Abstract
Although endodermal organs including the liver, pancreas, and intestine are of significant therapeutic interest, the mechanism by which the endoderm is divided into organ domains during embryogenesis is not well understood. To better understand this process, global gene expression profiling was performed on early endodermal organ domains. This global analysis was followed up by dynamic immunofluorescence analysis of key transcription factors, uncovering novel expression patterns as well as cell surface proteins that allow prospective isolation of specific endodermal organ domains. Additionally, a repressive interaction between Cdx2 and Sox2 was found to occur at the prospective stomach-intestine border, with the hepatic and pancreatic domains forming at this boundary, and Hlxb9 was revealed to have graded expression along the dorsal-ventral axis. These results contribute to understanding the mechanism of endodermal organogenesis and should assist efforts to replicate this process using pluripotent stem cells.
Keywords: Endoderm, organogenesis, flow cytometry, transcriptional profiling, axial patterning
Introduction
One of the first cell fate decisions made during vertebrate embryogenesis is the separation into germ layers: ectoderm, mesoderm and endoderm. The endoderm, which first forms on the outside of the embryo and is later internalized, gives rise to the digestive and respiratory tracts and their associated organs. Endodermally derived organs, including the liver, pancreas, lungs, intestine and stomach, are of considerable therapeutic interest, but much remains to be learned about how they first emerge from the primitive gut tube.
In the mouse, the definitive endoderm forms during gastrulation as a single-cell thick sheet of cells on the outside of the cup-shaped embryo. As gastrulation proceeds, this definitive endoderm migrates anteriorly, displacing the extraembryonic visceral endoderm (Lawson and Pedersen, 1987). The endodermal sheet subsequently begins to fold in at the anterior and posterior ends, forming the anterior and caudal intestinal portals (AIP and CIP respectively). The ventral lips of these tubes extend toward the center of the embryo, causing the sheet of endoderm to fold progressively into a tube, so that by the 13–15-somite stage (embryonic day 8.75 [E8.75] in the mouse), the entire endoderm has been internalized as a tube. Localized increased proliferation and/or migration instigated by mesodermal cues causes budding which is apparent by E9.5 in what will become the lung, liver, and dorsal and ventral pancreata. After E9.5, the specified endodermal organ domains as well as the distinct sections of the gut tube (esophagus, stomach, intestine) undergo morphogenesis and lineage diversification to create the functional architecture specific to each organ.
There is evidence suggesting that when endoderm is initially formed (between E6.5–E7.5 in the mouse), it is not yet committed to specific organ domains. When explanted mouse E7.5 anterior endoderm is placed in contact with posterior mesoderm, posterior endodermal genes can be activated and vice versa (Wells and Melton, 2000). Explants of mouse E8.25 ventral endoderm have the potential to activate pancreas-, liver-or lung-specific genes depending on their proximity to cardiac mesoderm or the concentration of fibroblast growth factors (Deutsch et al., 2001; Serls et al., 2005). Endoderm differentiation potential has been investigated further in chick embryos using heterotopic transplantation. In these experiments, 5–12-somite (equivalent to E8.25-E8.5) endoderm alters positional marker expression when transplanted more posteriorly yet retains positional markers when transplanted anteriorly (Kumar et al., 2003), and positional marker expression is committed by the 10–12-somite-stage (Kimura et al., 2007).
Clues to how the endoderm is patterned into organ domains have been uncovered but have not been pieced together to yield a coherent mechanism. In the mouse, genes that are eventually specific to the lungs, liver, and pancreas can be detected in small numbers of ventral endodermal cells at E8.5 (Serls et al., 2005), although detectable budding of these organ rudiments from the endodermal tube does not begin until one day later. Detailed transcription factor expression during this critical time period is lacking. Several signaling pathways, including those activated by retinoic acid (Stafford and Prince, 2002; Molotkov et al., 2005; Pan et al., 2007), bone morphogenetic protein (BMP) (Tiso et al., 2002; Shin et al., 2007), fibroblast growth factor (FGF) (Jung et al., 1999; Wells and Melton, 2000; Serls et al., 2005; Dessimoz et al., 2006), Wnt (Ober et al 2007, McLin et al 2007), Hedgehog (Hebrok et al., 1998) and Activin (Hebrok et al., 1998; Kim et al., 2000) families have been implicated in endoderm organ formation, yet little is known about direct transcriptional targets of these pathways. Additionally, adjacent tissues such as notochord (Kim et al., 1997), endothelium (Lammert et al., 2001; Matsumoto et al., 2001), cardiac mesoderm (Deutsch et al., 2001), septum transversum mesenchyme (Rossi et al., 2001), and lateral plate mesoderm (Kumar et al., 2003)are known to be important for endoderm organogenesis. Much work remains to fit these observations into a mechanistic model. One of the most basic facets of the organogenesis process that is poorly understood is the dynamics of expression of key transcription factors. Such knowledge could help to connect information about signaling pathways and inductive tissues to their specific transcriptional targets.
In order to begin to comprehend how the embryonic gut tube is subdivided into organ forming regions, we have focused on the expression of key transcription factors. These genes are, at a minimum, useful geographical markers and, in some cases, demonstrably play a role in specifying cell fates. We first performed microarrays of the endodermal progenitors in six organ domains to obtain an inclusive view of the factors expressed during organogenesis. We have followed up these microarrays by characterizing the dynamic cellular expression pattern for numerous genes that have restricted expression within the endodermal organs. These experiments provide a descriptive view of how the endoderm is patterned along its axes and where the organ domains form.
Materials and Methods
Animals and flow cytometry
For all experiments, outbred ICR mice were bred and maintained at the Harvard Biomedical Research Infrastructure. Embryos were considered to be E0.5 at noon of the day the plug was detected.
