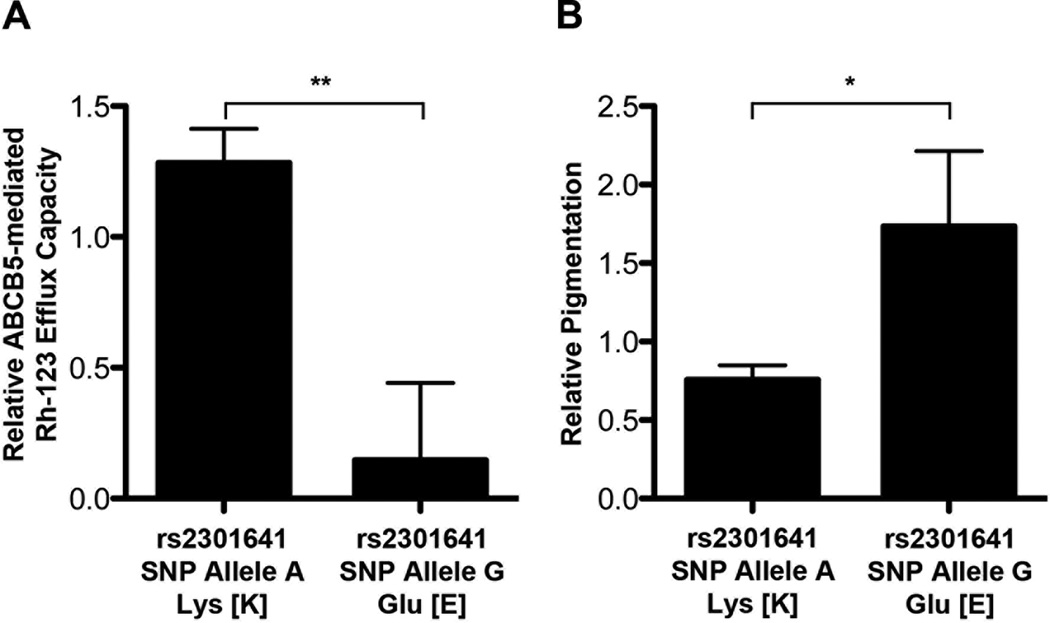
The SNP rs2301641 encodes a non-synonymous ABCB5 amino acid change (K115E). We further investigated its role in ABCB5 function, given its association with both melanoma risk and pigmentation. Efflux transport capacity for the dye Rh-123 is a hallmark function of ABCB5 [4], and was assessed for twelve rs2301641 SNP-genotyped melanoma cell lines by flow cytometry as described previously [4]. Four cell lines had the variant allele of which two had the genotype G/G (A375 and C8161) and two the genotype A/G (UACC257, SKMel30). The human melanoma specimens under study exhibited variable expression of cell surface ABCB5 as determined by flow cytometry (Fig.1A; Fig.1B shows flow cytometry staining examples for two of the twelve examined specimens). However, the rs2301641 ABCB5 genotype exhibited no correlation with levels of ABCB5 expression (Fig.1C). In a previous study, we had demonstrated in an ABCB5 gene transfection model that ABCB5 confers cellular Rh-123 efflux. We first confirmed that ABCB5 protein expression represents the principle mediator of Rh-123 efflux in human melanoma cells (Fig.2): Compared to isotype control mAb-treated melanoma cells, of which a subpopulation of 10.2% effluxed Rh-123 over a 120min incubation period at 37°C, Rh-123 efflux in ABCB5-blocking mAb-treated cultures (mAb clone 3C2-1D12 [4]) was inhibited by >95% (P<0.001) (Fig.2A; typical results are illustrated in Fig.2B). The effect of ABCB5 blockade on Rh-123 efflux was similar in magnitude and not significantly different from incubation of melanoma cells for 120min on ice, which blocks ATP hydrolysis and hence ABC transport function [23] (Fig.2A; typical results are illustrated in Fig.2B). Importantly, evaluation of relative Rh-123 efflux capacities in melanoma cell lines genotyped for the rs2301641 ABCB5 SNP (K115E) revealed that the SNP allele G (Glu [E]) was associated with markedly lower, 89%-reduced Rh-123 efflux capacity compared to the SNP allele A (Lys [K]) (P<0.01, Fig.3A). These functional studies revealed that the E form associated with lower melanoma risk is significantly associated with decreased ABCB5 function.
