SYNOPSIS
Malaria, caused by Plasmodia parasites, affects hundreds of millions of people. As purine auxotrophs, Plasmodia use transporters to import host purines for subsequent metabolism by the purine salvage pathway. Thus, purine transporters are attractive drug targets. All sequenced Plasmodia genomes encode four Equilibrative Nucleoside Transporters (ENTs). During the pathogenic intraerythrocytic stages, ENT1 is a major route of purine nucleoside/nucleobase transport. Another plasma membrane purine transporter exists because Plasmodium falciparum ENT1-knockout parasites survive at supraphysiological purine concentrations. The other three ENTs have not been characterized functionally. Codon-optimized P. falciparum and P. vivax ENT4 were expressed in Xenopus laevis oocytes and substrate transport determined with radiolabeled substrates. ENT4 transported adenine and 2′-deoxyadenosine at the fastest rate, with millimolar-range apparent affinity. ENT4-expressing oocytes did not accumulate hypoxanthine, a key purine salvage pathway substrate, or AMP. Micromolar concentrations of the plant hormone cytokinin compounds inhibited both Pf- and Pv-ENT4. In contrast to PfENT1, ENT4 interacted with the Immucillin compounds in the millimolar range and was inhibited by 10 μM dipyridamole. Thus, ENT4 is a purine transporter with unique substrate and inhibitor specificity. Its role in parasite physiology remains uncertain but is likely significant because of the strong conservation of ENT4 homologues in Plasmodia genomes.
Keywords: Plasmodium, adenine, adenosine, immucillin, metabolism
INTRODUCTION
Malaria affects hundreds of millions of people annually [1, 2]. Five species from the genus Plasmodium cause human malaria. Like many other protozoan parasites, they are unable to synthesize purines de novo. Rather, they rely on purine import from the host cell through transporters in the parasite plasma membrane and purine modification through the purine salvage pathway for nucleic acid synthesis [3, 4]. Thus, purine transporters are potential chemotherapeutic targets, and they may also mediate uptake of potential antimalarial compounds [5].
To date, the majority of Plasmodium purine transport studies have focused on PfENT1, also named PfNT1, (PF13_0252). Transport studies of PfENT1 heterologously expressed in Xenopus laevis oocytes show that PfENT1 is a low-affinity purine transporter with broad specificity for many nucleobases and nucleosides [6-10]. Other studies in Xenopus oocytes [11] and PfENT1-knockout parasites [12] have reported conflicting conclusions on the affinity and substrate specificity of PfENT1 based on radiolabeled substrate uptake experiments. However, it was demonstrated [9, 13] that some of the reported kinetic uptake data were strongly influenced by metabolism of the radiolabeled substrate in the oocyte and did not reflect transport as previously proposed. PfENT1 is expressed in blood-stage parasites at the parasite plasma membrane and is the main transporter mediating purine uptake for purine salvage pathways in cultured parasites [8]. Knockouts of the gene encoding PfENT1 in parasites indicate that PfENT1 is essential for parasite survival at physiological purine levels found in human serum [14, 15]. However, the parasite plasma membrane must contain alternate transport mechanisms, because PfENT1-knockout parasites survive in media containing supraphysiological purine levels. Furthermore, PfENT1-knockout parasites released from the host erythrocytes transport purine nucleobases and nucleosides [12, 14, 15]. Finally, adenosine monophosphate (AMP) and the Immucillin class of compounds, which are purine-analog compounds and are potent inhibitors of Plasmodium purine nucleoside phosphorylase (PNP), are not substrates of PfENT1 [4, 9].
P. falciparum and the related parasite P. vivax, which together account for the majority of malaria cases worldwide, encode four members of the Equilibrative Nucleoside Transporter (ENT) family [16]. These have been designated as PfENT1-4 or PvENT1-4 based on phylogenetic analysis [5]. Localization studies of epitope-tagged PfNT2 (MAL8P1.32 or PfENT2) in transgenic parasites show that its gene product is found in the parasite endoplasmic reticulum, however, its capacity to transport purines has not been demonstrated [17]. The localization and functions of PfENT3 (PF14_0662), PfENT4 (PfA0160c), and the ENTs from P. vivax remain unexplored. PfENT4 homologues with greater than 50% sequence identity are also present in the genomes of P. vivax (PVX_081595), P. knowlesi (PKH_021010), P. chabaudi (PCHAS_020830), P. yoelii (PY06526), and P. berghei (PBANKA_020990). Expression studies in synchronized parasite cultures show that PfENT4 mRNA has a maximum expression at 20-25 hours and then falls slightly as the parasites mature from the trophozoite to schizont stages [16, 18, 19]. PfENT4 mRNA is also expressed in blood stage parasites isolated from malaria patients but did not show significant differences amongst the three distinct transcriptional/metabolic states [20, 21]. Mass spectrometry based expression studies have identified PfENT4 peptide fragments in several lifecycle stages including trophozoites, early schizont, schizont rupture, in sporozoites and in late stage gametocytes [22, 23]. However, the levels of ENT4 protein expression during the intra-erythrocytic growth cycle are unknown.
This report describes the characterization of PfENT4 and PvENT4 expressed in Xenopus oocytes. Previous attempts to express PfENT4 from mRNA with the native P. falciparum codon usage in Xenopus oocytes were unsuccessful [8]. To obtain expression in oocytes, Xenopus codon-optimized versions of the PfENT4 and PvENT4 genes were synthesized. Our studies revealed that PfENT4 and PvENT4 have low affinity for adenine, adenosine and 2′-deoxyadenosine, and no capacity to transport hypoxanthine or AMP. [3H]Adenine transport was sensitive to protons and the ENT inhibitor dipyridamole. [14C]2′-deoxyadenosine uptake was inhibited by the plant hormone cytokinin molecules with micromolar apparent affinity. Competition studies with [3H]adenine showed that Immucillins and DADMe-Immucillins were recognized by PfENT4 and PvENT4, albeit with millimolar apparent affinity. Thus, PfENT4 and PvENT4 function as purine transporters.
MATERIALS AND METHODS
Reagents
[2,8-3H]-adenine (36.3 Ci/mmol), [2,8-3H]-adenosine (36.8 Ci/mmol), [8-3H]-guanine (10.7 Ci/mmol), [8-3H]-guanosine (9.1 Ci/mmol), [2,8-3H]-hypoxanthine (30.3 Ci/mmol), [2,8-3H]-inosine (7.9 Ci/mmol), [8-3H]-xanthine (19.4 Ci/mmol), [5-3H]-cytosine (26.1 Ci/mmol), [5-3H(N)]-cytidine (25.6 Ci/mmol), [2-14C]-thymine (52.0 mCi/mmol), [5′-3H]-thymidine (29.5 Ci/mmol), [5-3H]-uridine (17.3 Ci/mmol), and [2,8-3H]-adenosine-5′-monophosphate (10.8 Ci/mmol), were purchased from Moravek Biochemicals (Brea, CA). [1-3H]-D-ribose (15 Ci/mmol), [8-14C]-2′-deoxyadenosine (48.8 mCi/mmol), [8-14C]2′-deoxyguanosine (55 mCi/mmol), and [5,6-3H]-uracil (33.8 Ci/mmol) were purchased from Sigma Aldrich (St. Louis, MO). At the start of these experiments, the purity of each radiolabeled reagent was established by HPLC. Nitrobenzylmercaptopurineriboside (NBMPR, 10 mM), dipyridamole (10 mM), isopentenyl adenine (200 mM), trans-zeatin (200 mM), and cis-zeatin (200 mM) were purchased from Sigma Aldrich and were dissolved in DMSO as stock solutions. The Immucillins were generous gifts from Drs. Peter C. Tyler and Gary B. Evans at Industrial Research Ltd. (Lower Hutt, New Zealand); the compounds were dissolved in transport buffer before use. All other reagents were purchased from Sigma Aldrich or Thermo Fisher Scientific (Waltham, MA).
