Abstract
Rationale
Cardiomyocytes in adult mammalian hearts are terminally differentiated cells that have exited from the cell cycle and lost most of their proliferative capacity. Death of mature cardiomyocytes in pathological cardiac conditions and the lack of regeneration capacity of adult hearts are primary causes of heart failure and mortality. However, how cardiomyocyte proliferation in postnatal and adult hearts becomes suppressed remains largely unknown. The miR-17-92 cluster was initially identified as a human oncogene that promotes cell proliferation. However, its role in the heart remains unknown.
Objective
To test the hypothesis that miR-17-92 participates in the regulation of cardiomyocyte proliferation in postnatal and adult hearts.
Methods and Results
We deleted miR-17-92 cluster from embryonic and postnatal mouse hearts and we demonstrated that miR-17-92 is required for cardiomyocyte proliferation in the heart. Transgenic overexpression of miR-17-92 in cardiomyocytes is sufficient to induce cardiomyocyte proliferation in embryonic, postnatal and adult hearts. Moreover, overexpression of miR-17-92 in adult cardiomyocytes protects the heart from myocardial infarction-induced injury. Similarly, we found that members of miR-17-92 cluster, miR-19 in particular, are required for and sufficient to induce cardiomyocyte proliferation in vitro. We identified PTEN, a tumor suppressor, as a miR-17-92 target to mediate the function of miR-17-92 in cardiomyocyte proliferation.
Conclusions
Our studies therefore identify miR-17-92 as a critical regulator of cardiomyocyte proliferation and suggest this cluster of miRNAs could become therapeutic targets for cardiac repair and heart regeneration.
Keywords: miR-17-92, cardiomyocyte proliferation, myocardial infarction, cell cycle, PTEN, heart disease
INTRODUCTION
The adult mammalian heart has limited capability to regenerate itself after the lost of mature cardiomyocytes due to a variety of pathological conditions such as myocardial infarction. It is generally conceived that post-mitotic cardiomyocytes in adult hearts exit from the cell cycle and stop cell proliferation1, 2. However, the hearts of adult zebrafish can undergo cardiac regeneration without scar formation after resection of ventricle, primarily through cardiomyocyte proliferation3–6. Intriguingly, a recent report demonstrated that surgical resection of the ventricular apex in newborn mice stimulates the proliferation of cardiomyocytes and repairs the damaged heart but the mouse heart loses this regenerative potential within 7 days of its postnatal life and it is not clear how the regenerative potential is lost in the adult hearts7. To date, the molecular mechanism and regulatory pathways that control adult cardiomyocyte proliferation and cardiac regeneration remain largely unknown.
miRNAs are a class of ~22 nt non-coding RNAs that regulate the expression of protein-coding genes post-transcriptionally. More than 1,000 human miRNAs have been identified, however, the biological functions of many of them remain unknown. miR-17-92 cluster was initially reported as a human oncogene and named oncomir18, 9. Numerous reports have documented the expression of miR-17-92 in variety of human cancers and disorders10–12. Genetic studies demonstrated that miR-17-92 is indispensible for mouse development and cell proliferation and miR-17-92 mutant mice die postnatally, displaying defects in lung, hearts and others13–15. We hypothesized that miR-17-92 regulates the proliferation of cardiomyocytes. In this study, we tissue-specifically overexpress or delete the miR-17-92 cluster in cardiomyocytes in transgenic or knockout mice and we found that miR-17-92 participates in the regulation of cardiomyocyte proliferation in embryonic, postnatal and adult hearts.
METHODS
Cell culture, quantitative RT-PCR analyses, Western blot analyses, histology and immunochemistry were performed according to routine protocols. Details of materials and methods are provided in SI Appendix.
Statistics
Values are reported as means ± SEM unless indicated otherwise. The 2-tailed Mann-Whitney U test was used for comparing 2 means (Prism, GraphPad). Values of P<0.05 were considered statistically significant.
RESULTS
miR-17-92 is required for cardiomyocyte proliferation in embryonic and postnatal hearts
We crossed the miR-17-92flox/flox mice15 with Nkx2.5-Cre mice, in which the expression of Crerecombinase is under the control of the cardiac-specific Nkx2.5 gene, to delete miR-17-92 in embryonic, postnatal and adult hearts (Online Figure Ia). We found that cardiac-specific miR-17-92 mutant mice (named miR-17-92-cKO) are slightly under-representative at weaning age (20.3%), suggesting that cardiac-specific deletion of miR-17-92 resulted in partial embryonic lethality (Online Figure Ib). We confirmed that the expression of miR-17-92 miRNAs was significantly reduced in the hearts of mutant mice (Online Figure II).
The hearts of postnatal miR-17-92-cKO mice appear to be smaller than that of their littermate controls (Fig. 1a). We examined the proliferation of cardiomyocytes in miR-17-92-cKO hearts, using immunostaining for phosphorylated histone H3 (pH3) which marks mitosis, and we found that there is less proliferating cardiomyocytes in postnatal hearts of miR-17-92-cKO mice (Fig. 1b, c; Online Figure III). Quantitative analyses confirmed substantial decrease in total numbers of pH3 positive cardiomyocytes in miR-17-92-cKO hearts (Fig. 1d). We have also observed a decrease in cardiomyocyte proliferation in miR-17-92 heterozygous hearts (Fig. 1d). We asked whether loss of miR-17-92 affected the survival of cardiomyocytes. We performed TUNEL assay to measure apoptosis and we observed no change in TUNEL signals in postnatal hearts of miR-17-92-cKO mice when compared with controls (Online Figure IV).