Dissected tissues pooled from 10–12 embryos per sample were dissociated in 0.25% Trypsin (Invitrogen, Carlsbad, CA) for 2–5 minutes at 37 degrees and, after neutralization and centrifugation, were stained on ice for 15 minutes in DMEM/F12 (Invitrogen, Carlsbad, CA) with 2% FBS (Hyclone, Logan, UT) and 2 mM EDTA. Before flow cytometric sorting, cells were resuspended in staining media with calcein blue AM (Invitrogen, Carlsbad, CA) and sorted using a FACSAria (Becton Dickinson, San Jose, CA).
Electroporation experiments were performed according to published protocols (Pierreux et al., 2005). Full-length transcription factor cDNA was cloned into the pCIG vector, which has a chicken beta actin (CAGGS) promoter driving expression of a gene followed by an IRES driving GFP expression. Embryos were bathed in 1 μg/μL plasmid solution and electroporated using a T820 Electro Square Porator (BTX, Holliston, MA) using three 12-Volt pulses with 50 μs interval. Embryos were cultured for 24 hours in 1:1 DMEM:F12 (Invitrogen, Carlsbad, CA): rat serum (Valley Biomedical, Winchester, VA).
Microarrays
Sorted cell populations were collected in Trizol (Invitrogen, Carlsbad, CA), and RNA was isolated according to the manufacturer’s instructions. RNA was amplified and biotinylated cRNA probe generated using the Ambion Illumina TotalPrep RNA Amplification Kit (Applied Biosystems/Ambion, Austin, TX). Biotinylated cRNA was hybridized onto Illumina MouseRef-8 v2 microarrays (Illumina, San Diego, CA) according to the manufacturer’s instructions. Microarrays were scanned using Beadstation (Illumina, San Diego, CA), and data was analyzed using BeadStudio (Illumina, San Diego, CA). Three biological replicates were performed for each tissue analyzed.
Antibodies and immunostaining
The following primary antibodies were used: G8.8 (anti-EpCAM), F6A11 (anti-Pdx1), 74.5A5 (anti-Nkx2-2; Developmental Studies Hybridoma Bank, Iowa City, IA); goat anti-Foxa2 (M-20), goat anti-Sox2 (Y-17), rabbit anti-Hnf6 (H-100), rabbit anti-Hnf4a (H-171; Santa Cruz Biotechnology, Santa Cruz, CA); rat anti-Liv2, rat anti-Dlk1 (MBL International, Woburn, MA); biotinylated DBA (Vector Labs, Burlingame, CA); mouse anti-Prox1, rabbit anti-Otx2 (Millipore, Billerica, MA); mouse anti-Cdx2 (Biogenex, San Ramon, CA); rabbit anti-Hlxb9, rat anti-Pax9 (Abcam, Cambridge, MA); guinea pig anti-Pdx1 (gift from Christopher Wright); rabbit anti-Tbx1, mouse anti-Titf1 (Invitrogen, Carlsbad, CA); sheep anti-Cdcp1, rat anti-Rae1 (R and D Systems, Minneapolis, MN); mouse anti-Tcf2 and FITC-conjugated rat anti-CD26 (BD Biosciences, San Jose, CA). Alexa Fluor 488, 594, 647 and Pacific Blue secondary antibody conjugates (Invitrogen, Carlsbad, CA) as well as biotin, PE and APC conjugates (Jackson ImmunoResearch, West Grove, PA) were used for secondary detection.
For wholemount immunostaining, whole embryos were blocked for 1 hour in PBS with 1% Tween-20, 20% donkey serum (Jackson ImmunoResearch, West Grove, PA), and when mouse primary antibodies were used, Vector M.O.M. Blocking Reagent (Vector, Burlingame, CA). Primary and secondary antibody stainings were performed rocking overnight at 4 degrees in PBS with 1% Tween-20 and 5% donkey serum, and embryos were washed rocking for at least 5 hours at 4 degrees in PBS with 1% Tween-20.
After staining, wholemount embryos were transferred to 1:1 glycerol:PBS with 1% Tween and placed between two coverslips. Confocal imaging was performed using an LSM 510Meta confocal microscope (Carl Zeiss Inc., Germany). Pictures of whole embryos were taken using a fluorescent dissecting microscope (Leica Microsystems, Germany).
Quantitative image analysis
Total nuclear transcription factor expression was measured using Imaris 6.0.1 software (Bitplane, St. Paul, MN). The program reconstructed individual samples in three dimensions from confocal optical sections. The Surpass “add new spots” automated function was used to approximate nuclei as solid spheres of diameter 5 μm in user-defined regions of interest. Spots were filtered and manually edited to obtain one spot centered in each endoderm nucleus. When necessary, nuclei were divided into subgroups using the “Duplicate selection to new spots object” function. Intensity mean was taken to reflect the gene expression and exported to Microsoft Excel 2007 (Microsoft, Redmond, WA), and statistical analysis was performed in Excel as required.
Results
Microarray analysis of endodermal organ progenitors
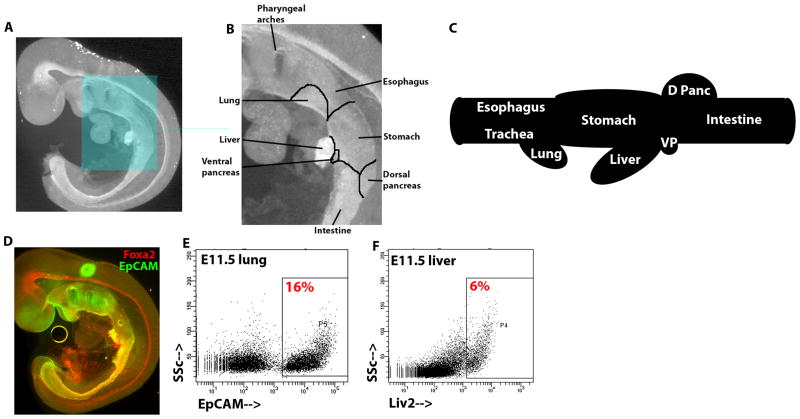
To study gene expression during endodermal organogenesis, we sought to identify genes expressed in restricted domains during organogenesis. By E9.5, the endoderm is a tube that spans the embryo from anterior to posterior. All endodermal cells, as well as the notochord and floor plate of the neural tube, express Foxa2 (Ang et al., 1993; Monaghan et al., 1993; Ruiz i Altaba et al., 1993); Figure 1a). Nascent organs are apparent as slight protrusions from the endoderm (Figure 1b). These protrusions bud outward and gain a defined shape between E9.5–11.5 (Figure 1c). The cell surface protein EpCAM is expressed most strongly in endoderm in the early mouse embryo (Sherwood et al., 2007), and EpCAM is expressed throughout the endoderm after organogenesis as well (Figure 1d). Between E9.5–E10.5, the hepatic endoderm loses expression of EpCAM (data not shown) but can be identified by expression of the cell surface protein Liv2 (Nierhoff et al., 2005).