Synthetic gene construct of PfENT4 and PvENT4 as a template for mRNA synthesis
The coding sequence of hypothetical proteins PfA0160c and Pv081595 from the PlasmoDB were optimized for codon usage in X. laevis using Gene Designer® software (DNA2.0, Menlo Park, CA). The construct included a BamHI restriction site at the 5′ end, a Kozak consensus sequence, the optimized gene, and a 3′ EcoRI restriction site (See Supplementary Figure 1). The construct was subcloned into the pXOON plasmid using the BamHI/EcoRI restriction enzymes and T4 DNA ligase. The pXOON plasmid, which is the same as the pXOOM plasmid [24] except for a different multiple cloning site, contains the 5′ and 3′ untranslated regions for the Xenopus beta-globin gene which enhances heterologous expression in oocytes. The pXOON plasmid containing either PfENT4 or PvENT4 were linearized with the NheI restriction enzyme. Capped mRNAs were synthesized with the mMESSAGE mMACHINE kit (Ambion, Austin, TX) and purified using the MEGAClear kit (Ambion). Recovered mRNA was dissolved in diethylpyrocarbonate (DEPC; Sigma) treated water and diluted to a concentration of 1 μg/μl.
Xenopus oocyte expression of PfENT4 and PvENT4 and radiolabeled flux assay
Female X. laevis toads were obtained from Nasco (Fort Atkinson, WI) and approved for use by the Albert Einstein College of Medicine Institutional Animal Care and Use Committee (Protocol #20110905). Stage V or VI defolliculated Xenopus oocytes were prepared as described elsewhere [10] and were injected with 23 nl of DEPC-treated water (herein referred to as “H2O-injected oocytes”) or mRNA for PfENT4 or PvENT4 (23 ng). Radiolabeled substrate uptake assays were performed 3 or 4 days after injection. For each experiment, radiolabeled substrate uptake by H2O-injected control oocytes was performed with the same batch of oocytes and at the same time as uptake into mRNA injected oocytes was measured. Thus, each experiment included its own H2O-injected control oocytes. For the assay, between 4 to 10 oocytes were incubated in 480 μl E1 transport buffer (concentrations in mM: 140 NaCl, 2.8 KCl, 2 MgCl2, 1 CaCl2, 10 HEPES; pH 7.4 with NaOH) plus radiolabeled substrate at room temperature. Other experimental conditions are noted as necessary. In the experiments measuring substrate uptake at pH 6.5, 6.0 and 5.5, the E1 buffer was made with 10 mM MES rather than HEPES. In the experiments measuring uptake in sodium-free buffer, the E1 buffer was made with 140 mM choline chloride rather than NaCl (pH 7.4 with KOH).
Substrate flux was terminated by washing the oocytes five times in 500 μl ice-cold E1 buffer. The washes were completed in 15 s. Oocytes were transferred to individual scintillation vials and solubilized in 300 μl of 5% SDS. Some competition experiments were performed by pooling eight oocytes together into one vial and solubilizing them in 1 ml of 5% SDS; they are noted in the text. Radioactivity was determined using a Tri-Carb 2910TR liquid scintillation counter (PerkinElmer, Waltham, MA). All uptake experiments were repeated in at least two batches of oocytes from separate frogs.
Uptake into H2O-injected oocytes was often subtracted from the uptake into oocytes expressing PfENT4 or PvENT4 and these subtracted values are referred to as “background-subtracted.” For the 60 min accumulation experiment (in Figure 2), AMP uptake was measured in the presence and absence of the 5′-ectonucleotidase inhibitor, α,β-methyleneADP (250 μM). For the competition experiments (in Figure 9) the oocytes were incubated with 150 nM [3H]adenine plus the indicated concentration of compound. DADMe-ImmG contained 2 molar equivalents of HCl and the 10 mM HEPES transport buffer was insufficient for buffering. Thus, experiments with DADMe-ImmG were performed in an isoosmotic buffered solution containing 30 mM HEPES with the pH adjusted to 7.4 and a compensating reduction in the NaCl concentration.
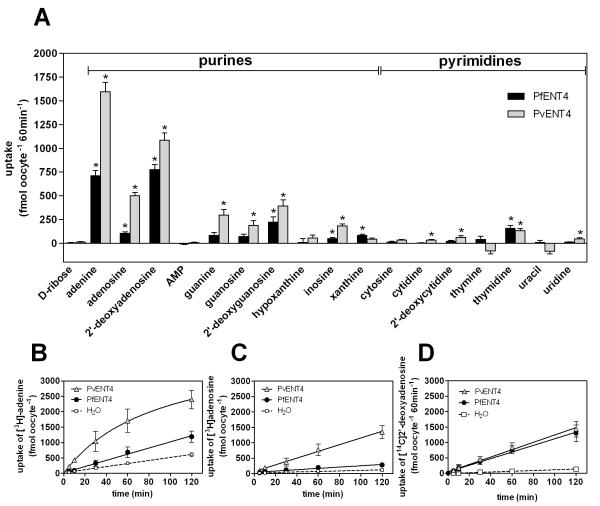
FIGURE 2. Uptake of nucleobases and nucleosides by oocytes expressing PfENT4 or PvENT4.
(A) Uptake of 1.5 μM radiolabeled substrates by oocytes expressing PfENT4 (black bars) or PvENT4 (gray bars) following a 60 min incubation. Bars represent the background-subtracted mean ± SEM uptake of 9 to 38 oocytes. These oocytes were from at least three different oocyte isolations, and all points are pooled to calculate the mean. The average values obtained from H2O-injected oocytes have been subtracted (unsubtracted values can be found in Supplementary Table 1 along with uptake by H2O-injected oocytes). We cannot distinguish whether the differences in uptake between PfENT4 and PvENT4 expressing oocytes are due to differences in the level of protein expression or to intrinsic differences in the transport properties of the two transporters. Bars marked with (*) indicate uptake values that are statistically significantly different than uptake into H2O-inejcted oocytes (p<0.01) by one-way ANOVA, Dunnett post-hoc analysis. (B, C, D) Uptake time course of [3H]adenine (B), [3H]adenosine (C), and [14C]2′-deoxyadenosine (D) in oocytes expressing PvENT4 (gray triangles), PfENT4 (black circles), or injected with H2O (white squares). Points represent the mean ± SD of uptake from 8 oocytes from one representative experiment.
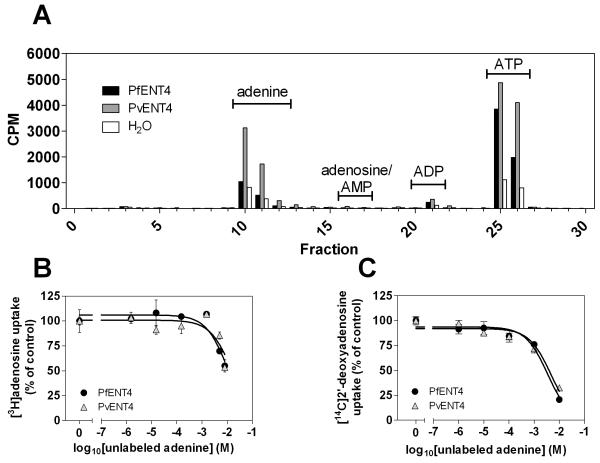
FIGURE 9. Inhibition of adenine uptake by various compounds.
