Abstract
Purpose
This study assessed whether Myristoylated Alanine Rich C-Kinase Substrate (MARCKS) can regulate glioblastoma (GBM) growth, radiation sensitivity and clinical outcome.
Experimental Design
MARCKS protein levels were analyzed in five GBM explant cell lines and eight patient-derived xenograft tumors by immunoblot, and these levels were correlated to proliferation rates and intracranial growth rates, respectively. Manipulation of MARCKS protein levels was assessed by lentiviral-mediated shRNA knockdown in the U251 cell line and MARCKS over-expression in the U87 cell line. The effect of manipulation of MARCKS on proliferation, radiation sensitivity and senescence was assessed. MARCKS gene expression was correlated with survival outcomes in the Repository of Molecular Brain Neoplasia Data (REMBRANDT) Database and The Cancer Genome Atlas (TCGA).
Results
MARCKS protein expression was inversely correlated with GBM proliferation and intracranial xenograft growth rates. Genetic silencing of MARCKS promoted GBM proliferation and radiation resistance, while MARCKS overexpression greatly reduced GBM growth potential and induced senescence. We found MARCKS gene expression to be directly correlated with survival in both the REMBRANDT and TCGA databases. Specifically, patients with high MARCKS expressing tumors of the Proneural molecular subtype had significantly increased survival rates. This effect was most pronounced in tumors with unmethylated O6-methylguanine DNA methyltransferase (MGMT) promoters, a traditionally poor prognostic factor.
Conclusions
MARCKS levels impact GBM growth and radiation sensitivity. High MARCKS expressing GBM tumors are associated with improved survival, particularly with unmethylated MGMT promoters. These findings suggest the use of MARCKS as a novel target and biomarker for prognosis in the Proneural subtype of GBM.
INTRODUCTION
Glioblastoma multiforme (GBM) represents the most common and deadly form of glioma (1). The current mainstay of treatment for GBM is surgical resection followed by radiation with concurrent and adjuvant chemotherapy with an alkylating agent. Indeed, the most significant developments in recent years were the improvement in survival with the addition of temozolomide to treatment regimens (2), and the recognition that the O-6-methylguanine-DNA methyltransferase (MGMT), a DNA repair protein encoded by the MGMT gene, is a key prognostic variable in glioma. The MGMT protein can effectively reverse the predominant DNA lesion produced by temozolomide chemotherapy, that of DNA methylation at the O-6 position of guanine (3). MGMT protein expression can be regulated through epigenetic silencing of the MGMT promoter through methylation. Therefore, methylated MGMT (often called hypermethylated MGMT) results in a silencing of MGMT transcription. Tumors with methylated MGMT (~33–45% of GBM) have a better prognosis overall and predict for improved response to temozolomide and radiation therapy (3, 4). The improvement in median survival was modest, however, from 12.1 to 14.6 months post-diagnosis (2). Conversely, unmethylated MGMT tumors have an intact MGMT DNA repair mechanism that yields poorer survival and earlier treatment failure. There are currently no proven alternative treatment options for those patients with un-methylated MGMT promoter status.
The MGMT DNA repair mechanism is merely one of many processes that contribute to poor survival in GBM. It is well known that several different mutations in oncogenes and loss of tumor suppressors may contribute to the pathogenesis of GBM, and these aberrations differ from patient to patient. This would suggest that effective treatment regimens for GBM should be tailored toward the particular pathogenesis of that patient’s neoplasm. Over the past several years, there have been many attempts to generate molecular profiles to better understand GBM and the prognostic factors that influence survival and response to therapy. Resources such as the Repository of Molecular Brain Neoplasia Data (REMBRANDT) database and The Cancer Genome Atlas (TCGA) Research Network have provided insight into the pathogenesis of GBM through allowing researchers to correlate gene expression with clinical outcome. Recently, genomic analyses of TCGA GBM samples lead to the identification of molecular subtypes, namely Classical, Mesenchymal, Proneural, and Neural. Indeed, abnormalities in several oncogenes and tumor suppressors were identified that are characteristic of each subtype (5, 6). Moreover, treatment efficacy differs among the subtypes, indicating that future clinical approaches will depend on subtype specificity (6).
One of the most common genetic alterations observed in approximately 90% of GBM is loss of heterozygosity (LOH) of chromosome 10q (7). This alteration often occurs in conjunction with mutation of the tumor suppressor gene Phosphatase and Tensin Homolog (PTEN) in up to 60% of GBMs with LOH (8, 9). PTEN executes its tumor suppressor function by antagonizing signaling through the phosphatidylinositol 3-kinase (PI3K)/Akt pathway. Activation of the PI3K/Akt pathway begins when the phospholipid, phosphatidylinositol (4,5) bisphosphate (PIP2) is phosphorylated by PI3K to phosphatidylinositol (3,4,5)-triphosphate (PIP3). PIP3 can then recruit the kinase Akt to the plasma membrane where it is phosphorylated and activated leading to changes in migration, invasion, angiogenesis, survival and proliferation. PIP3 can also be converted back to PIP2 by PTEN. Therefore, mutations or deletions of PTEN in gliomas lead to an over-activation of the Akt pathway and have been associated with a worse clinical outcome (10–13). This would indicate that the availability of the precursor phospholipid PIP2 is a critical factor for modulating activation of Akt and potentially, clinical outcome.
MARCKS is a protein that is capable of regulating PIP2 availability by sequestration via an electrostatic mechanism. MARCKS was identified over 20 years ago as a major protein substrate for protein kinase C (PKC) in brain synaptosomes (14). Since then, its major role has been defined as an integrator of PKC and Calmodulin signals into control of the actin cytoskeleton (15). It performs this function with its poly-basic effector domain that cross-links actin filaments into bundles and sequesters PIP2 with favorable stoichiometry via an electrostatic mechanism (16, 17). Phosphorylation of the effector domain by PKC or binding by Ca2+-Calmodulin leads to a local release of PIP2 and actin bundles; resulting in alterations in the actin cytoskeleton. This is the mechanism by which MARCKS controls cellular structural changes such as the extension of growth cones in neurons and the promotion of chemotaxis in neutrophils (18, 19). MARCKS has also been implicated in the pathogenesis of various malignancies. A particular mutation which truncates the protein within the effector domain is common in small intestinal adenocarcinomas (20). Levels of MARCKS were found to be inversely correlated with increased proliferation in melanoma cells, suggesting that MARCKS may function as a “tumor suppressor” (21). Interestingly, a recent study on MARCKS in GBM implicated this protein as a mediator of attachment and invasion, suggesting that higher levels of MARCKS would lead to a more aggressive disease (22). It is clear that the function of this protein in the pathogenesis of malignancy is multi-faceted and complex.
Due to the MARCKS protein’s ability to sequester PIP2, we hypothesized that MARCKS may be able to regulate growth and radiation sensitivity in GBM through regulation of the PI3K/Akt pathway. GBM cell lines and tumors with higher levels of MARCKS would have reduced availability of PIP2 for conversion to PIP3 and thus reduced activation of the PI3K/Akt pathway. This would lead to reduced growth rates and resistance to radiation; which could in turn lead to improved clinical outcomes. We investigated this in several established GBM cell lines as well as a patient-derived tumor xenograft mouse model. We confirmed the effect of MARCKS in the regulation of proliferation and radiation sensitivity with molecular manipulation and also identified a novel role in the regulation of senescence. To more closely evaluate the MARCKS protein in a clinical context, we then correlated MARCKS gene expression with molecular subtypes and clinical outcome. We found MARCKS expression to be particularly protective in the Proneural subtype of GBM, indicating that this protein may be a novel biomarker and therapeutic strategy for these patients.