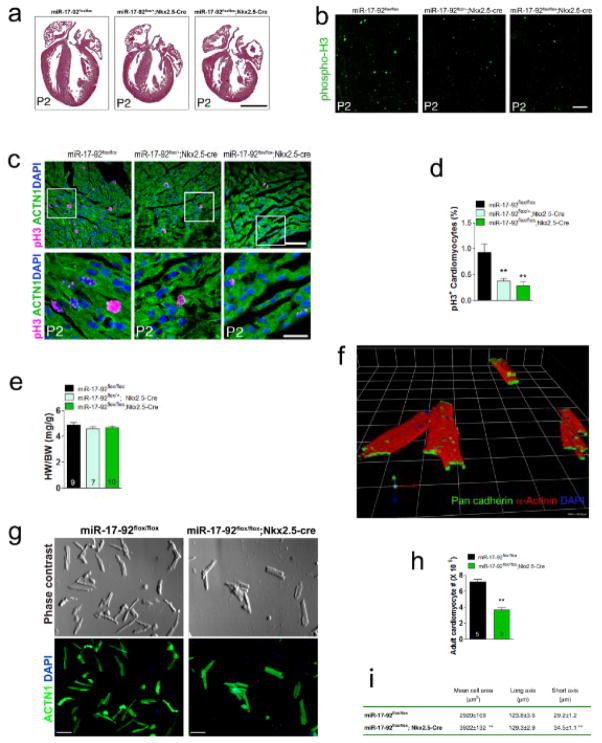
Figure 1. miR-17-92 is required for cardiomyocyte proliferation in embryonic, postnatal and adult hearts.
(A)Haematoxylin and Eosin (H&E) staining of sagittal sections of hearts from 2 days old wild type, heterozygote and mutant miR-17-92-KO mice. Bar = 1 mm.(B) Immunohistochemistry of phosphorylated histone H3 (phospho-H3, pH3) on sagittal sections of hearts from 2 days old wild type, heterozygote and mutant miR-17-92-KO mice. Bar = 100 μm. (C) Immunohistochemistry of sagittal sections of hearts from 2 days old wild type, heterozygote and mutant miR-17-92-KO mice. White boxes are enlarged in lower panels. pH3 labels proliferating cells; α-actinin (ACTN1) marks cardiomyocytes; DAPI labels nuclei. Bars = 50 μm in upper panel and 20 μm in lower panel. (D) Quantification of pH3 positive cardiomyocytes in wild type, heterozygote and mutant miR-17-92-KO hearts from 2 days old mice. **P<0.01 between genetic groups. (N=4~5 each group). (E)Heart weight (HW) to body weight (BW) ratios of 1 year old wild type, heterozygote and mutant miR-17-92-KO mice. N of each genotype group listed. (F)3D reconstruction from confocal images showing intact isolated adult mouse cardiomyocytes. Pan-cadherin stains desmonsomes to demonstrate intact cardiomyocytes (green). α-actinin marks intact sarcomeres (red). DAPI labels intact nuclei. 1 unit = 42.45 μm. (G)Morphology of freshly isolated adult cardiomyocytes from hearts of miR-17-92-KO and control mice. ACTN1 marks rod shaped cardiomyocytes (lower panel). Bars = 250 μm. (H)Quantification of total isolated adult cardiomyocytes from hearts of miR-17-92-KO and control mice. N of each genotype group is listed.(I)Quantitative measurement of the size of freshly isolated adult cardiomyocytes from hearts of miR-17-92-KO and control mice. One hundred individual cells from three different hearts were analyzed per group. ** P< 0.01 between genetic groups.
Most miR-17-92-cKO mice survived to adulthood and we next investigated miR-17-92 loss-of-function phenotype in adult hearts. There was no obvious difference in the gross cardiac morphology of the miR-17-92-cKO and their littermate control mice (Online Figure Va). The heart weight (HW) to body weight (BW) ratio was not altered in miR-17-92-cKO mice (Fig. 1e). However, there appeared to exhibit a compensatory cardiomyocyte hypertrophy in these hearts (Online Figure Vb). We used Langendorf perfusion method to isolated cardiomyocytes from adult hearts. Freshly isolated adult cardiomyocytes were stained with α-actinin to label cardiomyocytes; pan-cadherin to recognize desmosomes. Recostructed confocal images clearly indicate that the majority of isolated cardiomyocytes were intact (Fig. 1f). Quantitatively analysis indicated a substantial decrease in the total numbers of cardiomyocytes in the hearts of miR-17-92-cKO mice (Fig. 1g, h). Quantitative measurement of the size of isolated adult cardiomyocytes showed that the size of cardiomyocytes was increased in the heart of miR-17-92-cKO mice (Fig. 1i), consistent with the idea that increased size of cardiomyocytes compensates for the reduction of total numbers of cardiomyocytes in mutant hearts. TUNEL assays detected no change in apoptosis in the heart of 8 month-old miR-17-92-cKO mice when compared with controls (Online Figure IV). We examined the cardiac function, using echocardiography, and we found decreased in ventricle wall thickness, increased in ventricle systolic diameter and decreased in cardiac function in miR-17-92-cKO mice when compared with their littermate controls (Online Figure VI and Online Table I). Together, these results indicate that miR-17-92 is required for cardiomyocyte proliferation and normal cardiac function in postnatal and adult hearts.