Oocytes expressing PfENT4 (black bars) or PvENT4 (gray bars) were incubated in buffer for 60 min in the presence of 150 nM [3H]adenine and a competing compound at the indicated concentration. Uptake was normalized to the uptake in oocytes expressing PfENT4 or PvENT4 in the presence of no competitor (100%) and H2O-injected oocytes (0%). The bars represent the mean ± SEM of at least 12 oocytes from three separate experiments, except for 2 mM DADMeImmG which is the average of two experiments.
Metabolic labeling and determination of adenine metabolites in cells
Metabolic labeling was performed in Xenopus oocytes, human erythrocytes, P. falciparum infected erythrocytes and saponin-released P. falciparum parasites. Five oocytes injected with mRNA for PfENT4, PvENT4, or H2O were incubated in 150 nM [3H]adenine for 60-min, washed five times with ice-cold buffer and the samples were frozen until further use. Parasites were cultivated in RPMI media containing 370 μM hypoxanthine and Albumax II (Invitrogen) at a 4% hematocrit, in 75 cm2 tissue culture flasks in a 5.05% CO2, 4.93% O2 and 90.2% N2 gas mixture at 37 °C. Parasite growth was monitored by microscopic evaluation of Giemsa-stained thin smears. Human erythrocytes were drawn from healthy donors under the Albert Einstein College of Medicine Committee on Clinical Investigations Protocol M-1063. Metabolic studies were performed with parasites in the late trophozoite stage synchronized by two treatments with 5% (w/v) d-sorbitol solution in water. Parasites were magnet purified (MidiMACS™ Separator, Miltenyi Biotec) and the recovered infected erythrocytes were split in two sets. One set of parasites were released from the erythrocytes (free-parasites) by incubating pelleted infected-erythrocytes with 0.03% saponin in Dulbecco’s phosphate-buffered saline (DPBS, CellGro; 2.7 mM KCl, 1.5 mM KH2PO4, 136.9 mM NaCl, 8.1 mM Na2HPO4, pH 7.4) to lyse the red blood cells, followed by washing in DPBS as we have done previously [4]. The other set was used as intact infected-erythrocytes. Erythrocytes, infected erythrocytes and saponin-released parasites were placed in purine-free media for 30 min, then incubated with 50 nM [3H]adenine for 2 h, washed twice with ice-cold buffer and the samples were frozen until further use. Purine metabolites were extracted, fractionated, and detected as described in detail elsewhere [4, 9]. Briefly, following uptake, cells were homogenized in H2O and treated with perchloric acid to remove nucleic acids and proteins. Purines were extracted using a YM10 Centricon spin column (Amicon, Houston, TX) and analyzed using a reverse-phase ion-pair HPLC system (Luna C18(2), 150 mm×4.6 mm, 3 μm, Phenomenex, Torrance, CA). Metabolite analysis was performed using HPLC as described elsewhere [4]. The eluant was monitored at 254 nm and the flow rate was 1 ml/min. Fractions were collected every minute for 30 min and subjected to liquid scintillation counting. Radiolabeled peaks were identified based on UV detection of purine standards.
Data analysis
Statistical tests and curve-fitting used GraphPad Prism version 5.02 for Windows (La Jolla, CA). Nonlinear regression analyses included Michaelis-Menten kinetics and single- and two-site sigmoidal dose-response curves to determine IC50’s. Ki’s were determined using the method of Cheng and Prusoff [25]. Data are presented as mean + standard error of the mean (SEM).
RESULTS
Functional characterization of PfENT4 and PvENT4
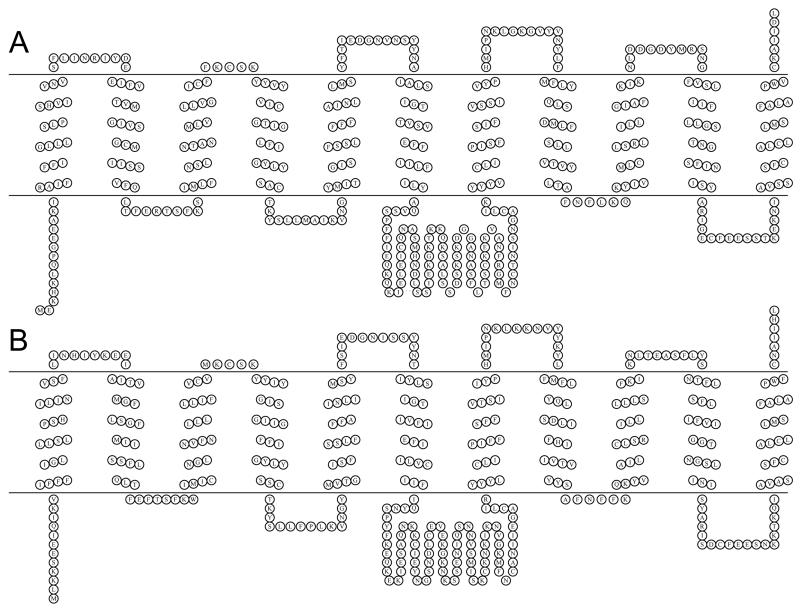
The predicted transmembrane topology of PfENT4 and PvENT4 is characteristic for equilibrative nucleoside transporter family members with eleven transmembrane (TM) spanning segments and a large cytosolic loop between TM 6 and 7 (Figure 1A and B). X. laevis oocytes provide an excellent expression system for nucleoside/nucleobase transport studies [26, 27]. To assess whether oocytes injected with PfENT4 or PvENT4 mRNA transport nucleosides and nucleobases, we performed radiolabeled substrate uptake assays. As an initial screen, oocytes were incubated in the presence of 1.5 μM radiolabeled compounds and substrate accumulation was measured after 60 min (Figure 2A, Supplementary Table 1). Oocytes injected with PfENT4 or PvENT4 mRNA had greater uptake of the purine nucleobase adenine and the purine nucleosides adenosine, 2′-deoxyadenosine, 2′-deoxyguanosine and inosine than H2O-injected oocytes. There was no observed uptake of hypoxanthine (Figure 2A, Supplementary Table 1). Oocytes have a 5′-nucleotidase which can convert AMP into adenosine [4]. This 5′-nucleotidase activity can be blocked by the addition of 250 μM α,β-methyleneADP. AMP transport was not observed in the presence of α,β-methyleneADP (Figure 2A, Supplementary Table 1). Oocytes expressing PfENT4 and PvENT4 displayed uptake of thymidine. However, PvENT4 expressing oocytes exhibited less accumulation of thymine and uracil than H2O-injected oocytes, though this trend is not statistically significant.
FIGURE 1. Schematic of the predicted transmembrane topology of ENTs.
(A) PvENT4. (B) PfENT4. The topology prediction was performed by the ConpredII algorithm [42] and the schematic image was generated using the TOPO2 program [43].
Adenine, adenosine, and 2′-deoxyadenosine uptake was time-dependent, with accumulation increasing up to 120 min (Figure 2B, 2C, 2D). Adenine and adenosine uptake was greater in PvENT4 expressing oocytes than PfENT4 expressing oocytes; 2′-deoxyadenosine uptake was similar for both transporters (Figure 2B, 2C, 2D).
Secondary active transport proteins use Na+ or H+ concentration gradients as the driving force for substrate transport. To determine whether PfENT4 or PvENT4 are secondary active transporters, [3H]adenine uptake was measured at different pH values (Figure 3A, Supplementary Table 2) or in a solution in which choline chloride replaced sodium chloride (Figure 3B, Supplementary Table 3). Adenine transport was inhibited at lower pH values, and the effect was more pronounced for PfENT4 than PvENT4. A similar trend was observed when adenosine was used as the substrate (data not shown). This observation rules out cotransport of protons and purines. Although replacing sodium in the transport buffer with choline caused significant inhibition of adenine transport by PfENT4 and PvENT4 (26% and 18% inhibition, Student’s t-test p<0.01), we would have expected complete inhibition of purine uptake if PfENT4 or PvENT4 were sodium-coupled transporters.