Figure 1.
Diagrams and isolation strategy of endodermal organ domains. (A) E9.5 embryo wholemount immunofluorescence image of Foxa2 antibody staining. Boxed region is magnified in (B), and the nascent organ domains within the continuous endodermal epithelium are demarcated and labeled. (C) Schematic of the gut tube at E11.5 with the distinct organ domains labeled. (D) E9.5 embryo wholemount immunofluorescence image of Foxa2 (red) and EpCAM (green). (E and F) Flow cytometric analysis of live, dissected (E) E11.5 lung stained with EpCAM (X-axis) and (F) E11.5 liver stained with Liv2 (X-axis), plotted against side scatter (Y-axis). The percentage of cells within the boxed region is displayed.
For gene expression analysis, six morphologically distinct endodermal domains were dissected at E11.5: the esophageal region; the lung and distal tracheal region; the stomach region; the hepatic region; the dorsal and ventral pancreatic region; and the intestinal region. Through flow cytometric separation using EpCAM expression to distinguish endoderm from surrounding mesenchyme, pure populations of endoderm progenitors from the esophageal, lung, stomach, pancreatic, and intestinal regions were isolated (Figure 1e, data not shown). Expression of Liv2 was used to isolate a pure population of hepatic endoderm progenitors (Figure 1f).
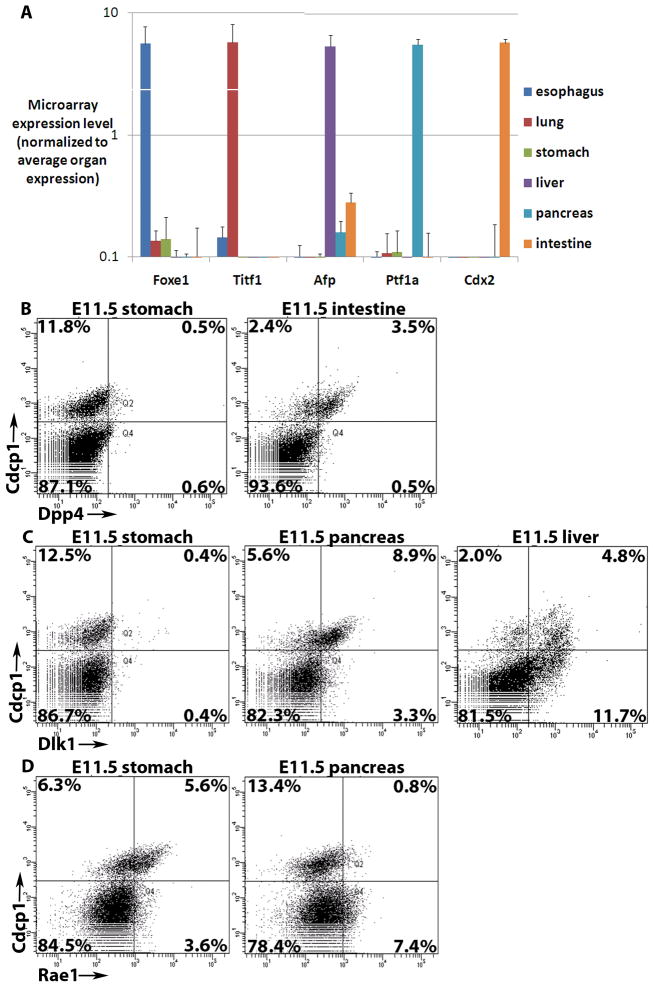
Microarray analysis was performed on the six endodermal organ progenitor populations using Illumina whole genome arrays. Three biological replicates of each sample were obtained, and average expression among replicates was used to compare expression in tissues. To confirm the validity of the dissection and flow cytometric isolation of each domain, expression values of genes reported to be expressed in specific endodermal organs were analyzed. The esophagus-enriched Foxe1 (Dathan et al., 2002), lung-enriched Titf1 (Lazzaro et al., 1991), hepatic-enriched Afp (Shiojiri, 1981), pancreas-enriched Ptf1a (Krapp et al., 1998) and intestine-enriched Cdx2 (Beck et al., 1995)were highly enriched in the appropriate microarray samples (Figure 2a). Definitive endoderm from E7.75, E8.25 (4–6-somite stage), and E8.75 (13–15-somite stage) were also collected and profiled using high expression of EpCAM and lack of DBA expression (Sherwood et al 2007). These data are available online at http://www.syscode.org/news.phpand at GEO (reference GSE13040).
Figure 2.
Results of microarray analysis of endodermal organs. (A) Graph of microarray expression values of five genes reported to be specific to endodermal organ domains. Arbitrary expression values are normalized to the average expression among the six organ domains profiled and are displayed on a log scale with standard deviation of three replicates. (B–D) Flow cytometric analysis of live, dissected (B) E11.5 stomach (left) and intestine (right) stained with Dpp4 (X-axis), (C) E11.5 stomach (left), pancreas (middle) and liver (right) stained with Dlk1 (X-axis), and (D) E11.5 stomach (left) and pancreas (right) stained with Rae1 (X-axis), all plotted against pan-endodermal Cdcp1 (Y-axis). The percentage of cells within each quadrant is displayed.