miR-17-92 is sufficient to induce cardiomyocyte proliferation in embryonic and postnatal hearts
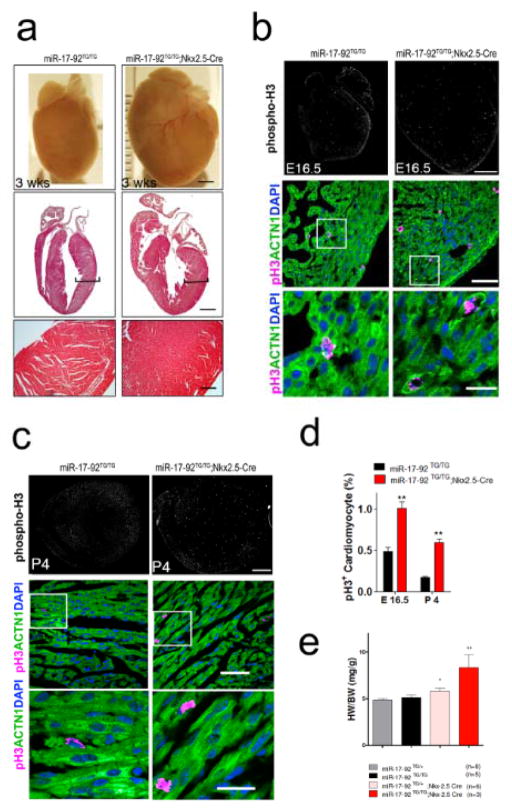
Having demonstrated that miR-17-92 is required for cardiomyocyte proliferation in embryonic and postnatal hearts, we next tested whether overexpression of miR-17-92 was sufficient to induce cardiomyocyte proliferation. We generated cardiac-specific conditional transgenic mice to overexpress miR-17-92 in the heart. We first bred the “floxed miR-17-92 knock-in allele”, in which a loxP-flanked Neo-STOP cassette was inserted upstream of the bicistronic human miR-17-92 cluster and knocked into the Rosa26 locus (named miR-17-92-KI)16, with the Nkx2.5-Cre mice to achieve the overexpression of miR-17-92 in embryonic and postnatal hearts (Online Figure VII). The hearts of cardiac-specific miR-17-92 transgenic mice (named miR-17-92-TGnkx2.5) were dramatically enlarged (Fig. 2a). Histological section revealed that the ventricle wall was substantially thickened in the hearts of miR-17-92-TGnkx2.5 mice (Fig. 2a). The heart was hyperplasia and trabeculae were highly condensed and packed (Fig. 2a, lower panels). There was no evidence of cardiomyocyte hypertrophy and the size of cardiomyocytes is comparable between miR-17-92-TGnkx2.5 and control mice. The increase of cardiomyocyte numbers resulted from an increase in cardiomyocyte proliferation. We used phosphorylated histone H3 (pH3) to mark proliferating cardiomyocytes and found that overexpression of miR-17-92 is sufficient to enhance cardiomyocyte proliferation in both embryonic and postnatal hearts (Fig. 2b, c, d; Online Figure VIII). Concordantly, there was a significant increase in heart/body weight ratios in miR-17-92-TGnkx2.5 mice (Fig. 2e).
Figure 2. miR-17-92 induces cardiomyocyte proliferation in embryonic and postnatal hearts.
(A)Gross morphology of hearts of 3 weeks old control and miR-17-92-TGnkx2.5 mice (upper panel, bar = 1 mm). H&E staining of sagittal sections of 3 week old control and miR-17-92-TGnkx2.5 mice (middle panel, bar = 1 mm); higher magnification of ventricle myocardium (lower panel, bar = 250 μm). (B) Immunohistochemistry of phospho-H3 on sagittal sections of embryonic 16.5 (E16.5) control and miR-17-92-TGnkx2.5 hearts (upper panel, bar = 500 μm); Immunohistochemistry of pH3 on sagittal sections of E16.5 wild type and miR-17-92-TGnkx2.5 hearts, ACTN1 marks cardiomyocytes; DAPI labels nuclei (middle panel, bar = 50 μm); white boxes are enlarged in lower panels (lower panel, bar = 20 μm). (C)Immunohistochemistry of phospho-H3 on sagittal sections of postnatal day 4 (P4) control and miR-17-92-TGnkx2.5 hearts (upper panel, bar = 500 μm); Immunohistochemistry of pH3 on sagittal sections of P4 control and miR-17-92-TGnkx2.5 hearts, ACTN1 marks cardiomyocytes; DAPI labels nuclei (middle panel, bar = 50 μm); white boxes are enlarged in lower panels (lower panel, bar = 20 μm). (D)Quantification of percentages of pH3+ cardiomyocytes in E16.5 and P4 control and miR-17-92-TGnkx2.5 hearts. N = 3 for each group. (E)The Heart Weight/Body Weight (HW/BW) ratio of 3-week-old miR-17-92 transgenic mice (miR-17-92TG/TG; Nkx2-5Cre/+) and their control littermates were shown. N of each genotype was indicated. *: P<0.05; **: P<0.01.
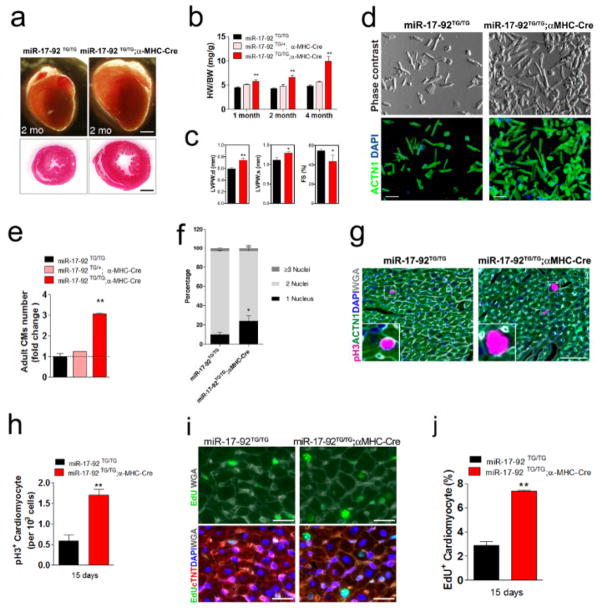
Next, we generated transgenic mice to overexpress miR-17-92 predominantly in postnatal and adult cardiomyocytes. We bred miR-17-92-KI mice with alpha-MHC-Cre transgenic mice, in which the expression of Crerecombinase is directed by the cardiac-specific alpha-MHC (Myh6) promoter, to generate cardiac-specific miR-17-92 transgenic mice (named miR-17-92-TGMHC). We observed about 2–5 folds higher of miR-17-92 expression in transgenic hearts (Online Figure IX). Most miR-17-92-TGMHC mice survived to adulthood without overt abnormality (Online Figure X). The hearts of the miR-17-92-TGMHC mice were substantially enlarged (Fig. 3a), and the heart/body weight ratio was significantly increased in these mice (Fig. 3b). Cardiac-specific overexpression of miR-17-92 increased the thickness of ventricle wall in transgenic mice (Fig. 3c; Online Table II). Quantitative measurement of cardiomyocyte cell size and cell number of the miR-17-92-TGMHC hearts demonstrated a substantial increase in the cell number in the heart of miR-17-92-TGMHC mice, whereas the size of cardiomyocyte was not changed (Online Figure XI). We isolated cardiomyocytes from adult hearts using Langendorf isolation method and determined that there was substantial increase in total cardiomyocyte numbers in miR-17-92-TGMHC hearts (Fig. 3d, e). Intriguingly, we found that there is an increase in total numbers of mono-nucleus cardiomyocytes and decrease in bi-nuclei cardiomyocytes in miR-17-92-TGMHC hearts (Fig. 3f; Online Figure XII). Consistent with the increase in the cell numbers, the proliferation of cardiomyocytes, marked by phosphorylated histone H3 (pH3), was enhanced in miR-17-92-TGMHC hearts (Fig. 3g, h; Online Figure XIII). Increased cell proliferation in miR-17-92-TGMHC hearts was further confirmed by 5-ethynyl-2′-deoxyuridine (EdU) incorporation and quantification (Fig. 3i, j). Finally, we stained cardiomyocytes with aurora B to detect cytokinesis. We observed an increase in aurora B signals in postnatal days 5 and 15 hearts of the miR-17-92-TGMHC mice (Online Figure XIV). Z-stack confocal images confirmed that positive aurora B signals are located in cardiomyocytes (Online Figure XIV).