Figure 1.
ABCB5 surface expression and rs230641 genotype. A, ABCB5 expression determined by flow cytometry across 12 melanoma cell lines. B, Representative flow cytometry plots from results in A are illustrated for a clinical specimen (left panels) and a melanoma cell line (A375, right panels). C, Percentage 21 ABCB5 expression (means±SE) across the three ABCB5 rs230641genotypes: A/A (n=8), A/G (n=2), G/G (n=2).
Figure 2.

Flow cytometric analysis of ABCB5-mediated Rh-123 efflux function in G3361 melanoma cells. A, Percent Rh-123-effluxing cells (means±SE) under conditions of isotype control mAb treatment or ABCB5-blocking mAb treatment with incubation at 37°C, or with incubation on ice to block ATP hydrolysis and hence ABC-mediated efflux transport. B, Illustrated are representative flow cytometry plots from the results summarized in A.
Figure 3.
Association between rs2301641 genotype and ABCB5-mediated Rh-123 efflux capacity and pigmentation. A, Relative Rh-123 efflux capacities in melanoma cell lines genotyped for the rs2301641 ABCB5 SNP (K115E) (means±SE; **P<0.01). B, Relative pigmentation in melanoma cell lines genotyped for the rs2301641 ABCB5 SNP (K115E) (means±SE; *P<0.05).
Association between rs2301641 genotype and pigmentation
Given the additional association of the rs2301641 ABCB5 SNP with non-red compared to red hair color (OR: 0.38, 95% CI: 0.14–1.03) in control subjects, we measured melanin concentration, a measure of pigmentation, in eleven human melanoma cell lines genotyped for this SNP (K115E). We found that the SNP allele G (Glu [E]), associated with non-red hair and lower ABCB5 transport function, was associated with higher pigmentation compared to the SNP allele A (Lys [K]), associated with red hair and higher ABCB5 function (relative pigmentation 0.76±0.09 vs. 1.74±0.48, respectively, mean±SE, P<0.05) (Fig.3B).
Discussion
In this study, we have identified three SNPs of the ABCB5 gene associated with melanoma risk, and shown for one SNP (rs2301641), which encodes a nonsynonymous ABCB5 amino acid change (K115E), that the variant genotype that confers decreased melanoma risk is associated with lower ABCB5 protein function. Moreover, this variant is associated with enhanced pigmentation. There are several important implications of our study:
First, our results establish for the first time an association of a functionally significant ABCB5 gene polymorphism with human pathology, i.e. the risk of developing melanoma. Bioinformatic in silico analyses of ABCB5 gene polymorphisms had previously predicted the K115E amino acid substitution encoded by the rs2301641 SNP, which changes a positively charged amino acid to a negatively charged one and localizes to an extracellular loop of the ABCB5 protein [4], to be potentially deleterious to ABCB5 molecular function [17], a notion now for the first time experimentally supported by our study. Furthermore, our identification that genetically determined ABCB5 functionality is associated with melanoma risk is consistent with the established molecular role of ABCB5 in driving tumorigenic growth [16] and the recent demonstration by Bertolotto and colleagues in a 2011 Nature study that MITF mutation-dependent induction of ABCB5 expression can predispose to clinical melanoma development. Our results therefore point to a functional role of ABCB5 in melanomagenesis, and provide an intriguing first example that functional variation in a prospective cancer stem cell marker [5; 24] can be associated with disease risk for the corresponding malignancy. Our results therefore provide a rationale to further assess the correlations of ABCB5 rs2301641 SNP variability with cancer risk in additional malignancies where ABCB5 is overexpressed on cancer stem cells [16; 25], to potentially improve personalized risk stratification and appropriate screening strategies and preventive medicine measures.
Second, our findings show that genetically determined ABCB5 functionality based on rs2301641 SNP variability correlates with pigmentation phenotype, in addition to melanoma risk. This finding adds ABCB5 as a new gene to the well-established multiplicity of genetic links between pigmentation phenotype and melanoma risk, whereby individuals with lighter pigment and increased sun sensitivity are at higher risk of developing melanoma [3]. In our study, we noted a strong association between genetically determined impaired ABCB5 functionality based on rs2301641 SNP variability and increased melanin production, consistent with the previously observed inverse relationships between ABCB5 expression and pigmentation phenotype [5]. This association is further corroborated by ethnic variation in the rs2301641 SNP. In SNP databases with >2000 samples including ABI, Perlegen, and SNP 500 Cancer, the frequency of the variant allele G of rs2301641 associated with lower ABCB5 function is much higher in African and African-American populations as compared to Caucasian and Asian populations. Our finding that a lower functioning ABCB5 allele is associated with both higher pigmentation and decreased melanoma risk is therefore consistent with established observations of lower melanoma risk in individuals with darker skin.