Statistical criteria were used to identify transcription factors with a restricted expression pattern in the endoderm. A total of 93 transcription factors were found to be expressed non-ubiquitously in E11.5 endoderm organs (Table 1). Several known and novel transcription factors were specifically expressed in a single organ domain (Table 1). For example, the pancreas specifically expresses the known transcription factors Myt1, Neurog3, Nkx2-2, Nkx6-1, and Ptf1a as well as the novel factors Rxrg and Tcf15. The majority (55/93) of the organ-expressed transcription factors are expressed in multiple domains. Some are expressed in anterior-posterior segments. Hoxa1, Hoxa2, Irx3, Irx5, Sox2, and Sox21 are expressed in the anterior three organs (esophagus, lung and stomach), and Dmrt3 and Tbx1 are expressed in the esophagus and lung. Conversely, Foxa3 and Gata6 are expressed in the five most posterior organs (lung, stomach, liver, pancreas and intestine), Hnf4a and Onecut2 in the stomach, liver, pancreas and intestine, and Nr5a2 in the liver, pancreas and intestine (Table 2). However, there are a significant number of distinct expression patterns such as those of Osr2, which is expressed in the esophagus and intestine, and Mrg1, which is expressed in the esophagus, stomach and pancreas (Supplemental Table 1). Thus, while not unexpected, there is no simple relationship of a transcription factor or set of factors that accurately and uniquely marks the emergence of an organ in the endoderm.
Table 1. Transcription factors exclusive to one organ within the endoderm.
Expression of these transcription factors is >3-fold higher in one organ than in all other organs, and expression is not detected significantly above background in other organs.
| Esophagus | Lung | Stomach | Liver | Pancreas | Intestine |
|---|---|---|---|---|---|
| Dlx3 | Foxp2 | Odd1 | Cebpa | Myt1 | Cdx2 |
| Erf | Irx1 | Creb3l3 | Neurog3 | Evx1 | |
| Foxe1 | Titf1 | Klf2 | Nkx2-2 | Hoxa7 | |
| Nfix | Nkx6-1 | Hoxa9 | |||
| Nrl | Ptf1a | Hoxb6 | |||
| Otx1 | Rxrg | Hoxb7 | |||
| Pltx1 | Tcf15 | Hoxb9 | |||
| Tcfap2c | Hoxd10 | ||||
| Twist1 | Hoxd3 | ||||
| Zfpn1a2 | Hoxd4 | ||||
| Hoxd8 | |||||
| Hoxd9 | |||||
| Pltx2 | |||||
| Tff3 |
Table 2. Transcription factors expressed in anterior-posterior blocks of the endoderm.
Expression of these transcription factors is >3-fold higher in the organs indicated than in all other organs, and expression is not detected significantly above background in other organs. Gray shading represents expression of the noted transcription factors.
| Esophagus | Lung | Stomach | Liver | Pancreas | Intestine |
|---|---|---|---|---|---|
| Dmrt3, Tbx1 | |||||
| Hoxa1, Hoxa2, Irx3, Irx5, Sox2, Sox21 | |||||
| Foxa3, Gata6 | |||||
| Hnf4a, Onecut2 | |||||
| Onecut3 | |||||
| Cited1, Hhex, Prox1 | |||||
| Nr5a2 | |||||
The Hox family of transcription factors has been implicated in a wide array of embryonic patterning events. Endodermal expression of Hox genes has been reported (Manley and Capecchi, 1995; Beck et al., 2000); however, a systematic understanding of their expression and role in endoderm development is lacking. We found 19 of the 39 Hox genes to be expressed in the endoderm. Because anterior boundary of Hox expression has been demonstrated to be of major functional significance in other germ layers (Duboule and Morata, 1994), this boundary was analyzed in the endoderm. Of the 19 endoderm-expressed Hox genes, nine have an anterior boundary within the esophageal endoderm, although their actual anterior boundary may be in the more anterior pharyngeal endoderm, and ten are exclusively expressed in the intestine (Supplemental Table 1), suggesting that Hox genes may play more essential roles in pharyngeal and intestinal development than in the formation of the other major endodermal organs.
While they are not believed to be as integral to cell fate specification as transcription factors, cell surface proteins are useful in allowing for isolation of specific populations by flow cytometry, so differentially expressed cell surface proteins were sought. A number of cell surface proteins expressed specifically in a single organ domain were identified (Supplemental Table 2). Only a small subset of these proteins have commercially available antibodies suitable for flow cytometry, so these antibodies as well as antibodies to some cell surface proteins expressed in combinations of organs were tested by flow cytometry. By flow cytometric co-staining with the pan-endodermal Cdcp1 (Sherwood et al 2007), it was found that Dpp4 is specifically expressed in intestinal endoderm (Figure 2b), Dlk1 is expressed in pancreatic and hepatic endoderm (Figure 2c), and Rae1 is expressed most strongly in stomach endoderm (Figure 2d).
An Endoderm Transcription Factor Map at E9.5
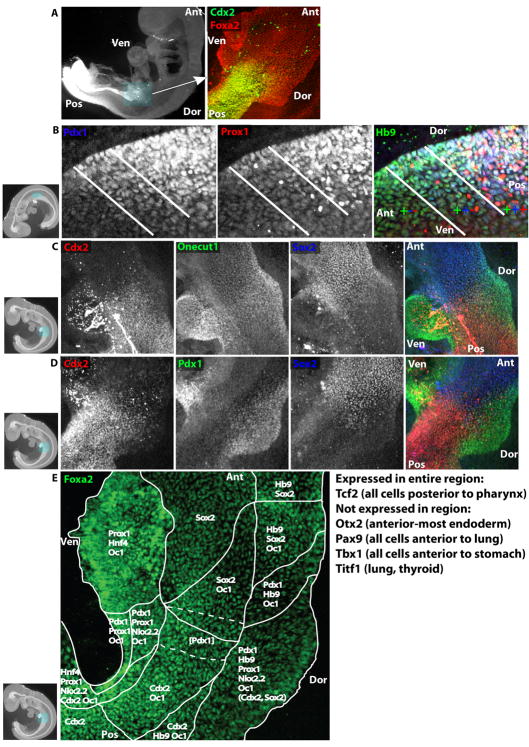
To examine the expression of key genes throughout the endoderm at cellular resolution, immunofluorescence staining was performed on whole embryos at E9.5 and analyzed using confocal microscopy to achieve three-dimensional cellular resolution. This method allows annotation of expression of transcription factors such as the intestine-specific Cdx2 in the embryo as a whole and with sufficient resolution to analyze expression in individual endodermal cells (Figure 3a).