Figure 3. miR-17-92 induces cardiomyocyte proliferation in postnatal and adult hearts.
(A)Gross morphology of hearts of 2 months old control and miR-17-92-TGMHC mice (upper panel). H&E staining of transverse sections of 2 months old control and miR-17-92-TGMHC hearts (lower panels). Bars = 1 mm. (B)Heart weight (HW) to body weight (BW) ratios of 1-, 2-, and 4-months old wild type, heterozygote and homozygote miR-17-92-TGMHC mice. N = 5 for each group.(C)Echocardiography analyses of cardiac function of 40 day-old miR-17-92-TGMHC mice and their control littermates. N = 3 for each group. (D)Morphology of freshly isolated adult cardiomyocytes from hearts of miR-17-92-TGMHC and control mice. ACTN1 marks rod shaped cardiomyocytes (lower panel). Bars = 250 μm. (E)Quantification of total isolated adult cardiomyocytes from 2 months old hearts of miR-17-92-TGMHC and control mice. N=3 for each genetic group. (F)Distribution of isolated adult cardiomyocytes with one (1), two (2) or three (3) and more nuclei from 2 months old hearts of miR-17-92-TGMHC and control mice. (G)Immunohistochemistry of pH3 on transverse sections of 15 days old control and miR-17-92-TGMHC hearts. ACTN1 marks cardiomyocytes and DAPI labels nuclei. Wheat germ agglutinin (WGA) staining marks cell surface (white). White boxes are enlarged in insets. (H)Quantification of percentages of pH3+ cardiomyocytes of 15 days old control and miR-17-92-TGMHC hearts. N=4 for each genetic group.(I)Immunohistochemistry of EdU incorporation on transverse sections of 15 days old control and miR-17-92-TGMHC hearts. Cardiac troponin T (cTNT) marks cardiomyocytes and DAPI labels nuclei. Wheat germ agglutinin (WGA) staining marks cell surface (white). (J)Quantification of percentages of EdU positive cardiomyocytes of 15 days old control and miR-17-92-TGMHC hearts. N=3 for each genetic group.
miR-17-92 induces cardiomyocyte proliferation in adult hearts and in response to injury
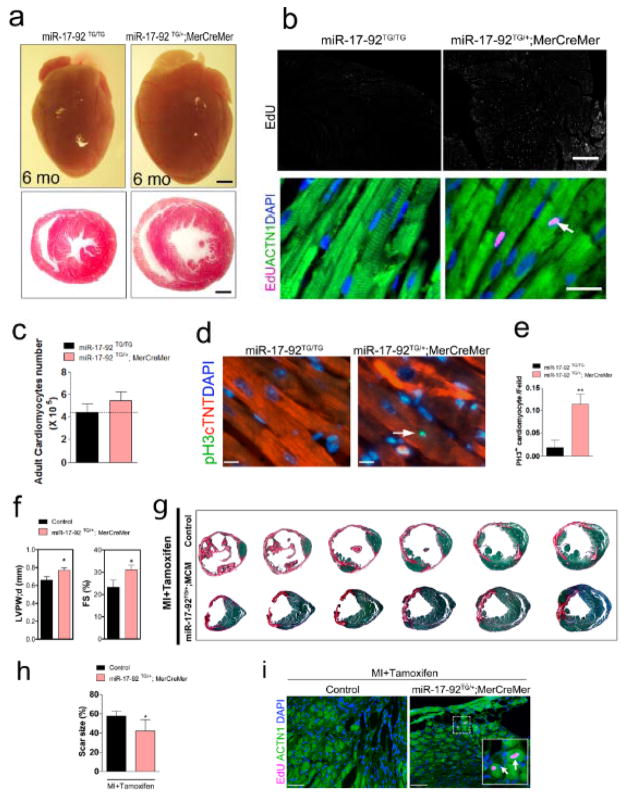
The above data indicate that overexpression of miR-17-92 was sufficient to induce cardiomyocyte proliferation in embryonic, neonatal and adult hearts. To further determine the function of miR-17-92 in the proliferation of postmitotic cardiomyocytes in adult hearts more definitely, we utilized an inducible system to overexpress miR-17-92 in cardiomyocytes of four-month-old mice. We bred the miR-17-92-KI mice with the alpha-MHC-MerCreMer transgenic mice in which the Myh6 promoter directs the expression of a tamoxifen-inducible Crerecombinase in cardiomyocytes. We induced miR-17-92 overexpression in adult cardiomyocytes by activating tamoxifen-inducible Crerecombinase (see Materials and Methods). We confirmed cardiac-specific overexpression of members of the miR-17-92 cluster in the hearts of miR-17-92 transgenic mice (named miR-17-92-TGMerCreMer) after tamoxifen administration (Online Figure XV). The hearts of miR-17-92-TGMerCreMer mice were substantially larger than that of the littermate controls after miR-17-92 overexpression (Fig. 4a). Tissue sections revealed an increase in wall thickness and left ventricle dimension (Fig. 4a).