MATERIALS AND METHODS
Xenograft passaging and processing
All animal studies were carried out in accordance with the policies set by the UAB IACUC. Dr. C. David James (UCSF, San Francisco, CA) and Dr. Jann Sarkaria (Mayo Clinic, Rochester, MN) have developed several partially characterized human glioblastoma xenograft lines through serial passage within athymic nude mice (23–25). Eight of these human glioblastoma xenograft lines were provided to Dr. Gillespie (UAB) and have been used for this study. Athymic nu/nu mice were obtained from National Cancer Institute Mouse Repository (NCI-Frederick, MD). The following primary glioblastoma xenograft lines were used for this study: GBM 6, GBM 10, GBM 12, GBM 14, GBM 15, GBM 22, GBM 39, and GBM 59 (23–25). Tumors xenografts were serially passaged in the mice (flanks) by harvesting and mechanically disaggregating the cells prior to injection as has been described before (23–25). Gross tumor is macrodissected from normal tissue and snap frozen for kinomic profiling and immunoblotting as detailed below. Intracranial tumor growth was determined by measuring the time from intracranial implantation to the day the mice begin to show neurological impairment.
Immunoblotting
Immunoblotting was performed by standard protocols with the following primary antibodies: rabbit anti-MARCKS, Epitomics, catalog # 1944-1; rabbit anti-MARCKS pS158/162, Epitomics, catalog # 2156-1; rabbit anti-Akt, Cell Signaling, catalog # 4691; rabbit anti-Akt pSer473, Cell Signaling, catalog # 4060S; rabbit anti-phospho DNA-PKcs T2609, Genetex, catalog #GTX-24194; rabbit anti-phospho H2AX Ser139, Millipore, catalog #MI-07-164; rabbit anti-LCB3, Novus Biologicals, catalog #NB600-1384; rabbit anti-Caspase 3, Cell Signaling, catalog #9662S; rabbit anti-GAPDH, Santa Cruz, catalog # sc-25778; mouse anti-αTubulin, Santa Cruz, catalog #sc-5286. A detailed description of the protocol is contained in the Supplementary Methods.
Plasmid constructs
A set of five MARCKS shRNA lentiviral transfer vector plasmids (catalog # RHS4533-NM_002356) and an empty vector control plasmid (catalog # RHS4080) were obtained from Thermo Biosystems. The psPAX2 packaging plasmid (Addgene plasmid 12260) and pCMV-VSV-G envelope plasmid (Addgene plasmid 8454) were obtained from Addgene (26). Detailed description of the construction of the pLenti4-MARCKS and pLenti4-DELTA plasmids is contained in the Supplementary Methods.
Lentiviral vector production
Lentivirus was generated by co-transfection of 293FT cells with pCMV-VSVG, psPAX2, and the appropriate lentiviral transfer vector with Lipofectamine. A detailed description of the production of lentivirus is contained in the Supplementary Methods.
Cell culture and stable line selection
U87, U251, U373, U118, D54 and D32 malignant glioma cell lines (American Type Culture Collection) and 293FT human embryonic kidney cells (Invitrogen) were cultured in DMEM with 10% FBS and 1% Pen-Strep at 37°C in 5% CO2. A detailed description of stable cell line selection is contained in the Supplementary Methods.
Proliferation assay
2×103 cells were seeded in 96-well plates in 100μL of media. Cells were allowed to grow for 5 days and were then harvested with the ATPlite Luminescence Assay System (catalog # 6016949, Perkin-Elmer) or WST-1 assay System (Calbiochem) per manufacturer’s instructions. Fluorescence was measured on a Synergy H1 Hybrid plate reader (Biotek) and absorbance was read on a Vmax Kinetic Microplate Reader (Molecular Devices). All measurements were performed in quadruplicate.
Clonogenic survival assay
U251 control and MARCKS knockdown cells grown to 80% confluency were trypsinized, counted with a hematocytometer, and serially diluted to defined concentrations. Three different defined numbers of cells were plated per dose of radiation in duplicate. Eight hours later, the cells were treated with 0, 3, 5, or 8 Gy radiation. Fourteen days later, cells were fixed and stained with 6.0% glutaraldehyde and 0.5% crystal violet. Colonies were counted in quadruplicate for each condition and a surviving fraction (S.F.) was calculated by using the equation (number of colonies formed/number of cells plated)/(number of colonies for sham irradiated group/number of cells plated). The results were then plotted as mean and standard error of the mean in a semi-logarithmic format using Microsoft Excel software. Dose enhancement ratio (DER) is calculated as the dose (Gy) for control divided by the dose for MARCKS knockdown treated cells (normalized for plating efficiency at 0 Gy) for which a S.F.=0.2 is achieved.
Clinical data
Clinical and gene expression data was obtained from the REMBRANDT database and TCGA Project. Gene expression values from the Affymetrix U133 Plus 2.0 Array Platform for MARCKS were downloaded for all glioma samples in the REMBRANDT database through the Advanced Search function. The clinical data was downloaded for these samples in BRB format through the Download function. For TCGA analysis, we used 192 out of the 202 patients that did not have prior glioma. Gene expression data from three separate microarray platforms was summarized by gene across three different microarray platforms using a method described elsewhere (6). The gene-summarized gene expression values are contained in the supplementary materials in (Verhaak et al., 2010) (6) for 11,861 genes from the TCGA samples. Basic sample characteristics such as counts by subtype, MGMT status, etc., are given elsewhere (6). A detailed description of the survival analysis is included in the Supplementary Methods.
Statistical analysis for in vitro data
The in vitro data were analyzed via analysis of variance (ANOVA) followed by a Bonferroni post-test using GraphPad Prism version 4.02 (GraphPad Software, San Diego, CA). Data is presented as average +/− standard error of mean.
RESULTS
MARCKS Regulates GBM Growth Rates in Cell Lines and in Human Xenografts
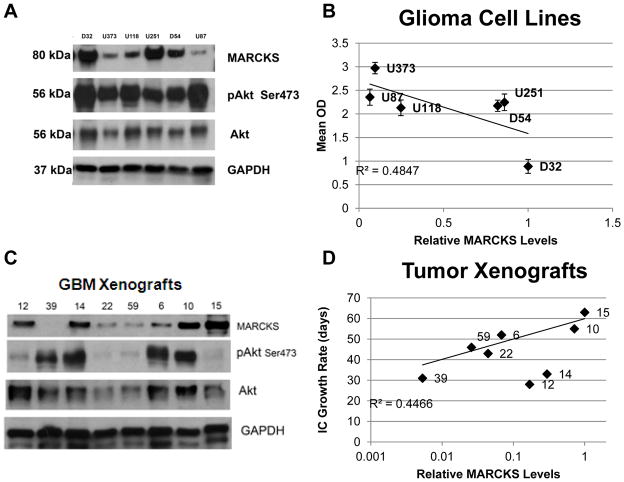
Previous studies have implicated a potential tumor suppressor role for the MARCKS protein (21). We thus hypothesized that GBM cell lines with higher levels of the MARCKS protein would have slower growth rates. To test our hypothesis, we measured MARCKS protein expression (normalized to GAPDH) in several well characterized immortalized GBM cell lines (Fig. 1A) demonstrating a spectrum of proliferation rates (Supplementary Fig. S1). We found variable MARCKS expression levels across these cell lines and that MARCKS levels inversely correlated (R2= 0.4847) with GBM cell proliferation (Fig. 1B). Despite the extensive characterization of these cell lines, however, it has been realized that they harbor genetic alterations relative to the original tumor (27). We thus performed a similar analysis with a more clinically relevant xenograft model.