Figure 3.
A high-resolution transcriptional map of the E9.5 stomach-intestine border. (A) E9.5 embryo wholemount immunofluorescence image of Cdx2 antibody staining (left). Boxed region is magnified in right panel, a wholemount confocal immufluorescence image of the stomach-intestine border co-stained with Cdx2 (green) and Foxa2 (red). (B–E) E9.5 embryo wholemount confocal immunofluorescence images. (B) Dorsal pancreatic region co-stained with Pdx1 (left and blue in merged image on right), Prox1 (middle, red), and Hlxb9 (abbreviated as Hb9, green). White lines demarcate regions of single expression of Hlxb9, co-expression of Hlxb9 and Pdx1, and co-expression of Hlxb9, Pdx1 and Prox1. (C) Stomach-intestine border co-stained with Cdx2 (left, red), Onecut1 (middle, green), and Sox2 (right, blue) and merged image (far right). (D) Stomach-intestine border co-stained with Cdx2 (left, red), Pdx1 (middle, green), and Sox2 (right, blue) and merged image (far right). (E) Stomach-intestine border stained with Foxa2. White lines demarcate regions of expression of transcription factors listed in each region. Dotted white lines demarcate a sub-region where, in addition to all other factors listed in the two solid-line regions, Pdx1 (in parentheses) is expressed weakly and is intermixed with Pdx1− cells. In the dorsal Pdx1-expressing region, Sox2 and Cdx2 (in parentheses) are expressed atE8.75, but they recede anteriorly and posteriorly respectively and become entirely excluded by E9.5. In thumbnail images, boxes highlight displayed region, and axes of embryo are labeled. Ant=Anterior, Pos=Posterior, Dor=Dorsal, Ven=Ventral.
Endodermal expression patterns of 15 transcription factors were investigated at E9.5. Our analysis was limited by the commercial availability of specific antibodies suitable for immunofluorescence. The region in which the liver and pancreas bud was of special interest, and as indicated by the microarray analysis, restricted expression of Cdx2, Hlxb9 (Hb9), Hnf4a, Nkx2-2, Onecut1, Pdx1, Prox1, and Sox2 was detected in this region. While expression of these genes in this region has been reported previously, the wholemount confocal microscopy technique allowed resolution of their domains at significantly higher resolution and allowed analysis of co-expression, which has not been rigorously examined. Although this region will eventually become the stomach, pancreas, liver and intestine, comparative analysis revealed multiple distinct expression patterns such as in the dorsal pancreatic region where Hlxb9 is expressed throughout the dorsal endoderm, and Pdx1 extends more broadly than Prox1 (Figure 3b), yet all are co-expressed in much of the dorsal pancreatic region and are necessary for normal pancreatic development (Jonsson et al., 1994; Offield et al., 1996; Harrison et al., 1999; Li et al., 1999; Wang et al., 2005). Sox2 is expressed in most anterior endodermal cells, although it is excluded from certain regions including the nascent lung (data not shown; (Que et al., 2007)). At the anterior-posterior level of the dorsal and ventral pancreatic buds, Sox2 expression ceases and forms a border with Cdx2, which is expressed in all posterior endoderm cells (Figure 3c). Neither Cdx2 nor Sox2 is expressed on the dorsal or ventral edges of the endoderm in this region. Onecut1 expression spans this Cdx2-Sox2 boundary and is additionally expressed in the dorsal and ventral hepatopancreatic regions (Figure 3c), consistent with results from in situ hybridization (Landry et al., 1997; Rausa et al., 1997). Pdx1 expression is mostly exclusive with Sox2 and Cdx2, as it is expressed dorsally and in the small ventral pancreatic region (Figure 3d). From this analysis, a cellular map of domains of transcription factor expression within the region spanning the nascent stomach, pancreas, liver and intestinal endoderm at E9.5 can be constructed (Figure 3e).
Anterior-Posterior Patterning Dynamics
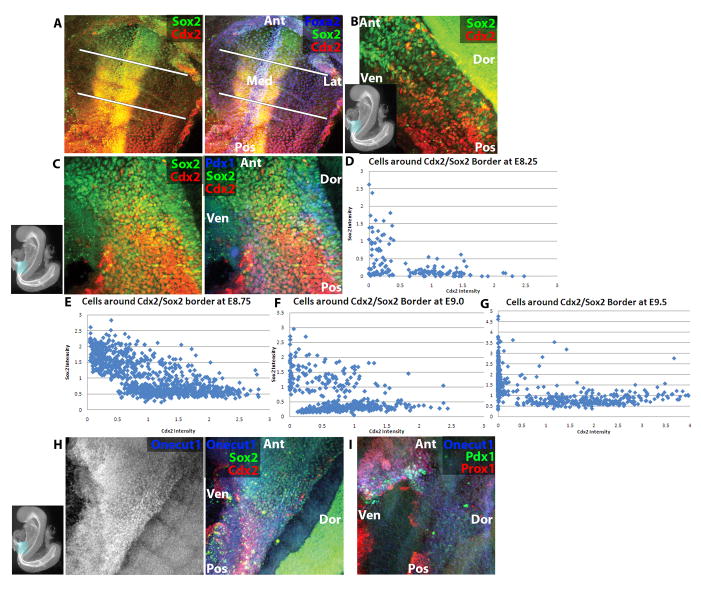
Because the transcription factor expression patterns hint at developmental mechanism, the dynamic expression of transcription factors with distinct anterior-posterior boundaries was analyzed in detail. Endodermal Sox2 and Cdx2 can be detected at E7.75 in the most anterior and posterior cells, respectively, significantly prior to Onecut1, Pdx1 or Prox1 (data not shown). Between E8.0–E8.5, the expression domain of Sox2 extends progressively posteriorly, and Cdx2 extends progressively anteriorly (Figure 4a–b). By E8.75 (13–15-somite stage), theSox2 and Cdx2 expression boundaries meet slightly posterior to the foregut-midgut junction, and there is co-expression of these genes in cells at the boundary (Figure 4c). This co-expression recedes between the 13–15 somite stage and E9.0, and as noted above, the reciprocal Sox2-Cdx2 expression pattern remains at E9.5, except that some dorsal and ventral cells expressing pancreatic and hepatic genes express neither Sox2 nor Cdx2 (Figure 3c–d). To provide a quantitative view of this border formation, average nuclear intensities of Sox2 and Cdx2 were calculated from cells spanning this border at several time points. At the 6–8-somite-stage, before Cdx2 and Sox2 form a visible border, the expected distribution of cells expressing either Sox2 alone, Cdx2 alone or neither is found (Figure 4d). At the 13–15-somite stage, there is a population of cells expressing both factors (Figure 4e). At E9.0, there are still a few cells that co-express Cdx2 and Sox2 yet most have resolved to express either Cdx2 or Sox2 (Figure4f), and by E9.5, the border is sharp with little to no co-expression (Figure 4g).