Figure 4. Control of cardiomyocyte proliferation by miR-17-92 in adult heart in response to myocardial infarction.
(A)Gross morphology of hearts of 6 months old miR-17-92-TGMerCreMer and control mice after tamoxifen administration (upper panels). H&E staining of transverse sections of control and miR-17-92-TGMerCreMer hearts (lower panels). Bars = 1 mm. (B)Immunohistochemistry of EdU incorporation on sagittal sections of 6 months old miR-17-92-TGMerCreMer and control mice after tamoxifen administration (upper panels, bar = 500 μm); Immunohistochemistry of EdU on sagittal sections of 6 months old miR-17-92-TGMerCreMer and control mice after tamoxifen administration. The arrow points to EdU positive signal in cardiomyocytes. ACTN1 marks cardiomyocytes; DAPI labels nuclei (lower panels, bar = 20 μm). (C)Quantification of total isolated adult cardiomyocytes from hearts of 6 months old miR-17-92-TGMerCreMer and control mice after tamoxifen administration. N = 3 for each group.(D)Immunohistochemistry of pH3 on sagittal sections of 6 months old miR-17-92-TGMerCreMer and control hearts after tamoxifen administration. The arrow points to pH3 positive signal in cardiomyocytes. cTNT marks cardiomyocytes; DAPI labels nuclei. Bars = 11 μm.(E)Quantification of percentages of pH3+ cardiomyocytes in 6 months old miR-17-92-TGMerCreMer and control hearts after tamoxifen administration. N = 3 for control group. N = 6 for miR-17-92-TGMerCreMer group. ** P< 0.01 between genetic groups. (F)Echocardiography analyses of cardiac function of miR-17-92TG/+; MerCreMer mice and their control littermates after MI and tamoxifen administration. N = 10 for miR-17-92TG/+; MerCreMer group and 8 for control group. (G)Representative images of series of transverse sections of 6 months old miR-17-92TG/+; MerCreMer and control mice after MI and tamoxifen administration. Sirius red/fast green collagen staining marks myocardium (green) and scar (red). Bars = 1 mm.(H)Quantification of the size of scar in the hearts of miR-17-92TG/+; MerCreMer (N=4) and control mice (N=5) after MI and tamoxifen administration. * P< 0.05 between genetic groups.(I)Immunohistochemistry of EdU on sagittal sections of 6 months old miR-17-92TG/+; MerCreMer and control mice after tamoxifen administration. White boxes are enlarged in insets and arrows point to EdU positive signal. ACTN1 marks cardiomyocytes; DAPI labels nuclei. Bars = 50 μm).
We examined whether overexpression of miR-17-92 in adult cardiomyocytes could induce cell proliferation. Using EdU incorporation assay, we found marked increase in the EdU incorporation in the cardiomyocytes of six months old miR-17-92-TGMerCreMer hearts (Fig. 4b, Online Figure XVI). We isolated cardiomyocytes from the hearts of miR-17-92-TGMerCreMer and control mice to determine the total numbers of cardiomyocytes and we found that overexpression of miR-17-92 substantially increased total cardiomyocyte number in adult hearts (Fig. 4c). Quantitative measurement of the size of isolated adult cardiomyocytes showed that the size of cardiomyocytes was reduced in the heart of miR-17-92-TGMerCreMer mice (Online Figure XVII). To further confirm the cardiomyocyte proliferation in the miR-17-92-TGMerCreMer hearts, we used phosphorylated histone H3 (pH3) to mark proliferating cardiomyocytes and found that overexpression of miR-17-92 is sufficient to enhance cardiomyocyte proliferation in adult hearts (Fig. 4d, e).
To test whether this cluster of miRNAs is involved in the regulation of cardiomyocyte proliferation and cardiac repair in response to injury, we introduced myocardial infarction (MI) by coronary artery occlusion (Online Figure XVIII). MI results in massive cardiomyocyte death, cardiac hypertrophy, fibrosis and cardiac remodeling. We found that overexpression of miR-17-92 in adult cardiomyocytes modestly protected the heart from MI-induced injury (Fig. 4f). There is an increase of cardiac function, as measured by echocardiography and documented as fractional shortening (FS), in miR-17-92-TGMerCreMer mice when compared with controls (Fig. 4f, g; Online Table III). Quantification confirmed the decrease in the size of scar in the hearts of miR-17-92-TGMerCreMer mice after MI (Fig. 4h). Using EdU incorporation assay, we found marked increase in the EdU incorporation in the cardiomyocytes of border zone of miR-17-92-TGMerCreMer hearts (Fig. 4i). We asked whether apoptosis is affected in miR-17-92 transgenic hearts. TUNEL assays showed no difference in apoptosis between miR-17-92 transgenic and control hearts in response to MI injury (Online Figure XIX). Similarly, we treated both miR-17-92 transgenic and control mice with doxorubicin, a cancer drug which cause heart failure as side-effect, to induce stress. We found that overexpression of miR-17-92 modestly induce cardiomyocyte proliferation upon doxorubicin treatment.