Figure 1.
Several GBM cell lines were grown under standard growth conditions and total protein lysates were prepared as described in the Methods and probed for MARCKS, phosphorylated Akt on serine 473 (pAkt Ser473), total Akt, and GAPDH (for protein normalization) by immunoblotting (A). Proliferation was determined for each cell line using a WST-1 cell proliferation assay (Supplementary Fig. S1) as described in the Methods. Densitometric quantitation of MARCKS levels were performed and normalized to GAPDH and relative levels of MARCKS were plotted vs. proliferation (Mean OD) with R2 correlation (B). Human GBM xenografts were harvested from athymic nude mice and total protein lysates were prepared as described in the Methods and probed for MARCKS, phosphorylated Akt on serine 473 (pAkt Ser473), total Akt, and GAPDH (for protein normalization) by immunoblotting (C). Densitometric quantitation of MARCKS levels were performed and normalized to GAPDH and relative levels of MARCKS were plotted vs. intracranial (IC) growth rate in days with R2 correlation (D).
The genetic makeup of the original tumor can be effectively preserved if the cells are maintained as xenolines in immunocompromised mice after direct implantation of patient tumor tissue (28). Human-derived GBM xenografts have been established by direct implantation of patient tumor tissue within immunocompromised mice and are serially passaged within these mice as xenograft lines. It has been shown that these xenografts do not demonstrate substantial alteration of genetic changes as compared to the original tumor (29). For this reason, these xenografts exhibit a wide range of intracranial growth rates and common genetic alterations that are seen in GBM (Supplementary Table 1). We performed similar measurements in the GBM xenografts (Fig. 1C) and correlated those levels with the average time of intracranial tumor growth for each tumor xenograft. This represents the average time from intracranial implantation to the time of passage (when mice demonstrate neurological decline) and thus is an inverse surrogate measure for their tumor proliferation rates (Supplementary Table 1). As in the GBM cell lines, there was an inverse correlation between the tumor xenograft intracranial growth rates and MARCKS expression levels (Fig. 1D; R2=0.4466), suggesting that MARCKS is an important regulator of GBM growth.
MARCKS Silencing Leads to Increased Growth Rates and Radiation Resistance
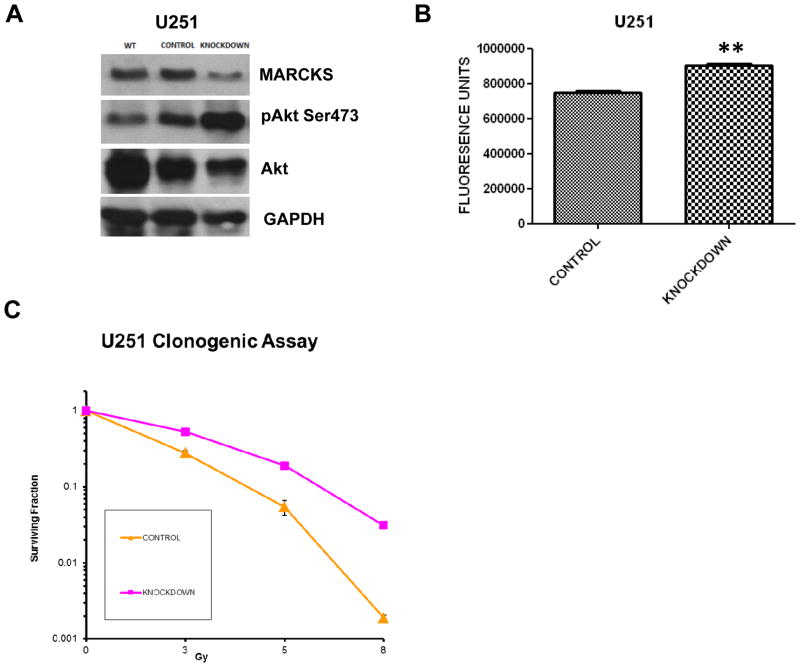
Our xenograft and cell culture data implicated MARCKS as a potential regulator of GBM growth. Therefore, we manipulated MARCKS expression in GBM cell lines to determine whether MARCKS directly affected GBM growth. We investigated MARCKS gene silencing using lentiviral-mediated MARCKS shRNA knockdown in U251 cells (high basal MARCKS expression). A control cell line was generated utilizing an empty vector control lentivirus. A stable 40% knockdown of the MARCKS protein in U251 cells (Fig. 2A) demonstrated a 26% increase in proliferation rate (p<0.01) (Fig. 2B). Because MARCKS can sequester PIP2, we suspected that knockdown of MARCKS would be associated with increased signaling through PI3K/Akt due to increased availability of PIP2 for conversion to PIP3 by PI3K. Akt is activated by phosphorylation of serine 473 after recruitment to the membrane by PIP3. We thus assessed our MARCKS knockdown for activation of Akt by immunoblotting for phosphoserine-473. Decreases in U251 MARCKS levels produced increased activation of Akt compared to control (Fig. 2A). To demonstrate that signaling through the PI3K/Akt pathway was responsible for these increases in proliferation we treated the MARCKS knockdown cells with the PI3K inhibitor LY294002 at 10μM. Treatment with LY294002 led to an 87% decreased activation of Akt as assessed by densitometry (Supplementary Fig. S2A) and subsequent decreases in proliferation (Supplementary Fig. S2B)(p = 0.0287). This suggests that decreased levels of MARCKS promote increased proliferation through enhanced signaling through PI3K/Akt.
Figure 2.
Parental U251 (WT=wild type) cells were infected with a control or MARCKS shRNA knockdown (MARCKS) lentiviral vector and selected for antibiotic resistance as described in Methods. The control and knockdown cell lines were probed for MARCKS, phosphorylated Akt on serine 473 (pAkt Ser473), total Akt, and GAPDH (for protein normalization) by immunoblotting (A). Proliferation was assessed using ATPlite Luminescence Assay System with mean and standard error shown (** = p<0.01) (B). Clonogenic survival assay was performed on U251 cells infected with either control or MARCKS shRNA (Knockdown) lentiviral vector and plotted in a semi-logarithmic fashion as mean surviving fraction (with standard error of the mean) vs. radiation dose in Gy (C). An upward and rightward shift indicates radiation protection.