FIGURE 3. Effect of extracellular cations on adenine transport.
(A) Uptake of 1.5 μM [3H]adenine by oocytes expressing PfENT4 or PvENT4 during a 60 min incubation at the indicated pH. Uptake at each pH was background-subtracted and normalized to the uptake at pH 7.4 (100%). Bars represent the normalized mean ± SEM of at least 12 oocytes pooled from three separate experiments. Bars marked with (*) indicate uptake values that are statistically significantly different than uptake at pH 7.4 (p<0.01) by one-way ANOVA, Dunnett post-hoc analysis. (B) Oocytes expressing PfENT4 or PvENT4 were incubated for 60 min in the presence of buffer containing either Na+ or choline+, pH adjusted to 7.4 with either NaOH or KOH, respectively. Uptake was background-subtracted and normalized to be a percentage of uptake in Na+ buffer (100%). Bars represent the mean ± SEM of 12 to 15 oocytes pooled from three separate experiments. Uptake values in the choline+ buffer are statistically significantly different from uptake values in the Na+ buffer (Student’s t-test, p<0.01). Non-background substracted values can be found in Supplementary Table 2 and 3.
To investigate the kinetic parameters of purine transport, we measured the concentration-dependent uptake of both adenosine and adenine (Figure 4) at 60 min, which is within the linear uptake range for both transporters and substrates. Uptake was not saturable at concentrations up to 10 mM, thus obtaining accurate Km values was not possible in this assay. Alternatively, we used unlabeled substrate to competitively inhibit radiolabeled substrate uptake to obtain estimates of the substrate Ki, which provided a measurement of a compound’s apparent affinity for transport (8). Unlabeled adenosine inhibited [3H]adenosine transport and the data were fit to a one-site competition model (Figure 5A and 5B) with mean Ki values ± SEM being 2.2 ± 1.0 mM (n=3) for PfENT4 and 1.0 ± 0.2 mM (n=3) for PvENT4 (Table 1). Similarly, unlabeled 2′-deoxyadenosine inhibited [14C]2′-deoxyadenosine uptake with mean Ki values of 1.5 ± 0.1 mM for PfENT4 and 4.3 ± 0.2 mM for PvENT4 (Table 1). In contrast, data for adenine competitively inhibiting [3H]adenine uptake was best fit using a two-site competition model with Ki values of 0.007 ± 0.002 and 3.6 ± 2.0 mM for PfENT4 and 0.006 ± 0.002 and 3.2 ± 0.5 mM for PvENT4 (Figure 5C and 5D and Table 1). The two sites are likely due to competition with both a metabolic enzyme and the transporter, where the site that is rate limiting in the uptake assay depends on the substrate concentration (8).
FIGURE 4. Concentration-dependent uptake of adenine and adenosine.
Michaelis-Menten analysis of adenosine (A, B) and adenine (C, D) uptake into oocytes expressing PfENT4 (black circles) or PvENT4 (gray triangles) or H2O-injected oocytes (white squares) during a 60 min incubation in the presence of varying concentrations of [3H]adenosine or [3H]adenine. For panels (B) and (D), transport of substrate via PfENT4 and PvENT4 was determined by subtracting the uptake into H2O-injected oocytes from the uptake into oocytes expressing PfENT4 or PvENT4. In these experiments, 8 oocytes were pooled together into one vial for scintillation counting, and each point is the amount of radioactivity detected divided by 8. The results from one representative experiment are shown.
FIGURE 5. Concentration-dependent inhibition of adenosine and adenine uptake.
(A, B) Competitive inhibition of 150 nM [3H]adenosine uptake for 60 min into oocytes expressing PfENT4 (black circles), PvENT4 (gray triangles), or H2O-injected oocytes (white squares) with increasing concentrations of unlabeled adenosine. The data for ENT4-expressing oocytes are fit with a single-site competition curve. (C, D) Competitive inhibition of 150 nM [3H]adenine uptake for 60 min into oocytes expressing PfENT4 (black circles), PvENT4 (gray triangles), or H2O-injected oocytes with increasing concentrations of unlabeled adenine. The data for ENT4-expressing oocytes are fit with a two-site competition curve. Panels (A) and (C) show the raw uptake values and panels (B) and (D) show the background-subtracted uptake into oocytes expressing PfENT4 and PvENT4 normalized to a % of uptake in the absence of unlabeled competitor. In these experiments, 8 oocytes were pooled together into one vial for scintillation counting, and each point is the amount of radioactivity detected divided by 8. The results from one representative experiment are shown.
Table 1. Summary of Inhibition Values.
Single IC50 or Ki values were obtained from individual experiments from a single batch of oocytes. The values in this table are the mean ± SEM of at least three independent determinations of the inhibition values except where noted.
| inhibitor | Tracer | IC50 or Ki (mM) | |
|---|---|---|---|
| PfENT4 | adenine | [2,8-3H]adenine | 0.007 ± 0.002, 3.6 ± 2.0a |
| [2,8-3H]adenosine | 9.0 ± 2.0b | ||
| [8-14C]2’-deoxyadenosine | 4.4 ± 0.8 | ||
| adenosine | [2,8-3H]adenosine | 2.2 ± 1.0 | |
| 2 ’-deoxyadenosine | [8-14C]2’ -deoxyadenosine | 1.5 ± 0.1 | |
| isopentenyl adenine | [8-14C]2’-deoxyadenosine | 0.0033 ± 0.0009 | |
| trans-zeatin | [8-14C]2’-deoxyadenosine | 0.020 ± 0.003 | |
| cis-zeatin | [8-14C]2’-deoxyadenosine | 0.190 ± 0.072 | |
| PvENT4 | adenine | [2,8-3H]adenine | 0.006 ± 0.002, 3.2 ± 0.5a |
| [2,8-3H]adenosine | 15.0 ± 0.5b | ||
| [8-14C]2’-deoxyadenosine | 4.9 ± 2.3 | ||
| adenosine | [2,8-3H]adenosine | 1.0 ± 0.2 | |
| 2 ’-deoxyadenosine | [8-14C]2’-deoxyadenosine | 4.3 ± 0.2 | |
| isopentenyl adenine | [8-14C]2’-deoxyadenosine | 0.0095 ± 0.0028 | |
| trans-zeatin | [8-14C]2’-deoxyadenosine | 0.193 ± 0.021 | |
| cis-zeatin | [8-14C]2’-deoxyadenosine | 0.284 ± 0.069 |
These data were fit to a two-site competition model.
These data are the mean from two experiments, and the error is range/2.
Adenine metabolism in oocytes and competitive inhibition of adenosine and 2′-deoxyadenosine transport with adenine
The competitive inhibition of [3H]adenine uptake with unlabeled adenine showed two components, suggesting competition for metabolism and transport. We tested whether [3H]adenine underwent metabolic transformation in oocytes. Oocytes expressing PfENT4 and PvENT4 were incubated in the presence of [3H]adenine for 60 min and then washed five times with ice-cold buffer to remove unincorporated labeled adenine. Oocyte cytoplasm was subjected to extraction and HPLC analysis. This demonstrated conversion of [3H]adenine to ATP (Figure 6A). A possible mechanism is adenine conversion to AMP by adenine phosphoribosyltransferase (APRT), with subsequent phosphorylation to ADP and ATP by adenylate kinase.
FIGURE 6. Adenine transport and metabolism in Xenopus oocytes.