Finally, given the demonstrated association of the rs2301641 SNP variability with ABCB5 function, and in light of the established role of ABCB5 as a drug resistance mechanism in multiple malignancies [12; 14; 15; 16; 25], it is likely that this ABCB5 SNP might also be associated with therapeutic drug responsiveness or resistance in human malignant melanoma and additional cancers where the drug resistance mediator ABCB5 is overexpressed at high levels on cancer stem cells [12; 14; 15; 16]. Our results therefore provide a rationale to further explore this potentially clinically relevant possibility.
Supplementary Material
Highlights.
Results establish a first association of an ABCB5 SNP with melanoma risk.
The lower melanoma risk allele is also associated with enhanced pigmentation.
The lower melanoma risk allele confers impaired ABCB5 protein function.
Functional variation in a cancer stem cell gene can correlate with tumorigenesis.
Results have implications for melanoma risk assessment and prevention.
Acknowledgements
This work was supported by funds from the NIH/NCI (grants R01CA113796, R01CA138231, R01CA158467 and P50CA093683 to MHF; R01CA122838, PO1CA87969, PO1CA055075 and RO1CA49449 to JH), and the U.S. Department of Veterans Affairs (BLR&D VA Merit Award 10688354 to NYF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schatton T, Frank MH. Cancer stem cells and human malignant melanoma. Pigment Cell Melanoma Res. 2008;21:39–55. doi: 10.1111/j.1755-148X.2007.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeh I, Bastian BC. Genome-wide associations studies for melanoma and nevi. Pigment Cell Melanoma Res. 2009;22:527–528. doi: 10.1111/j.1755-148X.2009.00622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature. 2007;445:843–850. doi: 10.1038/nature05660. [DOI] [PubMed] [Google Scholar]
- 4.Frank NY, Pendse SS, Lapchak PH, Margaryan A, Shlain D, Doeing C, Sayegh MH, Frank MH. Regulation of progenitor cell fusion by ABCB5 P-glycoprotein, a novel human ATP-binding cassette transporter. J Biol Chem. 2003;278:47156–47165. doi: 10.1074/jbc.M308700200. [DOI] [PubMed] [Google Scholar]
- 5.Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, Fuhlbrigge RC, Kupper TS, Sayegh MH, Frank MH. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Setia N, Abbas O, Sousa Y, Garb JL, Mahalingam M. Profiling of ABC transporters ABCB5, ABCF2 and nestin-positive stem cells in nevi, in situ and invasive melanoma. Mod Pathol. 2012 doi: 10.1038/modpathol.2012.71. [DOI] [PubMed] [Google Scholar]
- 7.Frank NY, Schatton T, Kim S, Zhan Q, Wilson BJ, Ma J, Saab KR, Osherov V, Widlund HR, Gasser M, Waaga-Gasser AM, Kupper TS, Murphy GF, Frank MH. VEGFR-1 expressed by malignant melanoma-initiating cells is required for tumor growth. Cancer Res. 2011;71:1474–1485. doi: 10.1158/0008-5472.CAN-10-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kupas V, Weishaupt C, Siepmann D, Kaserer ML, Eickelmann M, Metze D, Luger TA, Beissert S, Loser K. RANK is expressed in metastatic melanoma and highly upregulated on melanoma-initiating cells. J Invest Dermatol. 2011;131:944–955. doi: 10.1038/jid.2010.377. [DOI] [PubMed] [Google Scholar]
- 9.Linley AJ, Mathieu MG, Miles AK, Rees RC, McArdle SE, Regad T. The Helicase HAGE Expressed by Malignant Melanoma-Initiating Cells Is Required for Tumor Cell Proliferation in Vivo. J Biol Chem. 2012;287:13633–13643. doi: 10.1074/jbc.M111.308973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma J, Lin JY, Alloo A, Wilson BJ, Schatton T, Zhan Q, Murphy GF, Waaga-Gasser AM, Gasser M, Stephen Hodi F, Frank NY, Frank MH. Isolation of tumorigenic circulating melanoma cells. Biochem Biophys Res Commun. 2010;402:711–717. doi: 10.1016/j.bbrc.2010.10.