Increased signaling through Akt has been implicated in therapeutic resistance due to increased DNA repair and decreased apoptosis (30, 31). We thus hypothesized that MARCKS knockdown would lead to increased radiation resistance. We assessed the radiation sensitivity of our knockdown cell line by a clonogenic assay and found a significant increase in radiation resistance with lower levels of MARCKS compared to control (Dose enhancement ratio = 0.75; 3 Gy p<0.0001, 5 Gy p=0.0002, 8 Gy p<0.0001) (Fig. 2C and Supplementary Fig. S2C). Radiation induces cell death by multiple mechanisms such as apoptosis, autophagy, mitotic catastrophe, and senescence. It is thought that cell death occurs due to failure of the cell to adequately repair the DNA damage induced by irradiation. Activation of PI3K/Akt has been demonstrated to lead to decreased apoptosis as well as increased DNA repair by a non-homologous end joining (NHEJ) mechanism (32). We therefore investigated how knockdown of the MARCKS protein might affect these forms of cell death.
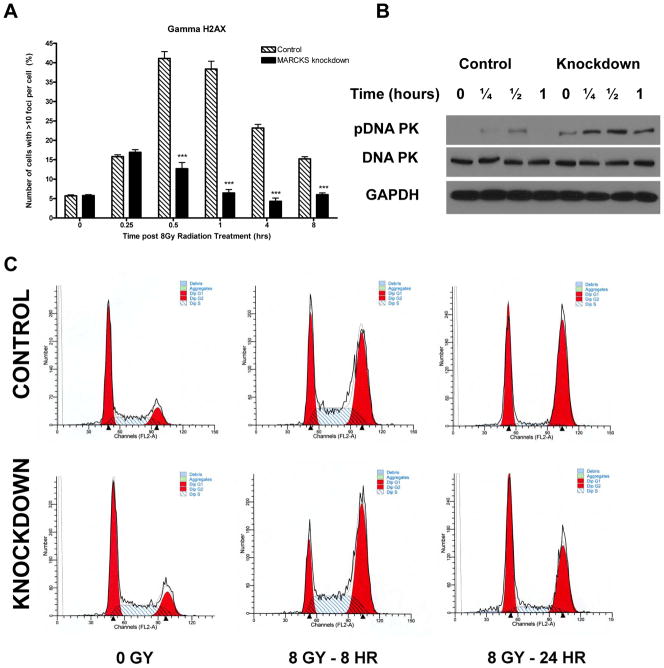
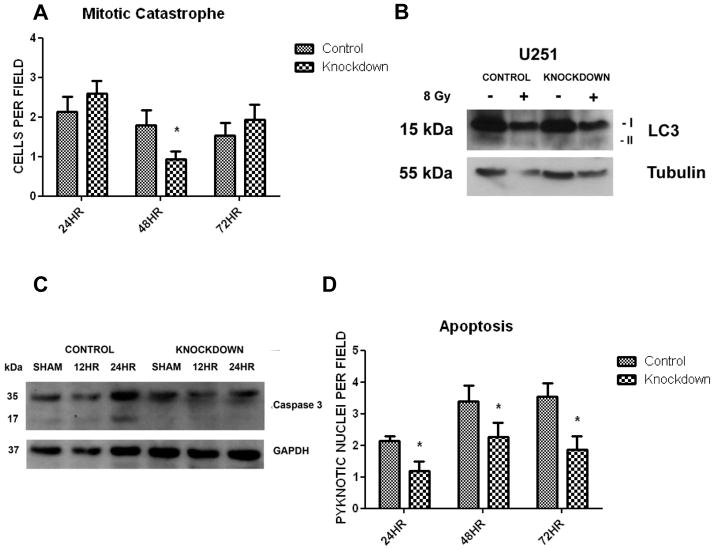
To investigate the mechanism of DNA repair, we quantified DNA double strand breaks (DSBs) over time after radiation. The DSB is the most lethal lesion inflicted by radiation. Upon sensing a DSB, the cell will phosphorylate histone H2A on serine 139 (γH2AX). Thus, staining for γH2AX foci as a marker for DNA DSB over time is a common method of investigating the rate of DNA repair. We noted a significantly decreased presence of DNA DSBs over time after radiation in our knockdown cell line (p<0.001 at 30 minutes, 1 hour, 4 hours and 8 hours)(Fig. 3A). To demonstrate that this effect could be due to increased DNA repair by NHEJ, we immunoblotted for phosphorylated DNA-dependent protein kinase, catalytic subunit (DNA-PKcs) in our control and knockdown cell lines after irradiation. We found that MARCKS knockdown lead to a 4-fold increase in peak DNA-PKcs activation after 8 Gy as assessed by densitometry (Fig 3B). Treatment of the MARCKS knockdown cell line with LY294002 at 10μM for 1 hour prior to radiation abrogated activation of DNA-PKcs, suggesting that this increase in DNA repair is mediated through PI3K/Akt signaling (Supplementary Fig. S3A). This increased DNA repair mechanism would suggest that manipulation of MARCKS levels would alter the various forms of cell death induced after irradiation. We therefore assessed the effect of MARCKS knockdown on cell cycle, mitotic catastrophe, autophagy, senescence and apoptosis. Cell cycle analysis was performed in the control and knockdown cell line after 8 Gy. We found no difference in the basal cell cycle distribution, however; knockdown of the MARCKS protein lead to a more rapid recovery toward the basal cell cycle distribution at 24 hours after 8 Gy (Fig. 3C and Supplementary Fig. S3B, C). This is consistent with more rapid DNA repair and recovery from cell cycle arrest. Mitotic catastrophe was assessed after 8 Gy at 24, 48, and 72 hours by DAPI staining and quantification of multi-nucleated cells (Supplementary Fig. S4A). We found a decrease in mitotic catastrophe with MARCKS knockdown at 48 hours compared to control (p=0.0242), but no difference at 24 or 72 hours (Fig. 4A). Senescence was assessed by staining for senescence associated β-galactosidase (SA-β-gal), a widely used biomarker of senescent cells (33). We did not detect senescence in either the control or MARCKS knockdown cell line after 8 Gy up to 72 hours (data not shown). Autophagy was quantified by immunoblotting for cleavage of the LC3-I protein to LC3-II, which is associated with completed autophagasomes and is often used as a marker for autophagy. We found a 2.95-fold increase (normalized to α-Tubulin) in LC3-II with knockdown of MARCKS (Fig. 4B), consistent with previous reports that the PI3K/Akt pathway negatively regulates autophagy (34). We assessed apoptosis by immunoblotting for cleaved caspase 3 and quantification of pyknotic nuclei with DAPI stain after 8 Gy treatment. We found an increase in cleaved caspase 3 at 24 hours (Fig. 4C), as well as an increased presence of pyknotic nuclei at 24, 48 and 72 hours in the control cell line (Fig. 4D and Supplementary Fig. S4A) (p<0.05). These results suggest that knockdown of MARCKS leads to increased resistance to radiation by increased DNA repair. This causes a more rapid recovery from cell cycle arrest, decreased apoptosis, and decreased mitotic catastrophe.
Figure 3.
Quantification of γH2AX foci was performed at the indicated time points after 8 Gy radiation (*** = p<0.001) (A). DNA repair by non-homologous end joining (NHEJ) was assessed by immunoblotting for phosphorylated DNA-PK at the indicated time points after treatment with 8 Gy (B). The distribution of the cell cycle at 8 hours and 24 hours after 8 Gy was assessed by propidium iodide staining and flow cytometry. Representative histograms for the control and MARCKS knockdown cell line at each time point are shown (C). Mean values for each phase of the cell cycle at each time point and p values for control versus MARCKS knockdown are available in Supplementary Fig. S3B and S3C.