(A) Adenine metabolism in oocytes expressing PfENT4, PvENT4 or injected with H2O. Oocytes were incubated for 60 min in 150 nM [3H]adenine. The metabolic fate of the imported adenine was analyzed by HPLC. Fractions were collected and analyzed for the presence of radioactivity. The retention times for adenine, adenosine, adenosine 5′-mono- (AMP), di- (ADP) and tri-phosphate (ATP) are indicated. (B, C) Competition of increasing concentrations of unlabeled adenine on the uptake of 150 nM [3H]adenosine (B) or 1.5 μM [14C]2′-deoxyadenosine (C) into oocytes expressing PfENT4 (black circles) or PvENT4 (gray triangles). Competition data was background-subtracted and normalized to a % of uptake in the absence of unlabeled competitor. In panel (B) each point represents the average from two separate experiments, one experiment in which individual oocytes were solubilized and counted and the other in which 8 oocytes were pooled together and counted as one sample. In panel (C), each point represents the normalized mean uptake of 15 oocytes derived from 3 separate experiments. Data are fit with a single site competition curve.
To investigate which affinity component represents inhibition at the site of transport, we measured the influx of either [3H]adenosine or [14C]2′-deoxyadenosine into oocytes in the presence of increasing concentrations of unlabeled adenine. By using [3H]adenosine or [14C]2′-deoxyadenosine as the labeled substrate, we could estimate the Ki for adenine transport because adenine, adenosine and 2′-deoxyadenosine compete for transport at ENT4 but not for metabolism via APRT. We previously showed that in oocytes adenosine is phosphorylated by adenosine kinase and then phosphorylated by adenylate kinase to ADP and ATP [9]. We observed inhibition of [3H]adenosine uptake by increasing concentrations of unlabeled adenine in the low millimolar range (Figure 6B). The calculated adenine Ki values were 9 ± 2 mM for PfENT4 and 15.0 ± 0.5 mM for PvENT4. Similarly, analysis of unlabeled adenine inhibition of [14C]2′-deoxyadenosine uptake was best fit with a single-site competition model, with apparent Ki values of 4.4 ± 0.8 mM for PfENT4 and 4.9 ± 2.3 mM for PvENT4 (Figure 6C).These relatively high Ki values suggest that the low apparent affinity component of adenine uptake was attributable to transport and the component with an apparent Ki less than 10 μM was likely due to competition at the metabolic enzyme, APRT.
Adenine metabolism in parasites
The fate of [3H]adenine in blood-stage P. falciparum, was evaluated by comparing metabolic labeling of uninfected erythrocytes, infected erythrocytes, and saponin-released parasites. Cells were incubated in purine-free media for 30 min, and then 50 nM of [3H]adenine was added for 2 h. Cells were then washed twice with ice-cold buffer to remove unincorporated [3H]adenine. Purines were extracted and analyzed by HPLC. Uninfected erythrocytes completely metabolized adenine to phosphorylated nucleotides, mainly ATP, through the action of APRT and adenylate kinase (Figure 7A). This is expected since normal uninfected erythrocytes contain ~2 mM ATP. Infected erythrocytes metabolized adenine predominantly to phosphorylated nucleotides as well as to other purines such as hypoxanthine and inosine (Figure 7B). Of note, in the infected erythrocytes much of the metabolized adenine was present as AMP and ADP compared to the uninfected erythrocytes where the predominant nucleotide was ATP (compare Figures 7A and B).
FIGURE 7. Qualitative analysis of adenine metabolism in erythrocytes, infected erythrocytes, and erythrocyte-free P. falciparum.
Erythrocytes (A), infected erythrocytes (B), and saponin-released parasites (C) were placed in purine-free media for 30 min. [3H]adenine was then added to the culture for 2 h. Purine metabolites were extracted and identified by HPLC and liquid scintillation spectrometry. Bars represent the mean ± SD of triplicate experiments. CPM, counts per minute. This is a qualitative profiling of the metabolic fate of the [3H]adenine, thus the absolute numbers between panels cannot be compared.
Isolated parasites accumulated [3H]adenine mostly as unmetabolized adenine with small amounts of adenine nucleotides (Figure 7C). Thus, adenine was not rapidly incorporated into the nucleoside/nucleotide pool within the parasites in the absence of the erythrocyte. The presence of small amounts of tritium label in other purine metabolites in the saponin-released parasites may result from a small percentage of unlysed infected erythrocytes contaminating the free parasites. Alternatively, there might be a low-efficiency P. falciparum APRTase activity, although no P. falciparum APRT gene has been identified [28].
Inhibition of [14C]2′-deoxyadenosine uptake with modified adenine compounds
The apparent affinities of adenine, adenosine, and 2′-deoxyadenosine were low relative to other members of the ENT family of proteins (reviewed in [29]). To investigate if other compounds were recognized by ENT4 with higher affinity, we tested the ability of modified adenine compounds, namely the cytokinin plant hormones isopentenyl adenine, trans-zeatin, and cis-zeatin, to compete with [14C]2′-deoxyadenosine uptake. All three compounds inhibited [14C]2′-deoxyadenosine uptake (Figure 8, Table 1). Isopentenyl adenine inhibited with the highest apparent affinity, Ki’s of 3.3 ± 0.9 μM for PfENT4 and 9.5 ± 2.8 μM for PvENT4. Trans-zeatin had a higher affinity for PfENT4 than for PvENT4, with Ki’s observed to be 20 and 193 μM, respectively. The apparent affinity of cis-zeatin for PfENT4 and PvENT4 appeared to be similar with observed Ki’s of 190 and 284 μM, respectively.
FIGURE 8. Concentration-dependent inhibition of 2′-deoxyadenosine uptake by plant cytokinins.
Inhibition of 1.5 μM [14C]2′-deoxyadenosine uptake into oocytes expressing PfENT4 (black circles), PvENT4 (gray triangles), or H2O-injected oocytes (white squares) with (A, B) isopentenyl adenine, (C, D) trans-zeatin, or (E, F) cis-zeatin. Each point in panels A, C, and E are the mean ± SD uptake of four oocytes from one experiment. Each point in panels B, D, and F are background-subtracted mean ± SD uptake into oocytes expressing PfENT4 and PvENT4 normalized to a % of uptake in the absence of unlabeled competitor. Data are fit with a single-site competition curve and IC50 values determined (see table 1).
Inhibition of adenine uptake by purine nucleoside phosphorylase transition state analogues and ENT inhibitors
We sought to determine whether other substrates interacted with PfENT4 or PvENT4 expressed in oocytes. The Immucillins are nucleoside analogue inhibitors of purine nucleoside phosphorylase, however they were not substrates for PfENT1 [9]. Competition experiments indicated that the Immucillins inhibited [3H]adenine uptake but only in the millimolar range, similar to the affinity for the other purines tested (Figure 9). Seven millimolar Immucillin-H caused significantly greater inhibition of PfENT4 than 2 mM, suggesting the dose-response relationship has a steep Hill slope coefficient if it were to be analyzed as a sigmoidal function. Plasma concentrations of uric acid, a product of human purine catabolism, is hypothesized to be increased during P. falciparum infection, creating a steep concentration gradient in the circulation, which could be potentially transported into parasites [30]. Urate, the conjugate base of uric acid, was not recognized by ENT4 because 500 μM urate did not compete for adenine transport (Figure 9). Adenine uptake was inhibited by ~80% by 10 μM dipyridamole, an inhibitor of other ENT proteins, but was insensitive to 10 μM NBMPR (Figure 9). Hypoxanthine did not inhibit adenine uptake, confirming that hypoxanthine is not transported or recognized by PfENT4 or PvENT4 (Figure 2A).