Figure 4.
Anterior-posterior transcriptional dynamics at the stomach-intestine border. (A) E8.25 (4–6-somite stage) embryo wholemount confocal immunofluorescence image of Sox2 (green), Cdx2 (red) and Foxa2 (blue, right only). White lines demarcate region of endoderm expressing neither Cdx2 nor Sox2. (B) E8.5 (9–11-somite stage) embryo wholemount confocal immunofluorescence image of Sox2 (green) and Cdx2 (red). (C) E8.75 (13–15-somite stage) embryo wholemount confocal immunofluorescence image of Sox2 (green), Cdx2 (red) and Pdx1 (blue, right only). (D–G) Graphs plotting mean nuclear intensity ratio of Cdx2 (X-axis) to Sox2 (Y-axis) in populations of cells spanning the stomach-intestine border at E8.25 6–8-somite stage (D), E8.75 (E), E9.0 (F), and E9.5 (G). Each dot represents one nucleus, and intensity values of Cdx2 and Sox2 are normalized as ratios to Foxa2 expression. (H) E8.5 (6–8-somite stage) embryo wholemount confocal immunofluorescence image of Sox2 (green, right only), Cdx2 (red, right only) and Onecut1 (blue). (I) E8.75 (11–13-somite stage) embryo wholemount confocal immunofluorescence image of Pdx1 (green), Prox1 (red) and Onecut1 (blue). In thumbnail images, boxes highlight displayed region, and axes of embryo are labeled. Ant=Anterior, Pos=Posterior, Med=Medial, Lat=Lateral, Dor=Dorsal, Ven=Ventral.
Because Cdx2 and Sox2 form a border in the middle of the pancreatic and hepatic domains at E9.5, the dynamics of pancreatic and hepatic transcription factors were studied in conjunction with Cdx2 and Sox2. Of the pancreatic and hepatic transcription factors analyzed, Onecut1 is expressed earliest, with expression starting in a stripe of foregut and midgut cells that covers the entire dorsal-ventral endoderm at the 6–8-somite stage and spans both the Sox2 and Cdx2 expression domains (Figure 4h). While the Sox2 and Cdx2 expression domains converge, Onecut1 remains expressed in a stripe spanning these two domains through E9.5 (Figure 3c). Prox1, which has been shown by in situ hybridization to be expressed in the nascent pancreas and liver (Burke and Oliver, 2002)becomes expressed in the ventral Onecut1-expressing cells by the 9–11-somite stage, and a subset of these cells ventral cells begins to express Pdx1 at the 11–13-somite stage (Figure 4i). Dorsal Prox1 and Pdx1 expression do not appear until the 13–15-somite stage, and these cells co-express Onecut1 and either Sox2 or Cdx2 (Figure 4c, data not shown), although Sox2 and Cdx2 are lost in this dorsal Pdx1-expressing domain by E9.5 (Figure 3d).
To begin to link these expression patterns with the mechanism of anterior-posterior patterning in the endoderm, the effect of ectopic expression of transcription factors was studied. Plasmids carrying Cdx2 or Pdx1 DNA driven by a constitutive chicken beta-actin promoter were electroporated into the E8.25 foregut (anterior) endoderm (Pierreux et al., 2005). After electroporation, embryos were cultured for 24 hours and analyzed by wholemount immunofluorescence and confocal microscopy. Mean nuclear intensity of Sox2 in cells ectopically expressing Cdx2 or Pdx1 to their neighbors was determined. Cells ectopically expressing Cdx2 have on average 50% as much Sox2as their untransfected neighbors (Figure 5a–b), whereas Pdx1 has no effect on Sox2 expression (Figure 5c–d). Conversely, Sox2 was transfected into the posterior endoderm, and cells ectopically expressing Sox2 have 80% as much Cdx2 as their untransfected neighbors (Figure 5e–f). Thus, Cdx2 appears capable of downregulating Sox2 expression, providing a feasible explanation for stomach-intestine border formation.
Figure 5.