Figure 4.
Mitotic catastrophe was quantified at 24, 48, and 72 hours after 8 Gy radiation by DAPI stain and counting the number of multi-nucleated cells per field (40×)(* = p<0.05)(A). An example of mitotic catastrophe is available in Supplementary Fig. S4. Autophagy was quantified at 72 hours after 8 Gy radiation by immunoblotting for conversion of LC3-I to LC3-II with normalization to α-Tubulin (B). Apoptosis was quantified after 8 Gy radiation by immunoblotting for cleaved caspase 3 at 12 and 24 hours (C), and by quantification of pyknotic nuclei per field (40×) at 24, 48, and 72 hours (* = p<0.05)(D). An example of a pyknotic nucleus is available in Supplementary Fig. S4.
MARCKS Over-expression Leads to Decreased Growth Rates and Senescence
Since a reduction in MARCKS levels led to increased growth and resistance to radiation, we hypothesized that MARCKS overexpression would lead to decreased growth and increased sensitivity to radiation. MARCKS overexpression was achieved in U87 GBM cells (low basal MARCKS expression) using lentiviral infection, with an empty vector lentivirus as a control. Expectedly, 6.3-fold over-expression of the MARCKS protein within U87 GBM cells (Fig. 5A) showed a 52% decrease in proliferation rate compared to control (p=0.0003)(Fig. 5B). We attempted to generate a stable overexpressing cell line, but we were unable to do so as these cells did not increase in confluence over a period of 21 days, while the control cell line was passaged three times during the same period. Because these cells did not proliferate, the effect on radiation sensitivity due to MARCKS overexpression could not be assessed.
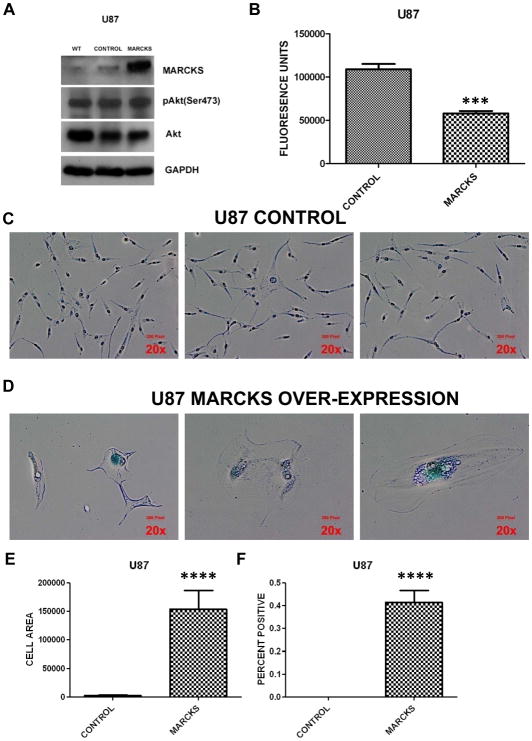
Figure 5.
Parental U87 (WT=wild type) cells were infected with a control or MARCKS expression (MARCKS) lentiviral vector and selected for antibiotic resistance as described in Methods. The control and MARCKS overexpressing cell lines were probed for MARCKS, phosphorylated Akt on serine 473 (pAkt Ser473), total Akt, and GAPDH (for protein normalization) by immunoblotting (A). Proliferation was assessed using ATPlite Luminescence Assay System with mean and standard error shown (*** = p<0.001)(B). Representative photos showing phase contrast images of Control (C) or MARCKS over-expressing (D) cells at 20× magnification. The center image of the MARCKS over-expressing cells demonstrates a multi-nucleated cell. The blue color indicates positive staining for Senescence associated β-galactosidase (SA-β-gal). Cell area of the MARCKS over-expressing and control cell line was measured using ImageJ as described in the Supplementary Methods and mean cell area with standard error of the mean is plotted (**** = p<0.0001)(E). Senescence was quantified by counting the percent of cells staining positive for SA-β-gal at 10× magnification (**** = p<0.0001)(F).
We noted from imaging that the MARCKS overexpressing cells were adopting a flattened morphology and had a 50-fold increase in size compared to the control cell line (p<0.0001) (Fig. 5C, D, and E). These morphological changes are consistent with senescence, a state in which normal diploid cells lose their ability to divide (35, 36). The MARCKS-overexpressing U87 cells were stained for SA-β-gal to confirm senescence (Fig. 5C and D). The percent of cells staining positive for SA-β-gal was quantified and 41.5% of the MARCKS-overexpressing U87 demonstrated positive staining, while none of the control cells stained positive (Fig. 5F)(p<0.0001). Because SA-β-gal is a relatively insensitive marker of senescence, we also quantified the percent of cells demonstrating the morphologic changes consistent with senescence. 200 cells were imaged and 79.5% of the MARCKS-overexpressing cells demonstrated these changes compared to 1% of the control cells. We also noted the presence of multi-nucleated giant cells, which are morphologically similar to those found in a previously identified giant cell GBM subtype (37) (central panel, Fig. 5D). The multi-nucleated giant cells in giant cell GBM are halted in the early phases of mitosis and lack proliferative capacity (38). These results confirm MARCKS as a regulator of GBM growth and suggest a novel role in regulating senescence.
MARCKS Expression is a Favorable Prognostic Factor for Glioma Patients
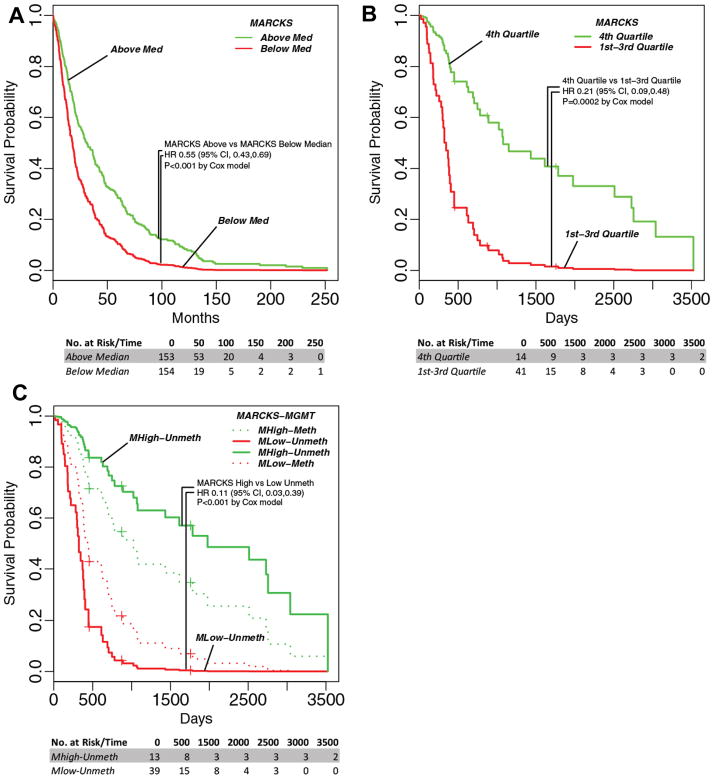
In light of our cell culture and xenograft model data, we anticipated that overexpression of MARCKS would be a favorable characteristic for survival. Thus we sought to evaluate MARCKS expression more closely in terms of clinical outcomes. We thus analyzed MARCKS gene expression in all glioma samples (low grade and high grade) within the REMBRANDT database for survival probability. As shown in Fig. 6A, we compared the survival of glioma patients with MARCKS expression above and below the median expression and found a statistically significant survival benefit for those patients with high MARCKS expression (HR = 0.55; 95% CI 0.43–0.69; p<0.001 by Cox model). This suggested that up-regulation of MARCKS is a favorable prognostic factor for patients with glioma.