miR-17-92 is sufficient to induce neonatal cardiomyocyte proliferation in vitro
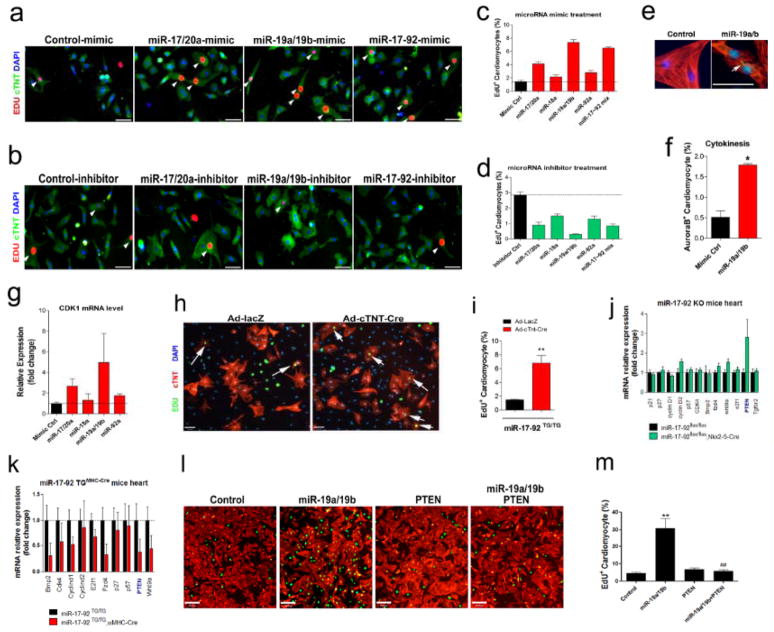
To test whether members of this cluster of miRNAs play a similar role in cardiomyocyte proliferation in vitro, we transfected neonatal rat cardiomyocytes with mimics or inhibitors of each member of the miR-17-92 cluster and assayed cardiomyocyte proliferation17, 18. We used cardiomyocytes isolated from postnatal day 1 (P1) hearts, in which cardiomyocytes are still undergoing active proliferation7. After transfected with miR-17-92 mimics (Fig. 5a) or inhibitors (Fig. 5b), the cell culture was then incubated with EdU to label DNA synthesis and cell proliferation. Indeed, we found that miR-17-92 mimics, especially miR-19a/b family, potently induce cardiomyocyte proliferation (Fig. 5a, c). Conversely, inhibition of members of the miR-17-92 cluster, in particular miR-19a/b, substantially reduced cardiomyocyte proliferation, evidenced by the decrease of EdU signal and the reduction of total cardiomyocyte numbers when compared with controls (Fig. 5b,d). We applied aurora B staining to detect cytokinesis in these cells (Fig. 5e). Quantitative analysis confirmed a significant increase in aurora B signal after miR-19a/19b treatment (Fig. 5f). Furthermore, we treated postnatal day 4 (P4) cardiomyocytes, in which cell proliferation starts to diminish, with miR-17-92 mimics and we found that miR-17-92 mimics, miR-19a/b in particular, significantly induced EdU incorporation (Online Figure XXa). This observation is further confirmed by quantitative analyses (Online Figure XXb). We examined the expression of cyclin-dependent kinase 1 (CDK1), a highly conserved serine/threonine kinase involved in cell cycle progression19, in miR-17-92 mimic treated cardiomyocytes. Consistent with the view that miR-17-92 induced cardiomyocyte proliferation, we found that miR-17-92 induced the expression of CDK1 in cardiomyocytes (Fig. 5g).
Figure 5. miR-17-92 regulates cardiomyocyte proliferation and represses the expression and function of PTEN.
(A)Primary neonatal (P1) rat cardiomyocytes were transfected with indicated miRNA mimics or control mimic and cells were incubated with EdU (10 μM). One day later, cultures were fixed and stained with antibodies for EdU. cTNT marks cardiomyocytes. DAPI stains nuclei. Arrowheads point to EdU positive cardiomyocytes. Bars = 70 μm. (B)P1 rat cardiomyocytes were transfected with indicated miRNA inhibitors or control inhibitor and cells were incubated with EdU. One day later, cultures were fixed and stained with antibodies for EdU. cTNT marks cardiomyocytes. DAPI stains nuclei. Arrowheads point to EdU positive cardiomyocytes. Bars = 70 μm. (C)Quantification of percentages of EdU+ cardiomyocytes in cultured neonatal rat cardiomyocytes after treated with miRNA mimics or control. (D)Quantification of percentages of EdU+ cardiomyocytes in cultured neonatal rat cardiomyocytes after treated with miRNA inhibitors or control. (E)P1 rat cardiomyocytes were transfected with miR-19a/b mimics or control mimic. One day later, cultures were fixed and stained with antibodies for aurora B (green). cTNT (red) marks cardiomyocytes. DAPI (blue) stains nuclei. Arrowheads point to aurora B positive signal. Bars = 50 μm. (F)Quantification of percentages of aurora Bpositive cardiomyocytes in cultured neonatal rat cardiomyocytes after treated with miR-19a/b mimics or control. *P < 0.01. (G)Quantification RT-PCR (qPCR) analyses of CDK1 expression in cultured neonatal rat cardiomyocytes after treated with miRNA mimics or mimic control. (H)Neonatal mouse cardiomyocytes were isolated from miR-17-92TG/TG mice and cultured in the presence of serum-free medium and EdU (10 μM). After transduced with Ad-cTNT-Cre (or Ad-lacZ in control), cell proliferation is determined by EdU incorporation. cTNT marks cardiomyocytes. DAPI stains nuclei. Arrows point to EdU positive cardiomyocytes. Bars = 70 μm. (I)Quantification of percentages of EdU+ cardiomyocytes in cultured neonatal mouse cardiomyocytes isolated from miR-17-92TG/TG mice and transduced with Ad-cTNT-Cre or Ad-lacZ in control. **P < 0.01. (J)qPCR analyses of the expression of putative miR-17-92 targets the hearts of 20 days old miR-17-92-KO and control mice.(K)Quantification RT-PCR (qPCR) analyses of the expression of putative miR-17-92 targets the hearts of 15 days old miR-17-92-TGMHC and control mice. (L)P1 rat cardiomyocytes were transfected with miR-19a/b mimics, control mimics, modify RNA for PTEN (modi-PTEN), or both miR-19a/b mimics and modi-PTEN and cells were incubated with EdU. One day later, cultures were fixed and stained with antibodies for EdU (green). cTNT (red) marks cardiomyocytes. DAPI (blue) stains nuclei. Bars = 150 μm. (M)Quantification of percentages of EdU+ cardiomyocytes in cultured neonatal rat cardiomyocytes after treated with miR-19a/b mimics, control mimics, modify RNA for PTEN (modi-PTEN), or both miR-19a/b mimics and modi-PTEN.