DISCUSSION
Resolving transport and metabolism
In this work we sought to identify the function and substrate specificity of ENT4 homologues from P. falciparum and P. vivax by heterologously expressing Xenopus codon-optimized ENT4 transcripts in Xenopus oocytes. Our results indicate that PfENT4 and PvENT4 are low affinity adenine, adenosine, and 2′-deoxyadenosine transporters (Figure 2, 4, 5). They can also transport some guanine derived nucleobases and nucleosides. Studies of nucleoside/nucleobase transport are complicated by the fact that many of these substrates are metabolized once inside the cell to potentially non-transportable compounds. This may affect kinetic analysis of the observed flux depending on whether transport or metabolism is the rate limiting step [9, 13]. Metabolic analysis of purines indicates that Xenopus oocytes expressing PfENT4 or PvENT4, transport adenine and metabolize it to ATP. The results are consistent with the sequential actions of APRT and adenylate kinase. For both PfENT4 and PvENT4, adenine inhibition of [3H]adenine uptake into oocytes had a biphasic inhibition profile: one component with a low micromolar apparent Ki and one component with a low millimolar apparent Ki. In contrast, adenine inhibition of [3H]adenosine or [14C]2′-deoxyadenosine uptake displayed only a single competition site in the millimolar range. The calculated apparent Ki for adenine varied by a factor of five depending on whether [3H]adenine or [3H]adenosine was used as the tracer (Table 1), however, using [14C]2′-deoxyadenosine yielded similar results as [3H]adenine. The difference in calculated Ki may relate to the transport rate differences between adenosine and 2′-deoxyadenosine, but what is clear is that adenine competes with adenosine at one site with a millimolar apparent Ki value that is attributable to transport and not metabolism. This implies that adenine has a low affinity for PfENT4 and PvENT4. Of note, none of the four purine uptake pathways defined by Quashie et al. [12] in isolated P. falciparum parasites have affinities consistent with those of PfENT4. As has been pointed out previously, it is likely that in their studies in P. falciparum parasites, the metabolic enzymes and not the transporters were rate limiting in uptake experiments; thus their calculated affinities are those of the various metabolic enzymes and not the transporters [9, 13].
For a number of the substrates tested, the uptake was greater in PvENT4 expressing oocytes than in PfENT4 expressing oocytes (Figure 2A). These differences could be due to differences in the level of expression of the two proteins. This might be due to differences in the codon-optimization process used to generate the DNA constructs or may be due to the differences in amino acid sequence between the two ENT4 homologues that are 64% sequence identical. Alternatively, they may represent differences in the substrate translocation rates for the two ENT4 homologues. Without being able to accurately quantify the levels of protein expression in each experiment we cannot distinguish between these two possibilities.
The rate of purine transport by oocytes expressing PfENT4 and PvENT4 was significantly slower than for oocytes expressing PfENT1. This could be due to differences in the level of protein expression and/or to differences in the rate of transport by the specific proteins. At present we cannot distinguish between these alternatives.
ENT4 specificity
For PfENT4, adenine and 2′-deoxyadenosine transport was similar and 7-, 9 -and 10-fold greater than adenosine, guanine and guanosine, respectively (Figure 2A). Purine transport studies in saponin-released ATP-depleted pfENT1Δ knockout parasites have demonstrated adenine uptake but no measurable uptake of adenosine, guanine or guanosine [15]. We infer that PfENT4 may account for the observed adenine uptake in the PfENT1-knockout parasites; but PfENT4 may be expressed at low levels in the parasites making the uptake of adenosine, guanine and guanosine undetectable under the experimental conditions.
Ion effects on ENT4 function
Adenine transport via PfENT4/PvENT4 was inhibited by decreasing pH. This implies that neither transporter is a proton-purine cotransporter. The pKa for this inhibition was between pH 5.5 and 6.0 (Figure 3A). This suggests that the imidazole moiety of a critical histidine residue(s) is being titrated across the range of pH values, thereby inhibiting transporter function. Another possibility is that this transporter is a proton antiporter, though we think this is less likely because we would have expected the PvENT4 transport process to be more sensitive to pH-inhibition because of its faster rates of adenine transport. The molecular basis of this pH-dependent inhibition is currently under investigation.
We observed that adenine uptake into oocytes bathed in a choline chloride buffer was less than in oocytes bathed in a sodium chloride buffer. This may reflect a weak interaction of choline with the ENT4 proteins resulting in inhibition of adenine uptake. Given that 140 mM choline only inhibited transport by 18-26% (Figure 3B) we did not investigate this effect in greater detail.
Inhibitor specificity of ENT4
In contrast to PfENT1, PfENT4 and PvENT4-mediated uptake of adenine was inhibited by the classical nucleoside transport inhibitor dipyridamole. Dipyridamole inhibits growth of chloroquine-resistant P. falciparum parasites in vitro, though the reported efficacy varies with IC50 values ranging from 30 nM [31] to 13.9 μM [32]. Dipyridamole inhibits recombinant P. falciparum phosphodiesterase-1, however, the mechanism of dipyridamole’s antiparasitic activity in vitro is not known [33]. Previous work from our lab and others have demonstrated that PfENT1 is insensitive to dipyridamole [6, 8, 10], thus dipyridamole’s inhibitory effect must be exerted through another mechanism, and ENT4 may be contributing to that mechanism. Similar to PfENT1, neither PfENT4 nor PvENT4 were inhibited by NBMPR.
Adenine metabolism in P. falciparum
Of the radiolabeled substrates tested, adenine and 2′-deoxyadenosine were transported the fastest by PfENT4 and PvENT4. It is not clear if either of these substrates play a role in purine salvage during the intraerythrocytic growth stages of P. falciparum. Human serum has low concentrations of 2′-deoxyadenosine and the erythrocyte lacks nuclei, so 2′-deoxyadenosine is not likely to be a significant nutrient for the intraerythrocytic parasites. Additionally, human erythrocytes have <0.4 μM adenine in their cytoplasm [34]. Our metabolic labeling data (Figure 7C) support the hypothesis that P. falciparum does not efficiently use adenine in purine salvage for nucleic acids. In the absence of the erythrocyte, adenine was only converted to nucleotides at a very low level and accumulated inside the parasites mainly as unmetabolized adenine [3]. We cannot rule out that the conversion of labeled adenine to nucleotides was due to a minor contamination of the isolated parasites by infected erythrocytes.
We previously demonstrated that P. falciparum is capable of salvaging AMP from the erythrocyte cytosol as an alternative purine source [4]. Thus, the metabolic pattern observed in infected erythrocytes likely due to erythrocytic APRT activity and AMP transport. Although previous reports described an adenine phosphoribosyl transferase-like activity in purified P. falciparum parasite lysates and saponin-released parasites, the observed purine incorporation occurs at rates consistent with minor contamination of the samples with erythrocyte cytoplasm [35, 36]. Genome-wide searches of apicomplexan parasites failed to find APRT genes, which are conserved across other species [28].
The results presented here suggest that purine substrates for the purine salvage pathway, through recognized and transported by PfENT4 and PvENT4, may not be the primary substrates for these transporters in vivo during the intraerythrocytic stages. Given the broad expression pattern of ENT4 during the Plasmodium life cycle, it may play a role in purine salvage, for example, during replication in hepatocytes or within the mosquito. Alternatively, other yet to be identified compounds may be the true substrates of ENT4.
Of note, the energetic state of infected erythrocytes appeared to be impaired compared to uninfected erythrocytes. In uninfected erythrocytes, adenine was almost completely converted to ATP (Figure 7A). In contrast, in infected erythrocytes about one-half of the radiolabeled adenine was present as AMP or ADP (Figure 7B). We infer that a lack of sufficient energy production in the trophozoite-infected erythrocyte cytoplasm diminished their ability to convert adenine to AMP and then to ADP.