Figure 6.
Kaplan-Meier curves for MARCKS expression above (Green) and below (red) the median in all glioma samples from the REMBRANDT database (46) (A). National Cancer Institute. 2005. REMBRANDT home page. <http://rembrandt.nci.nih.gov>. Accessed 2011 September 30. Kaplan-Meier curves (age-adjusted) for MARCKS expression dichotomized on the fourth quartile (4th Quartile=MHigh) vs. the lower three quartiles (1st – 3rd Quartile=MLow) in Proneural GBM samples from the TCGA database (B) and then further stratified by MGMT status (Meth=Methylated promoter; Unmeth=Unmethylated promoter) (C) (47) The number at risk at indicated time points is shown below each curve.
To confirm and expand the REMBRANDT data, we performed similar analyses using the TCGA database that was used to define GBM molecular subtypes (6). We analyzed MARCKS gene expression within the TCGA dataset of 192 GBM patients that had no prior glioma with defined molecular subtype information. We fit a Cox proportional hazards model as a function of age and MARCKS expression and discovered a protective influence of increased MARCKS expression on survival outcome (p=0.044, Supplementary Table 2). We also performed survival analyses for each subtype (Classical, Mesenchymal, Neural, and Proneural) and found that only the Proneural subtype retained statistical significance when age-adjusted (p=0.003594) (Supplementary Table 2). This analysis shows that the influence of MARCKS expression on survival outcome shows high subtype specificity with very strong effect in the Proneural subtype and almost no effect in the other subtypes, thus the full sample analysis effectively shows a dilution of the effect in the Proneural subtype. To observe the linearity of the effect of MARCKS gene expression on survival within Proneural individuals, we performed survival regression in which we categorized MARCKS expression by quartiles (4 levels), (Supplementary Table 2). The quartile-based analysis shows the effect of MARCKS to be highly nonlinear, with the 4th quartile (highest MARCKS expression) showing a strong protective influence compared with other quartiles (HR=0.28, p=0.01). In further analysis, we chose to dichotomize MARCKS at the 4th quartile for maximum discrimination (Median Survival of 47.2 months versus 12.2 months, HR=0.21; 95% CI 0.09–0.48; p<0.0002) as shown in Fig. 6B.
MGMT methylation status and age are the two most common factors in determining prognosis for GBM. We therefore performed survival analysis after adjusting for both MGMT status and age using the MARCKS quartile expression data (Fig. 6C and Supplementary Table 2). The MGMT term did not show significance (HR=1.48, p=0.24) as a main effect, but we suspected a possible interaction with MARCKS. We then stratified by MGMT status (methylated and unmethylated), and found that MARCKS protection was highly significant in the unmethylated individuals (Median Survival of 65.3 months versus 10.7 months, HR=0.11; 95% CI 0.03–0.39; p<0.001), but not significant in the methylated individuals (HR=0.39, p=0.19). This indicates that the patients that benefit to the highest degree from elevated MARCKS expression in GBM are those of the Proneural subtype with unmethylated MGMT status.
DISCUSSION
Our analysis of survival outcomes in the REMBRANDT and TCGA databases with respect to MARCKS expression in all samples indicated a direct correlation with patient survival (Fig. 6A and B, Supplementary Table 2). However, we suspect that the importance of MARCKS may ultimately depend on the particular cell signaling cascades that are active within the tumor. This is consistent with our data showing molecular subtype specificity in the TCGA analysis. Indeed, when we looked at MARCKS impact on survival based on molecular subtype, only the Proneural subtype retained significance (Supplementary Table 2). An even more striking result is observed when MARCKS expression is broken down into quartiles. Within the Proneural subtype, the highest quartile (4th quartile; highest MARCKS levels) demonstrates a significant survival benefit compared to the lower 3 quartiles; with 50% of patients surviving three years or more (Fig. 6B). Surprisingly, the importance of MARCKS was particularly evident in unmethylated MGMT tumors of the Proneural subtype. Patients with unmethylated MGMT are expected to suffer from especially poor outcomes. However, we noted that unmethylated MGMT tumors with high MARCKS expression had a much better prognosis than those with low MARCKS expression (Fig. 6C). Therefore, MARCKS expression has protective influence and prognostic value above and beyond the two most commonly used predictive factors for survival, age and MGMT status (HR=0.18, p=0.001) for those patients with GBM tumors of the Proneural subtype. However, the mode of action may be complex, as the protective influence seems to interact with MGMT status. Our results predict that individuals with unmethylated MGMT, which are considered of poorer prognostic status due to their decreased sensitivity to therapy, benefit to a much greater extent from higher MARCKS expression than their methylated counterparts. To our knowledge, this is the first prognostic factor for individuals with tumors with un-methylated MGMT that would outperform the methylated group in terms of survival.
Our results suggest that this survival benefit may be in part due to the MARCKS protein’s ability to regulate proliferation and radiation sensitivity. This likely occurs through modulation of PIP2-dependent signaling pathways such as cell division and the PI3K/Akt pathway; as well as a potential role in regulating senescence. PI3K mutations are a common occurrence in the Proneural subtype, and thus MARCKS expression would have a direct impact on signaling through this pathway in these tumors (6). PDGFR mutations are also frequent, and MARCKS expression could modulate this pathway by controlling the availability of PIP2 to cleavage by Phospholipase C (PLC) or conversion to PIP3 by PI3K. A link between MARCKS and other mutations such as IDH1 and TP53 that are characteristic of this subtype is less clear.
It should be noted that MARCKS expression did not have an impact on survival in the Classical subtype (Supplemental Table 2), for which EGFR mutations are relatively common and aberrant phospholipid signaling would be expected. A potential explanation for this might be the increased phosphorylation of MARCKS that was observed in EGFRvIII expressing cells that showed changes in invasion (22). In these cells a greater proportion of MARCKS is phosphorylated and thus unable to sequester PIP2 from participating in these signaling cascades. In the context of this cellular background, high MARCKS expression may have a reduced impact on modulating these pathways. It is also interesting that the U87 cell line enters a senescent state with over-expression of MARCKS, but U251 cells possess high levels of MARCKS and continue to proliferate. Senescence is known to be dependent on p53 and pRB DNA damage signaling pathways. U251 and U87 differ with respect to p53 status (U251 is mutant, U87 is wild type)(39), so it is also likely that the differing cell signaling backgrounds are responsible for this effect. This supports the concept that the effect of high MARCKS expression on clinical outcome would be highly dependent on the particular cell signaling background.