Next, we isolated neonatal cardiomyocytes from miR-17-92-KI mice. Cultured cardiomyocytes were infected with Ad-cTNT-Cre to induce the overexpression of miR-17-92 (Fig. 5h). We found that overexpression of miR-17-92 in mouse neonatal cardiomyocytes, but not the cells treated with the control Ad-lacZ, dramatically enhanced the incorporation of EdU, indicating an increase in cardiomyocyte proliferation (Fig. 5h, i). Together, our data demonstrate that miR-17-92 mimics induce, while miR-17-92 inhibitors reduce cardiomyocyte proliferation in vitro and ex vivo.
miR-17-92 represses PTEN to induce cardiomyocyte proliferation
We tested the expression of putative miR-17-92 targets that are known to play a role in cell proliferation20. We reasoned that the expression of these targets should be inversely correlated with the expression of miR-17-92, which is decreased in the hearts of miR-17-92-TG mice and increased in the hearts of miR-17-92-KO mice. Indeed, we found that the expression of several targets was elevated in the hearts of miR-17-92-KO mice (Fig. 5j), and repressed in the hearts of miR-17-92-TG mice (Fig. 5k). We focused on PTEN, a tumor suppressor and a member of family of protein tyrosine phosphatases21–23, which was most dramatically altered in the hearts of miR-17-92 transgenic and mutant mice (Fig. 5j, k). PTEN was reported a direct target of miR-19a/b24, the most potent member of the miR-17-92 cluster to induce tumor growth24 and to promote cardiomyocyte proliferation in our study (Fig. 5a–d). Deletion of PTEN led to axon regeneration in central neural system, further highlighting the role of PTEN in cell proliferation and regeneration25. We asked whether PTEN could mediate the function of miR-19a/b in cardiomyocyte proliferation, and more specifically, we tested whether overexpression of PTEN could suppress miR-19a/b-induced cardiomyocyte proliferation. We overexpressed PTEN in neonatal rat cardiomyocyte using a modified RNA approach26. We achieved dose-dependent overexpression of PTEN protein in transfected cells (Online Figure XXI). Overexpression of PTEN completely abolished miR-19a/b-induced cardiomyocyte proliferation (Fig. 5l, m).
DISCUSSION
In this report, our genetic studies using miR-17-92 knockout and transgenic mice, together with results of in vitro cell culture, demonstrated that members of the miR-17-92 cluster are required for and sufficient to induce cardiomyocyte proliferation. We found that PTEN is one of the miR-17-92 targets that mediate the function of this cluster of miRNAs, at least in vitro in cultured cardiomyocytes, to regulate cardiomyocyte proliferation.
In sharp contrast to embryonic cardiomyocytes, which exhibit strong proliferative activity, the rate of cardiomyocyte proliferation and turn over in adult hearts is very low and it is generally conceived that adult hearts retain very limited (if any) potential for regeneration. As the consequence, the intrinsic renewal rate is insufficient to reverse cardiomyocyte loss and to restore cardiac function under pathophysiological conditions27, 28. Numerous attempts have been developed to overcome this hurdle and one of the approaches is to induce cell cycle activity in the surviving cardiomyocytes27, 29. Previous reports indicate that targeted overexpression of members of the cyclin D, cyclin D2 in particular, is sufficient to induce cardiomyocyte cell cycle activity in adult hearts, resulting in improved cardiac function upon myocardial injury30–32. Despite the fact that we know the critical role of the cell cycle regulators in cardiomyocyte proliferation, the molecular pathways that diminish adult cardiomyocyte proliferation remain largely unknown. Our studies reported here, using genetic approaches, demonstrated that miRNAs are previously unidentified regulators of cardiomyocyte proliferation. Interestingly, a recent study reported that family of miR-15 inhibits cardiomyocyte proliferation in a manner that inhibition of it increased myocyte proliferation in adult hearts after myocardial infarction33. The potential of applying miRNAs to reconstitute lost cardiomyocytes in injured hearts could be of considerable therapeutic value for human cardiovascular disease.
Whereas miR-17-92 is required for normal animal development, cell proliferation and tumor growth, individual member of the miR-17-92 cluster appears to possess distinct function8, 14, 15. For example, miR-17 was shown to reduce cell proliferation. Transgenic overexpression of miR-17 resulted in growth retardation in animals 34. It was recently reported that overexpression of miR-92a blocks angiogenesis in vitro and in vivo35. Additional studies found that miR-19 is a key component of the miR-17-92 cluster to induce cell proliferation and oncogenic growth24. Our results demonstrate that miR-19a/miR-19b are sufficient and required for neonatal cardiomyocyte proliferation in vitro, consistent with the view that miR-19 is a key component of the miR-17-92 cluster in controlling cell proliferation. In the future, it will be important to determine whether miR-19a/miR-19b promote cardiomyocyte proliferation in vivo, in particular in postmitotic adult cardiomyocytes.
We found that PTEN is a functional target of miR-17-92 cluster. More specifically, we showed that PTEN mediates the function of miR-19a/19b in the regulation of cardiomyocyte proliferation. PTEN is a tumor suppressor which was previously shown to be repressed by miR-17-92 during tumor growth24. Genetic deletion of PTEN in the heart led to hypertrophic growth36. Addition investigation revealed that loss of PTEN protect the heart from the development of pathological hypertrophy and heart failure under biomechanical stress, further highlighting the critical involvement of PTEN in cardiac function and disease30. Our studies link the function of miRNAs and PTEN in cardiomyocyte proliferation. Future studies will be important to illustrate how the miR-17-92-PTEN axis participates in cardiac regeneration.