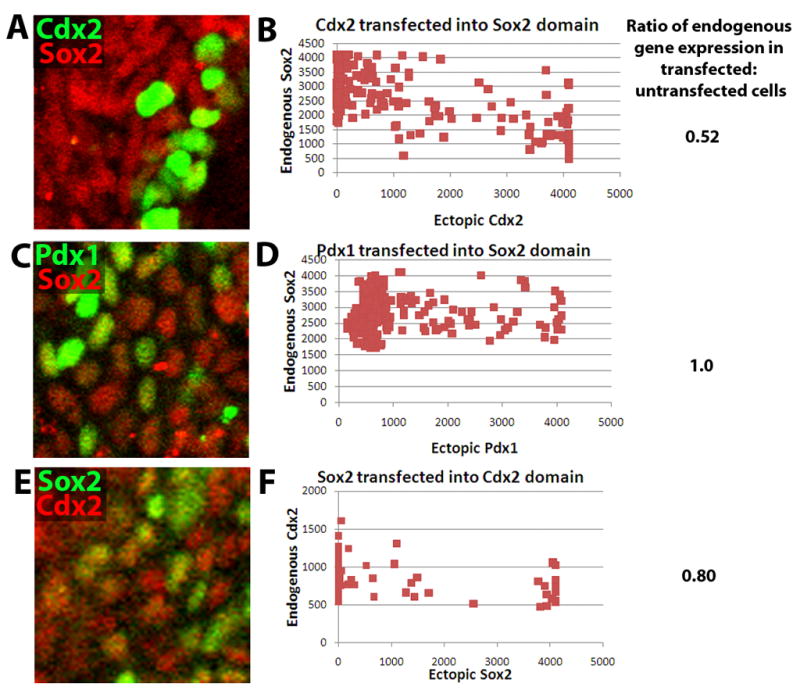
Dominance of Cdx2 over Sox2 in anterior-posterior patterning. (A, C, E) Wholemount confocal immunofluorescence of E8.25 embryos electroporated with (A) pCAGGS Cdx2 IRES GFP, (B) pCAGGS Pdx1 IRES GFP, or (C) pCAGGS Sox2 IRES GFP and cultured for 24 hours in 1:1 DMEM-F12:rat serum. In (A), Cdx2 is in green and Sox2 in red, in (B), Pdx1 is in green and Sox2 in red, and in (C), Sox2 is in green and Cdx2 in red. The endogenous gene is typically expressed in all nuclei within the imaged territory, so green nuclei are indicative of ectopic gene downregulation of the endogenous gene, whereas yellow nuclei are indicative of lack of downregulation. (B, D, F) Graphs plotting mean nuclear intensity ratio of the transfected factor (X-axis) to the endogenous gene (Y-axis). The average ratio of endogenous gene expression in transfected vs. untransfected cells is displayed to the right of each graph.
Hlxb9 forms a dorsal-ventral gradient at E9.5
While a large number of transcription factors have restricted anterior-posterior expression patterns, only one transcription factor, Hlxb9, which is expressed throughout the anterior-posterior axis of the endoderm, shows a dorsal-ventral asymmetry, as has been reported (Harrison et al., 1999; Li et al., 1999). Hlxb9 begins expression within the endoderm at E8.0 (data not shown). By E8.25, the endoderm is a single-cell thick layer with around 25 cells from left to right. Hlxb9 is expressed in the medial-most half of the endoderm surrounding the strongly Hlxb9-expressing notochord (Figure 6a). At E8.5, after the anterior endoderm has undergone tubulogenesis to form the AIP, Hlxb9 retains expression in the dorsal-most 10–15 cell layers, presumably the descendants of the medial-most cells (Figure 6b). At E9.5, expression is still detected in the dorsal half of the endoderm; however, expression appears to be strongest in the most dorsal endoderm and progressively weaker in more ventral cells (Figure 6c).
Figure 6.
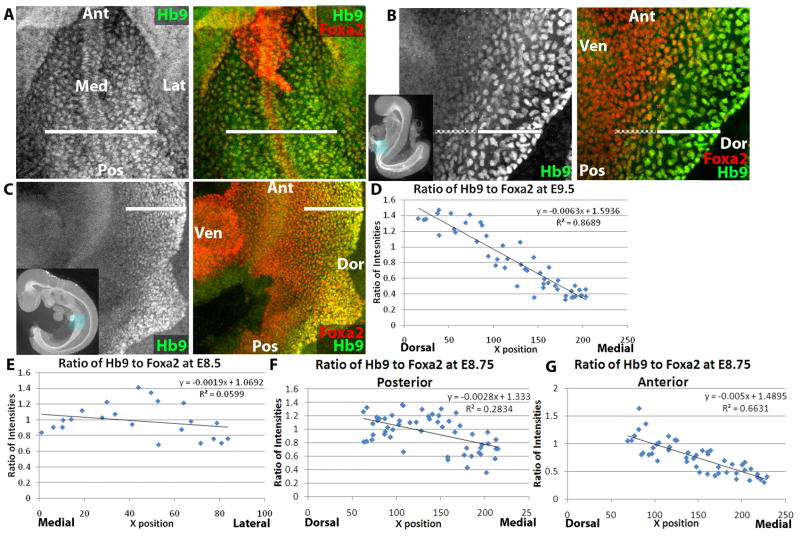
Hlxb9 forms a dorsal-ventral gradient within the endoderm. (A) E8.25 (4–6-somite stage) embryo wholemount confocal immunofluorescence image of Hlxb9 (abbreviated as Hb9, green) and Foxa2 (red, right only). White line demarcates region of endoderm expressing Hlxb9. (B) E8.5 (9–11-somite stage) embryo wholemount confocal immunofluorescence image of Hlxb9 (green) and Foxa2 (red, right only). Solid white line demarcates region of endoderm expressing Hlxb9 strongly and dashed white line demarcates region expressing Hlxb9 weakly. (C) E9.5 embryo wholemount confocal immunofluorescence image of Hlxb9 (green) and Foxa2 (red, right only). White line demarcates region of endoderm expressing Hlxb9, although levels of Hlxb9 expression decrease from dorsal to ventral. (D–G) Graphs plotting ratio of mean nuclear intensity of Hlxb9 to Foxa2 (Y-axis) in endodermal cells at E9.5 (D), E8.5 (E), and E8.75 (F–G). Cells are plotted by their position along the dorsal-ventral axis (X-axis) from dorsal to medial (D, F, G) or medial to lateral (E). Equation and R2 value are shown for each graph. In panels F–G, graphs are of posterior (F) and anterior (G) regions of the same embryo. In thumbnail images, boxes highlight displayed region, and axes of embryo are labeled. Ant=Anterior, Pos=Posterior, Med=Medial, Lat=Lateral, Dor=Dorsal, Ven=Ventral.