Cytokinins are ENT4 ligands
The plant cytokinin hormones, adenine derivatives, inhibited 2′-deoxyadenosine uptake with apparent affinities ten- to one thousand-fold higher than adenine, adenosine, or 2′-deoxyadenosine. We infer that the inhibition of 2′-deoxyadenosine uptake is due to transport of these adenine derivatives, but lacking radiolabeled compounds we could not demonstrate uptake of these substances into the oocytes. The interaction of these adenine derivative with Pf- and Pv-ENT4 was surprising, especially since hypoxanthine, which also differs at the C6 carbon position in the purine ring, was not recognized by PfENT4 or PvENT4 either by direct transport or in competition with adenine. This implies that substrate recognition by these transporters is sensitive to the chemical moiety at the purine C6 carbon position.
Although the physiological relevance of the relatively high affinity of PfENT4 and PvENT4 for the cytokinin compounds is not known, purine transporters in other organisms are involved in the translocation of these compounds [37-39]. Additionally, isopentenyl adenine was reported to be lethal to in vitro growth of P. falciparum FCR-3 strain parasites with an IC50 of 8.34 μM [40]. PfENT4 mediated transport to an intracellular target may give rise to the reported inhibition of growth.
AMP is not transported by ENT4
One motivation for expressing and characterizing PfENT4 and PvENT4 was to determine if these transporters are responsible for mediating uptake of AMP and/or the Immucillin class of compounds. Previous work showed that PfENT1 does not transport these compounds [4, 9]. Here, we show that AMP was not transported by PfENT4 or PvENT4. In contrast, the Immucillin compounds were recognized with millimolar affinity by these two transporters as evidenced by their ability to inhibit adenine uptake (Figure 9). Of note, Immucillin H had a stronger interaction with the PfENT4 transporter than with PvENT4. The basis for this is unclear but may relate to amino acid sequence differences between the two ENT4 homologues. It is not clear if the ENT4 transporters are the predominant route for the uptake of Immucillins to reach their target, purine nucleoside phosphorylase, in the parasite cytoplasm [41].
In conclusion, we showed that PfENT4 and PvENT4 are low-affinity adenine, adenosine and 2′-deoxyadenosine transporters. Their purine substrate specificity is distinct from PfENT1. Whether the physiological function of the ENT4 transporters is for purine uptake for the purine salvage pathway or for transport of other important substrates, such as the cytokinins, remains to be determined. If the primary substrate transported by parasite ENT4s is critical to the parasites growth and survival then that might explain why Plasmodium, and potentially other protozoan parasites, encode multiple nucleoside transporters. We speculate that nucleoside transporters may not only satisfy nutritional requirements for the parasite, but may be important proteins for translocating other potentially important molecules that are yet to be identified.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Peter C. Tyler and Gary B. Evans at Industrial Research Ltd. for providing the Immucillin compounds. We thank Julia Goldberg, Jon Hefter, and Asif Rahman for technical assistance. We thank Drs. Moez Bali, Keith Hazleton, Nicole McKinnon, Rishi Parikh and Paul Riegelhaupt for helpful discussions. Data in this paper are from a thesis to be submitted in partial fulfillment of the requirements for the Degree of Doctor of Philosophy in the Graduate Division of Medical Sciences, Albert Einstein College of Medicine, Yeshiva University.
FUNDING
I. J. Frame was supported in part by an NIH Medical Scientist Training Program grant [T32-GM007288]. This work was supported in part by a grant from the National Institutes of Health to VLS [RO1-AI049512].
Abbreviations used
- APRT
adenine phosphoribosyltransferase
- DADMeImm
4′-deaza-1′-aza-2′-deoxy-1′-(9-methylene) immucillin
- DEPC
diethylpyrocarbonate
- ENT
equilibrative nucleoside transporter
- HPLC
high performance liquid chromatography
- Imm
Immucillin
- NBMPR
nitrobenzylmercaptopurineriboside
- PfENT1
Plasmodium falciparum Equilibrative Nucleoside Transporter 1
- PfENT4
Plasmodium falciparum Equilibrative Nucleoside Transporter 4
- PNP
purine nucleoside phosphorylase
- PvENT1
Plasmodium vivax Equilibrative Nucleoside Transporter4.
Footnotes
AUTHOR CONTRIBUTIONS
I. J. Frame, Emilio Merino, Vern Schramm, María Cassera and Myles Akabas designed the experiments. Emilio Merino designed the codon-normalized constructs. I. J. Frame and Maria Cassera performed the experiments. I. J. Frame, Emilio Merino, Vern Schramm, María Cassera and Myles Akabas wrote and/or edited the manuscript.
REFERENCES
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO . World malaria report: 2011. World Health Organization; 2011. [Google Scholar]
- 3.Downie MJ, Kirk K, Mamoun CB. Purine salvage pathways in the intraerythrocytic malaria parasite Plasmodium falciparum. Eukaryot. Cell. 2008;7:1231–1237. doi: 10.1128/EC.00159-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassera MB, Hazleton KZ, Riegelhaupt PM, Merino EF, Luo M, Akabas MH, Schramm VL. Erythrocytic adenosine monophosphate as an alternative purine source in Plasmodium falciparum. J. Biol. Chem. 2008;283:32889–32899. doi: 10.1074/jbc.M804497200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldwin SA, McConkey GA, Cass CE, Young JD. Nucleoside transport as a potential target for chemotherapy in malaria. Curr. Pharm. Des. 2007;13:569–580. doi: 10.2174/138161207780162845. [DOI] [PubMed] [Google Scholar]
- 6.Parker MD, Hyde RJ, Yao SY, McRobert L, Cass CE, Young JD, McConkey GA, Baldwin SA. Identification of a nucleoside/nucleobase transporter from Plasmodium falciparum, a novel target for anti-malarial chemotherapy. Biochem. J. 2000;349:67–75. doi: 10.1042/0264-6021:3490067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Downie MJ, Saliba KJ, Broer S, Howitt SM, Kirk K. Purine nucleobase transport in the intraerythrocytic malaria parasite. Int. J. Parasitol. 2008;38:203–209. doi: 10.1016/j.ijpara.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Downie MJ, Saliba KJ, Howitt SM, Broer S, Kirk K. Transport of nucleosides across the Plasmodium falciparum parasite plasma membrane has characteristics of PfENT1. Mol. Microbiol. 2006;60:738–748. doi: 10.1111/j.1365-2958.2006.05125.x. [DOI] [PubMed] [Google Scholar]
- 9.Riegelhaupt PM, Cassera MB, Frohlich RF, Hazleton KZ, Hefter JJ, Schramm VL, Akabas MH. Transport of purines and purine salvage pathway inhibitors by the Plasmodium falciparum equilibrative nucleoside transporter PfENT1. Mol. Biochem. Parasitol. 2010;169:40–49. doi: 10.1016/j.molbiopara.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riegelhaupt PM, Frame IJ, Akabas MH. Transmembrane segment 11 appears to line the purine permeation pathway of the Plasmodium falciparum equilibrative nucleoside transporter 1 (PfENT1) J. Biol. Chem. 2010;285:17001–17010. doi: 10.1074/jbc.M110.115758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carter NS, Ben Mamoun C, Liu W, Silva EO, Landfear SM, Goldberg DE, Ullman B. Isolation and functional characterization of the PfNT1 nucleoside transporter gene from Plasmodium falciparum. J. Biol. Chem. 2000;275:10683–10691. doi: 10.1074/jbc.275.14.10683. [DOI] [PubMed] [Google Scholar]