In summary, our findings uncovered that miR-17-92 plays a key role in the regulation of cardiomyocyte proliferation in embryonic, postnatal and adult hearts, implying the therapeutic potential of miR-17-92 in human cardiomyocyte proliferation, cardiac repair, regeneration and related disorders.
Supplementary Material
Novelty and Significance.
What Is Known?
Loss of cardiomyocytes in adult hearts is one of the most common causes of heart failure.
Adult mammalian hearts have limited capacity to regenerate.
microRNAs (miRNAs) are a class of small non-coding RNAs that regulate a variety of biological processes.
It has been shown before that miR-17-92 regulates cell proliferation and tumorigenesis.
What New Information Does This Article Contribute?
miR-17-92 is a key regulator of cardiomyocyte proliferation.
Loss-of-function studies show that miR-17-92 is required for cardiomyocyte proliferation.
Gain-of-function studies demonstrate that miR-17-92 is sufficient to induce cardiomyocyte proliferation in embryonic, postnatal and adult hearts.
PTEN is one of the key targets of miR-17-92 that mediates cardiomyocyte proliferation.
The adult heart is primarily composed of terminally differentiated, mature cardiomyocytes that exit the cell cycle. The molecular mechanism that controls the cardiomyocyte proliferation and cardiac regeneration is not fully understood. We hypothesized that miRNAs are key regulators of cardiomyocyte proliferation. Herein we report that genetic deletion of miR-17-92 in cardiomyocytes reduced cardiomyocyte proliferation and total numbers of cardiomyocytes in the heart. Conversely, transgenic overexpression of miR-17-92 in cardiomyocytes stimulated cardiomyocyte proliferation and resulted in an increase of cardiomyocyte number. We found that miR-17-92 directly inhibited PTEN, a tumor suppressor, in cardiomyocytes. These findings suggest that miR-17-92 could be a potential therapeutic target for myocardial infarction and heart failure.
Acknowledgments
We thank Dr. Bernhard Kuhn for advice and stimulating discussion, Dr. John Mably for critical reading of the manuscript and discussion.
SOURCES OF FUNDING
This work was supported by the March of Dimes Foundation and the NIH. Masaharu Kataoka is supported by Banyu Life Science Foundation International. ZP Huang is a postdoctoral fellow and DZ Wang is an Established Investigator of the American Heart Association.
Nonstandard Abbreviations
- ACTN1
α-actinin
- BW
body weight
- CDK1
cyclin-dependent kinase 1
- cTNT
cardiac troponin T
- EdU
5-ethynyl-2′-deoxyuridine
- H&E
haematoxylin and eosin
- HW
heart weight
- MI
myocardial infarction
- pH3
phosphorylated histone H3
- PTEN
phosphatase and tensin homolog
- qPCR
quantitative polymerase chain reaction
Footnotes
DISCLOSURES
The authors have declared that no competing interests exist.
References
- 1.Kathiresan S, Srivastava D. Genetics of human cardiovascular disease. Cell. 2012;148:1242–1257. doi: 10.1016/j.cell.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mudd JO, Kass DA. Tackling heart failure in the twenty-first century. Nature. 2008;451:919–928. doi: 10.1038/nature06798. [DOI] [PubMed] [Google Scholar]
- 3.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 4.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 7.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 10.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, Van Haeringen A, Genevieve D, Goldenberg A, Oufadem M, Manouvrier S, Munnich A, Vidigal JA, Vekemans M, Lyonnet S, Henrion-Caude A, Ventura A, Amiel J. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, Hammond SM, MacPherson D. miR-17~92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 2011;25:1734–1745. doi: 10.1101/gad.17027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang DZ. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MG, Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987;327:31–35. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- 20.Mestdagh P, Bostrom AK, Impens F, Fredlund E, Van Peer G, De Antonellis P, von Stedingk K, Ghesquiere B, Schulte S, Dews M, Thomas-Tikhonenko A, Schulte JH, Zollo M, Schramm A, Gevaert K, Axelson H, Speleman F, Vandesompele J. The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol Cell. 2010;40:762–773. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, DePinho RA. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 23.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 24.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun F, Park KK, Belin S, Wang D, Lu T, Chen G, Zhang K, Yeung C, Feng G, Yankner BA, He Z. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011;480:372–375. doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubart M, Field LJ. Cardiac regeneration: repopulating the heart. Annu Rev Physiol. 2006;68:29–49. doi: 10.1146/annurev.physiol.68.040104.124530. [DOI] [PubMed] [Google Scholar]
- 28.Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87:521–544. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 30.Hassink RJ, Pasumarthi KB, Nakajima H, Rubart M, Soonpaa MH, de la Riviere AB, Doevendans PA, Field LJ. Cardiomyocyte cell cycle activation improves cardiac function after myocardial infarction. Cardiovasc Res. 2008;78:18–25. doi: 10.1093/cvr/cvm101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasumarthi KB, Nakajima H, Nakajima HO, Soonpaa MH, Field LJ. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ Res. 2005;96:110–118. doi: 10.1161/01.RES.0000152326.91223.4F. [DOI] [PubMed] [Google Scholar]
- 32.Soonpaa MH, Koh GY, Pajak L, Jing S, Wang H, Franklin MT, Kim KK, Field LJ. Cyclin D1 overexpression promotes cardiomyocyte DNA synthesis and multinucleation in transgenic mice. J Clin Invest. 1997;99:2644–2654. doi: 10.1172/JCI119453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, Mammen PP, Rothermel BA, Olson EN, Sadek HA. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A. 2013;110:187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee SP, Siragam V, Yang BB. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;11:1031–1038. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- 35.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 36.Crackower MA, Oudit GY, Kozieradzki I, Sarao R, Sun H, Sasaki T, Hirsch E, Suzuki A, Shioi T, Irie-Sasaki J, Sah R, Cheng HY, Rybin VO, Lembo G, Fratta L, Oliveira-dos-Santos AJ, Benovic JL, Kahn CR, Izumo S, Steinberg SF, Wymann MP, Backx PH, Penninger JM. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell. 2002;110:737–749. doi: 10.1016/s0092-8674(02)00969-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.