Our results have also identified a potential novel role for the MARCKS protein in promoting senescence and the formation of multi-nucleated giant cells. The large, flattened, multi-nucleated giant cells (central panel, Fig. 5D) found upon MARCKS over-expression are morphologically similar to the giant cell GBM subtype (37). This giant cell subtype of GBM strikes younger patients, has a better prognosis, and genetically resembles secondary disease (i.e. high incidence of TP53 mutations and a low incidence of EGFR aberrations) (9, 40, 41), similar to the recently identified Proneural subtype (6). It has been proposed that the formation of these multi-nucleated giant cells is the product of abnormal centrosome duplication resulting from the high incidence of TP53 mutations that is characteristic of the giant cell GBM subtype (40, 41). As mentioned above, the wild-type p53 status of U87 cells (39) suggests that high levels of the MARCKS protein may interfere with the function of the p53 protein leading to the formation of multi-nucleated giant cells. However, the formation of these cells may also be a result of the MARCKS protein’s role in regulating the actin cytoskeleton, as it has been previously shown that experimental sequestration of PIP2 leads to defects in cytokinesis and the formation of multi-nucleated cells (42, 43). We also noted that these giant cells were in a senescent state, indicated by positive staining for SA-β-gal (Fig. 5C and D). High levels of MARCKS have been previously associated with a quiescent state in fibroblasts (44), however, to our knowledge this is the first report that the MARCKS protein can induce a senescent state. The induction of such a state in GBM has obvious clinical implications.
To our surprise, the increases in PI3K/Akt signaling following MARCKS knockdown in U251 were not mirrored by MARCKS over-expression in U87 as we found no commensurate decrease in Akt activation. This is likely the result of changes in signaling that occur with senescence. It has been previously shown that Akt activity increases with adoption of the senescent state (45). Thus, any decreases in signaling through PI3K/Akt from MARCK over-expression may be eliminated by a compensatory up-regulation of Akt activity. These data suggest that alternative cell signaling pathways regulated by the MARCKS protein should not be excluded and warrant further investigation. For example, as a major substrate for PKC, higher levels of MARCKS might lead to decreased signaling through alternate PKC substrates. As a binding partner for Ca2+-Calmodulin, higher levels of MARCKS might lead to decreased signaling through alternate Calmodulin pathways. Thus, the impact that MARCKS expression has on these cell signaling pathways, and therefore patient survival, is likely due to a balance of the predominant cell signaling pathways that are characteristic of that particular disease background. As such, our results indicate that MARCKS is a novel biomarker and therapeutic target for a specific group of GBM patients, particularly those with Proneural tumors with un-methylated MGMT promoters. Efforts are currently underway to identify therapies that increase MARCKS expression in the context of this cellular background, as this may provide a significant survival benefit for these patients.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Glioblastoma multiforme (GBM) is the most common and deadly primary brain malignancy necessitating improved understanding of GBM biology. In this study, we explore the role of Myristoylated Alanine Rich C-Kinase Substrate (MARCKS) in the context of GBM. We have discovered that the MARCKS protein regulates GBM growth as well as response to radiation therapy through its effects on proliferation, senescence, and DNA repair based on our studies in cell culture and in patient-derived xenograft tumors implanted in mice. Importantly, our analysis of clinical patient data demonstrates that MARCKS is an independent predictor for outcome in GBM patients. Indeed, high MARCKS levels promoted improved outcomes, which was consistent across our model systems. Our results suggest that MARCKS may be a biomarker for prognosis as well as a potential target for therapy in GBM.
Acknowledgments
Didier Trono and Bob Weinberg for providing the psPAX2 and pCMV-VSV-G plasmids, respectively, via Addgene. Plasmid constructs were sequenced at the UAB Center for AIDS Research DNA Sequencing Core, supported by Core grant P30-AI-27767. D54 cells were obtained from Harald Sontheimer (UAB). U87 and U251 were obtained from Tika Benveniste (UAB). U373, U118, and D32 were obtained from Donald Buchsbaum (UAB); American Society for Radiation Oncology Junior Faculty Research Training Award (to C.D.W.); University of Alabama Health Services Foundation General Endowment Fund Scholar Award (to C.D.W.); IMPACT Award from the Department of Radiation Oncology, University of Alabama at Birmingham Comprehensive Cancer Center, University of Alabama at Birmingham School of Medicine (to E.S.Y.). This project has been funded in whole or in part with Federal Funds from the National Cancer Institute, National Institutes of Health, in the form of grant P50 CA 097247 (to G.Y.G.) and the associated UAB Brain SPORE pilot project program (to C.D.W.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
CONTRIBUTORS
CDW, JSJ, and JCA designed the study. CDW, JSJ, JCA, CWD, TM, SN, TM, ESY, and ROM did the investigation. Additional assistance was provided by PHH, ACW, TDR, SC, TRB, and GYG. CDW, JSJ, JCA, JAB, GYG, CWD, ROM, TM, and ESY interpreted the data, and CDW, JSJ, JCA, CWD, and ESY wrote the report with the help of all authors.
CONFLICTS OF INTEREST
We declare that there we have no conflicts of interest pertaining to the contents of this manuscript. Nevertheless, occasional honoraria from Bristol-Myers Squibb, ImClone Systems, Inc. and Eli Lilly (JAB) and Varian (CDW) are disclosed.
References
- 1.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin United States. 2010:166–93. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 4.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 5.Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, et al. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364:627–37. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fults D, Pedone C. Deletion mapping of the long arm of chromosome 10 in glioblastoma multiforme. Genes Chromosomes Cancer. 1993;7:173–7. doi: 10.1002/gcc.2870070311. [DOI] [PubMed] [Google Scholar]
- 8.Wang SI, Puc J, Li J, Bruce JN, Cairns P, Sidransky D, et al. Somatic mutations of PTEN in glioblastoma multiforme. Cancer Res. 1997;57:4183–6. [PubMed] [Google Scholar]
- 9.Kato H, Kato S, Kumabe T, Sonoda Y, Yoshimoto T, Han SY, et al. Functional evaluation of p53 and PTEN gene mutations in gliomas. Clin Cancer Res. 2000;6:3937–43. [PubMed] [Google Scholar]
- 10.Raffel C, Frederick L, O’Fallon JR, Atherton-Skaff P, Perry A, Jenkins RB, et al. Analysis of oncogene and tumor suppressor gene alterations in pediatric malignant astrocytomas reveals reduced survival for patients with PTEN mutations. Clin Cancer Res. 1999;5:4085–90. [PubMed] [Google Scholar]
- 11.Ermoian RP, Furniss CS, Lamborn KR, Basila D, Berger MS, Gottschalk AR, et al. Dysregulation of PTEN and protein kinase B is associated with glioma histology and patient survival. Clin Cancer Res. 2002;8:1100–6. [PubMed] [Google Scholar]