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schatton T, Schutte U, Frank NY, Zhan Q, Hoerning A, Robles SC, Zhou J, Hodi FS, Spagnoli GC, Murphy GF, Frank MH. Modulation of T-cell activation by malignant melanoma initiating cells. Cancer Res. 2010;70:697–708. doi: 10.1158/0008-5472.CAN-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chartrain M, Riond J, Stennevin A, Vandenberghe I, Gomes B, Lamant L, Meyer N, Gairin JE, Guilbaud N, Annereau JP. Melanoma chemotherapy leads to the selection of ABCB5-expressing cells. PLoS ONE. 2012;7:e36762. doi: 10.1371/journal.pone.0036762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reid AL, Millward M, Pearce R, Lee M, Frank MH, Ireland A, Monshizadeh L, Rai T, Heenan P, Medic S, Kumarasinghe P, Ziman M. Markers of circulating tumour cells in the peripheral blood of patients with melanoma correlate with disease recurrence and progression. Br J Dermatol. 2013;168:85–92. doi: 10.1111/bjd.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank NY, Margaryan A, Huang Y, Schatton T, Waaga-Gasser AM, Gasser M, Sayegh MH, Sadee W, Frank MH. ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res. 2005;65:4320–4333. doi: 10.1158/0008-5472.CAN-04-3327. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Anderle P, Bussey KJ, Barbacioru C, Shankavaram U, Dai Z, Reinhold WC, Papp A, Weinstein JN, Sadee W. Membrane transporters and channels: role of the transportome in cancer chemosensitivity and chemoresistance. Cancer Res. 2004;64:4294–4301. doi: 10.1158/0008-5472.CAN-03-3884. [DOI] [PubMed] [Google Scholar]
- 16.Wilson BJ, Schatton T, Zhan Q, Gasser M, Ma J, Saab KR, Schanche R, Waaga-Gasser AM, Gold JS, Huang Q, Murphy GF, Frank MH, Frank NY. ABCB5 identifies a therapy-refractory tumor cell population in colorectal cancer patients. Cancer Res. 2011;71:5307–5316. doi: 10.1158/0008-5472.CAN-11-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moitra K, Scally M, McGee K, Lancaster G, Gold B, Dean M. Molecular evolutionary analysis of ABCB5: the ancestral gene is a full transporter with potentially deleterious single nucleotide polymorphisms. PLoS One. 2011;6:e16318. doi: 10.1371/journal.pone.0016318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R. Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 19.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stram DO, Leigh Pearce C, Bretsky P, Freedman M, Hirschhorn JN, Altshuler D, Kolonel LN, Henderson BE, Thomas DC. Modeling and E-M estimation of haplotype-specific relative risks from genotype data for a case-control study of unrelated individuals. Hum Hered. 2003;55:179–190. doi: 10.1159/000073202. [DOI] [PubMed] [Google Scholar]
- 21.Pan T, Zhu J, Hwu WJ, Jankovic J. The role of alpha-synuclein in melanin synthesis in melanoma and dopaminergic neuronal cells. PLoS One. 2012;7:e45183. doi: 10.1371/journal.pone.0045183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nan H, Kraft P, Hunter DJ, Han J. Genetic variants in pigmentation genes, pigmentary phenotypes, and risk of skin cancer in Caucasians. Int J Cancer. 2009;125:909–917. doi: 10.1002/ijc.24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izawa A, Schatton T, Frank NY, Ueno T, Yamaura K, Pendse SS, Margaryan A, Grimm M, Gasser M, Waaga-Gasser AM, Sayegh MH, Frank MH. A novel in vivo regulatory role of P-glycoprotein in alloimmunity. Biochem Biophys Res Commun. 2010;394:646–652. doi: 10.1016/j.bbrc.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank NY, Schatton T, Frank MH. The therapeutic promise of the cancer stem cell concept. J Clin Invest. 2010;120:41–50. doi: 10.1172/JCI41004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung ST, Cheung PF, Cheng CK, Wong NC, Fan ST. Granulin-epithelin precursor and ATP-dependent binding cassette (ABC)B5 regulate liver cancer cell chemoresistance. Gastroenterology. 2011;140:344–355. doi: 10.1053/j.gastro.2010.07.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.