- 12.Quashie NB, Dorin-Semblat D, Bray PG, Biagini GA, Doerig C, Ranford-Cartwright LC, De Koning HP. A comprehensive model of purine uptake by the malaria parasite Plasmodium falciparum: identification of four purine transport activities in intraerythrocytic parasites. Biochem. J. 2008;411:287–295. doi: 10.1042/BJ20071460. [DOI] [PubMed] [Google Scholar]
- 13.Kirk K, Howitt SM, Broer S, Saliba KJ, Downie MJ. Purine uptake in Plasmodium: transport versus metabolism. Trends Parasitol. 2009;25:246–249. doi: 10.1016/j.pt.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 14.El Bissati K, Zufferey R, Witola WH, Carter NS, Ullman B, Ben Mamoun C. The plasma membrane permease PfNT1 is essential for purine salvage in the human malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9286–9291. doi: 10.1073/pnas.0602590103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Bissati K, Downie MJ, Kim SK, Horowitz M, Carter N, Ullman B, Ben Mamoun C. Genetic evidence for the essential role of PfNT1 in the transport and utilization of xanthine, guanine, guanosine and adenine by Plasmodium falciparum. Mol. Biochem. Parasitol. 2008;161:130–139. doi: 10.1016/j.molbiopara.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin RE, Henry RI, Abbey JL, Clements JD, Kirk K. The ‘permeome’ of the malaria parasite: an overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 2005;6:R26. doi: 10.1186/gb-2005-6-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downie MJ, El Bissati K, Bobenchik AM, Nic Lochlainn L, Amerik A, Zufferey R, Kirk K, Ben Mamoun C. PfNT2, a permease of the equilibrative nucleoside transporter family in the endoplasmic reticulum of Plasmodium falciparum. J. Biol. Chem. 2010;285:20827–20833. doi: 10.1074/jbc.M110.118489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 19.Bozdech Z, Zhu J, Joachimiak MP, Cohen FE, Pulliam B, DeRisi JL. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daily JP, Scanfeld D, Pochet N, Le Roch K, Plouffe D, Kamal M, Sarr O, Mboup S, Ndir O, Wypij D, Levasseur K, Thomas E, Tamayo P, Dong C, Zhou Y, Lander ES, Ndiaye D, Wirth D, Winzeler EA, Mesirov JP, Regev A. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007;450:1091–1095. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- 21.Lemieux JE, Gomez-Escobar N, Feller A, Carret C, Amambua-Ngwa A, Pinches R, Day F, Kyes SA, Conway DJ, Holmes CC, Newbold CI. Statistical estimation of cell-cycle progression and lineage commitment in Plasmodium falciparum reveals a homogeneous pattern of transcription in ex vivo culture. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7559–7564. doi: 10.1073/pnas.0811829106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silvestrini F, Lasonder E, Olivieri A, Camarda G, van Schaijk B, Sanchez M, Younis Younis S, Sauerwein R, Alano P. Protein export marks the early phase of gametocytogenesis of the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics. 2010;9:1437–1448. doi: 10.1074/mcp.M900479-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ, Jr., Treatman C, Wang H. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jespersen T, Grunnet M, Angelo K, Klaerke DA, Olesen SP. Dual-function vector for protein expression in both mammalian cells and Xenopus laevis oocytes. BioTechniques. 2002;32:536–538. 540. doi: 10.2144/02323st05. [DOI] [PubMed] [Google Scholar]
- 25.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 26.Broer S. Xenopus laevis Oocytes. Methods Mol. Biol. 2010;637:295–310. doi: 10.1007/978-1-60761-700-6_16. [DOI] [PubMed] [Google Scholar]
- 27.Yao SYM, Cass CE, Young JD. The Xenopus oocyte expression system for the cDNA cloning and characterization of plasma membrane transport proteins. In: Baldwin SA, editor. Membrane Transport. Oxford University Press Inc.; New York: 2000. pp. 47–76. [Google Scholar]
- 28.Chaudhary K, Darling JA, Fohl LM, Sullivan WJ, Jr., Donald RG, Pfefferkorn ER, Ullman B, Roos DS. Purine salvage pathways in the apicomplexan parasite Toxoplasma gondii. J. Biol. Chem. 2004;279:31221–31227. doi: 10.1074/jbc.M404232200. [DOI] [PubMed] [Google Scholar]
- 29.Young JD, Yao SY, Sun L, Cass CE, Baldwin SA. Xenobiotica; the fate of foreign compounds in biological systems. 2008;38:995–1021. doi: 10.1080/00498250801927427. [DOI] [PubMed] [Google Scholar]
- 30.Orengo JM, Leliwa-Sytek A, Evans JE, Evans B, van de Hoef D, Nyako M, Day K, Rodriguez A. Uric acid is a mediator of the Plasmodium falciparum-induced inflammatory response. PloS One. 2009;4:e5194. doi: 10.1371/journal.pone.0005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akaki M, Nakano Y, Ito Y, Nagayasu E, Aikawa M. Effects of dipyridamole on Plasmodium falciparum-infected erythrocytes. Parasitol. Res. 2002;88:1044–1050. doi: 10.1007/s00436-002-0690-8. [DOI] [PubMed] [Google Scholar]
- 32.Gero AM, Scott HV, O’Sullivan WJ, Christopherson RI. Antimalarial action of nitrobenzylthioinosine in combination with purine nucleoside antimetabolites. Mol. Biochem. Parasitol. 1989;34:87–97. doi: 10.1016/0166-6851(89)90023-6. [DOI] [PubMed] [Google Scholar]
- 33.Yuasa K, Mi-Ichi F, Kobayashi T, Yamanouchi M, Kotera J, Kita K, Omori K. PfPDE1, a novel cGMP-specific phosphodiesterase from the human malaria parasite Plasmodium falciparum. Biochem. J. 2005;392:221–229. doi: 10.1042/BJ20050425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 35.Queen SA, Vander Jagt DL, Reyes P. Characterization of adenine phosphoribosyltransferase from the human malaria parasite, Plasmodium falciparum. Biochim. Biophys. Acta. 1989;996:160–165. doi: 10.1016/0167-4838(89)90242-2. [DOI] [PubMed] [Google Scholar]
- 36.Mehrotra S, Bopanna MP, Bulusu V, Balaram H. Adenine metabolism in Plasmodium falciparum. Exp. Parasitol. 2010;125:147–151. doi: 10.1016/j.exppara.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Cedzich A, Stransky H, Schulz B, Frommer WB. Characterization of cytokinin and adenine transport in Arabidopsis cell cultures. Plant Physiol. 2008;148:1857–1867. doi: 10.1104/pp.108.128454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirose N, Makita N, Yamaya T, Sakakibara H. Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiol. 2005;138:196–206. doi: 10.1104/pp.105.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 2008;59:75–83. doi: 10.1093/jxb/erm157. [DOI] [PubMed] [Google Scholar]
- 40.Harmse L, van Zyl R, Gray N, Schultz P, Leclerc S, Meijer L, Doerig C, Havlik I. Structure-activity relationships and inhibitory effects of various purine derivatives on the in vitro growth of Plasmodium falciparum. Biochem. Pharmacol. 2001;62:341–348. doi: 10.1016/s0006-2952(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 41.Kicska GA, Tyler PC, Evans GB, Furneaux RH, Kim K, Schramm VL. Transition state analogue inhibitors of purine nucleoside phosphorylase from Plasmodium falciparum. J. Biol. Chem. 2002;277:3219–3225. doi: 10.1074/jbc.M105905200. [DOI] [PubMed] [Google Scholar]
- 42.Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T. ConPred II: a consensus prediction method for obtaining transmembrane topology models with high reliability. Nucleic Acids Res. 2004;32:W390–393. doi: 10.1093/nar/gkh380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johns SJ. TOPO2, Transmembrane protein display software. http://www.sacs.ucsf.edu/TOPO2/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.