- 12.Haas-Kogan D, Shalev N, Wong M, Mills G, Yount G, Stokoe D. Protein kinase B (PKB/Akt) activity is elevated in glioblastoma cells due to mutation of the tumor suppressor PTEN/MMAC. Curr Biol. 1998;8:1195–8. doi: 10.1016/s0960-9822(07)00493-9. [DOI] [PubMed] [Google Scholar]
- 13.Zhou YH, Tan F, Hess KR, Yung WK. The expression of PAX6, PTEN, vascular endothelial growth factor, and epidermal growth factor receptor in gliomas: relationship to tumor grade and survival. Clin Cancer Res. 2003;9:3369–75. [PubMed] [Google Scholar]
- 14.Albert KA, Nairn AC, Greengard P. The 87-kDa protein, a major specific substrate for protein kinase C: purification from bovine brain and characterization. Proc Natl Acad Sci U S A. 1987;84:7046–50. doi: 10.1073/pnas.84.20.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartwig JH, Thelen M, Rosen A, Janmey PA, Nairn AC, Aderem A. MARCKS is an actin filament crosslinking protein regulated by protein kinase C and calcium-calmodulin. Nature. 1992;356:618–22. doi: 10.1038/356618a0. [DOI] [PubMed] [Google Scholar]
- 16.Yarmola EG, Edison AS, Lenox RH, Bubb MR. Actin filament cross-linking by MARCKS: characterization of two actin-binding sites within the phosphorylation site domain. J Biol Chem. 2001;276:22351–8. doi: 10.1074/jbc.M101457200. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Gambhir A, Hangyás-Mihályné G, Murray D, Golebiewska U, McLaughlin S. Lateral sequestration of phosphatidylinositol 4,5-bisphosphate by the basic effector domain of myristoylated alanine-rich C kinase substrate is due to nonspecific electrostatic interactions. J Biol Chem. 2002;277:34401–12. doi: 10.1074/jbc.M203954200. [DOI] [PubMed] [Google Scholar]
- 18.Thelen M, Rosen A, Nairn AC, Aderem A. Regulation by phosphorylation of reversible association of a myristoylated protein kinase C substrate with the plasma membrane. Nature. 1991;351:320–2. doi: 10.1038/351320a0. [DOI] [PubMed] [Google Scholar]
- 19.Gatlin JC, Estrada-Bernal A, Sanford SD, Pfenninger KH. Myristoylated, alanine-rich C-kinase substrate phosphorylation regulates growth cone adhesion and pathfinding. Mol Biol Cell. 2006;17:5115–30. doi: 10.1091/mbc.E05-12-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel S, Kloor M, Singh S, Gdynia G, Roth W, von Knebel Doeberitz M, et al. Coding microsatellite instability analysis in microsatellite unstable small intestinal adenocarcinomas identifies MARCKS as a common target of inactivation. Mol Carcinog. 2010;49:175–82. doi: 10.1002/mc.20587. [DOI] [PubMed] [Google Scholar]
- 21.Brooks G, Brooks SF, Goss MW. MARCKS functions as a novel growth suppressor in cells of melanocyte origin. Carcinogenesis. 1996;17:683–9. doi: 10.1093/carcin/17.4.683. [DOI] [PubMed] [Google Scholar]
- 22.Micallef J, Taccone M, Mukherjee J, Croul S, Busby J, Moran MF, et al. Epidermal growth factor receptor variant III-induced glioma invasion is mediated through myristoylated alanine-rich protein kinase C substrate overexpression. Cancer Res. 2009;69:7548–56. doi: 10.1158/0008-5472.CAN-08-4783. [DOI] [PubMed] [Google Scholar]
- 23.Carlson BL, Grogan PT, Mladek AC, Schroeder MA, Kitange GJ, Decker PA, et al. Radiosensitizing effects of temozolomide observed in vivo only in a subset of O6-methylguanine-DNA methyltransferase methylated glioblastoma multiforme xenografts. Int J Radiat Oncol Biol Phys. 2009;75:212–9. doi: 10.1016/j.ijrobp.2009.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12:2264–71. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 25.Sarkaria JN, Yang L, Grogan PT, Kitange GJ, Carlson BL, Schroeder MA, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6:1167–74. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 26.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Witt Hamer PC, Van Tilborg AA, Eijk PP, Sminia P, Troost D, Van Noorden CJ, et al. The genomic profile of human malignant glioma is altered early in primary cell culture and preserved in spheroids. Oncogene. 2008;27:2091–6. doi: 10.1038/sj.onc.1210850. [DOI] [PubMed] [Google Scholar]
- 28.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol. 2005;7:164–76. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro Oncol. 2005;7:164–76. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao GD, Jiang Z, Fernandes AM, Gupta AK, Maity A. Inhibition of phosphatidylinositol-3-OH kinase/Akt signaling impairs DNA repair in glioblastoma cells following ionizing radiation. J Biol Chem. 2007;282:21206–12. doi: 10.1074/jbc.M703042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao R, Cooper GM. Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science. 1995;267:2003–6. doi: 10.1126/science.7701324. [DOI] [PubMed] [Google Scholar]
- 32.Mukherjee B, McEllin B, Camacho CV, Tomimatsu N, Sirasanagandala S, Nannepaga S, et al. EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69:4252–9. doi: 10.1158/0008-5472.CAN-08-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–34. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 35.Sugrue MM, Shin DY, Lee SW, Aaronson SA. Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53. Proc Natl Acad Sci U S A. 1997;94:9648–53. doi: 10.1073/pnas.94.18.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 37.Müller W, Slowik F, Firsching R, Afra D, Sanker P. Contribution to the problem of giant cell astrocytomas. Neurosurg Rev. 1987;10:213–9. doi: 10.1007/BF01782050. [DOI] [PubMed] [Google Scholar]
- 38.Maeda K, Mizuno M, Wakabayashi T, Takasu S, Nagasaka T, Inagaki M, et al. Morphological assessment of the development of multinucleated giant cells in glioma by using mitosis-specific phosphorylated antibodies. J Neurosurg. 2003;98:854–9. doi: 10.3171/jns.2003.98.4.0854. [DOI] [PubMed] [Google Scholar]
- 39.Ishii N, Maier D, Merlo A, Tada M, Sawamura Y, Diserens AC, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9:469–79. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer-Puttlitz B, Hayashi Y, Waha A, Rollbrocker B, Boström J, Wiestler OD, et al. Molecular genetic analysis of giant cell glioblastomas. Am J Pathol. 1997;151:853–7. [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez R, Roggendorf W, Baretton G, Klein R, Toedt G, Lichter P, et al. Cytogenetic and molecular genetic analyses of giant cell glioblastoma multiforme reveal distinct profiles in giant cell and non-giant cell subpopulations. Cancer Genet Cytogenet. 2007;175:26–34. doi: 10.1016/j.cancergencyto.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 42.Logan MR, Mandato CA. Regulation of the actin cytoskeleton by PIP2 in cytokinesis. Biol Cell. 2006;98:377–88. doi: 10.1042/BC20050081. [DOI] [PubMed] [Google Scholar]
- 43.Emoto K, Inadome H, Kanaho Y, Narumiya S, Umeda M. Local change in phospholipid composition at the cleavage furrow is essential for completion of cytokinesis. J Biol Chem. 2005;280:37901–7. doi: 10.1074/jbc.M504282200. [DOI] [PubMed] [Google Scholar]
- 44.Herget T, Brooks SF, Broad S, Rozengurt E. Expression of the major protein kinase C substrate, the acidic 80-kilodalton myristoylated alanine-rich C kinase substrate, increases sharply when Swiss 3T3 cells move out of cycle and enter G0. Proc Natl Acad Sci U S A. 1993;90:2945–9. doi: 10.1073/pnas.90.7.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyauchi H, Minamino T, Tateno K, Kunieda T, Toko H, Komuro I. Akt negatively regulates the in vitro lifespan of human endothelial cells via a p53/p21-dependent pathway. EMBO J. 2004;23:212–20. doi: 10.1038/sj.emboj.7600045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7:157–67. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.