As Hlxb9 appears to form a dorsal-ventral gradient of expression, statistical tools were employed to determine whether this expression is indeed graded. The uniform endodermal expression of Foxa2 was used to normalize differences in intensity based on position within the epithelium. Thus, the average ratio of nuclear intensity of Hlxb9:Foxa2 was calculated for a dorsal-ventral line of endoderm in several anterior-posterior positions. When plotted, normalized Hlxb9 expression at E9.5 forms a linear gradient of intensity (Figure 6d). This gradient forms between E8.5 and E9.5, as no linear decay of Hlxb9 can be observed along the dorsal-ventral axis at E8.5 (Figure 6e).
At E8.75, some detectable dorsal-ventral asymmetry appears but without a clear gradient. In more posterior regions, the dorsal-most half of the Hlxb9-expressing endoderm (which is itself the dorsal half of the endoderm) has significantly higher expression than the ventral Hlxb9-expressing half (Figure 6f). There is a sharp difference between the two halves, and there is no gradient or dorsal-ventral asymmetry within each half (Supplemental Figure 1). More anteriorly, the two discernable halves are still present, but a gradient begins to form in the ventral Hlxb9-expressing cells (Figure 6g, Supplemental Figure 1). Thus, Hlxb9 begins to form a gradient at E8.75, starting at the anterior end of the endoderm.
To determine whether a gradient forms by strengthening of Hlxb9 expression dorsally or weakening ventrally, the ratio of Hlxb9:Foxa2 expression was spatially and temporally compared. At all time points, regions of endoderm lacking visual Hlxb9 expression have a Hlxb9:Foxa2 ratio of 0.3 (data not shown), due to non-nuclear background. At E8.5, the ratio between Hlxb9 and Foxa2 averages 1, varying randomly with respect to the medial-lateral axis between 0.6 and 1.4. At E8.75, the dorsal half of the Hlxb9 positive region maintains an average expression ratio of 1, varying between 0.6 and 1.4, while the average ratio in the ventral half decreases to 0.7, varying between 0.4 and 1. While average Hlxb9 expression for each half at E8.75 does not change along the anterior-posterior axis, the ventral Hlxb9-positive half begins to take on a gradient, as noted above. At E9.5, at all anterior-posterior regions, the ratio decreases linearly from 1.4 to 0.3, suggesting that increased Hlxb9 expression in the most dorsal cells as well as decreases more ventrally contribute to gradient formation.
Discussion
During a 24 hour period during mouse organogenesis between E8.0–E9.0, the endoderm transforms from a flat sheet with little known heterogeneity into a tube in which the domains for all of the major organs have been defined. It has been appreciated that molecular heterogeneity precedes morphological differentiation, yet the transcriptional dynamics of endoderm organogenesis have not been well characterized. To begin to shed light on endodermal organogenesis, a combination of global transcriptional profiling and confocal imaging at cellular resolution has been applied.
Transcriptional profiling reveals the complexities of the organogenesis process. Few transcription factors are unique to individual organs. Organogenesis is likely to rely on multiple distinct signaling and transcriptional inputs, as is underlined by the fact that embryos lacking both Pdx1 and Ptf1a, the two transcription factors most vital to pancreatic development, still form a rudimentary pancreatic bud (Burlison et al., 2008) and ectopic expression of Pdx1 in the endoderm is insufficient to drive complete pancreatic morphogenesis (Grapin-Botton et al., 2001). Thus, these few organ-specific transcription factors are unlikely to be the sole effectors of organ morphogenesis. Instead, the large cohort of transcription factors expressed in multiple, but not all organ domains, are likely to make important, combinatorial inputs to organogenesis. Expression domains alone do not yield simple clues to mechanisms of organogenesis, as there are transcription factors expressed in many different combinations of organs; however, an increased understanding of mechanisms driving expression of these factors as well as their network interactions will be valuable.
Transcriptional profiling also allowed for the identification of cell surface proteins that can be used to isolate specific endoderm organ populations. Each organ has a signature set of cell surface proteins (Supplemental Table 2) that can be exploited to isolate organ progenitors as well as subsets of cells within each organ. Using commercially available antibodies, intestinal endoderm can be isolated by its expression of Dpp4, hepatic and pancreatic endoderm by their expression of Dlk1, and stomach endoderm by its strong expression of Rae1. The ability to isolate specific populations of endoderm organ progenitors will be useful for embryonic stem cell differentiation experiments, in which cell types are not identifiable by morphology.
As a complementary approach to the global transcriptional profiling, wholemount immunofluorescent staining followed by confocal imaging allowed elucidation of expression patterns at cellular resolution. This approach allows the inspection of transcriptional borders, which is difficult to do with wholemount in situ hybridization because of a lack of cellular resolution, or with section immunofluorescence, in which only a small region can be analyzed. Such analysis is vital to comprehending the mechanism of organogenesis, as the molecular markers analyzed are likely the downstream effectors of organ-forming signals and transcriptional pathways.
Observing expression of Cdx2 and Sox2 between E8.0–E9.5 reveals an interesting reciprocal relationship. Both genes become expressed in the endoderm before somitogenesis, when the entire endoderm is a flat sheet (summarized in Figure 7a). Cdx2 expression begins in the most posterior endoderm and extends progressively anteriorly, while Sox2 expression begins in the most anterior cells and extends progressively posterior during early somite-stages as the endoderm beings to form a tube at both ends. By the 13–15-somite-stage, the Sox2 and Cdx2 boundaries meet in the midgut and some cells at this border co-express these two genes. This co-expression resolves, as by E9.5, while the cellular border remains, cells at the border express either Sox2 or Cdx2.
Figure 7.
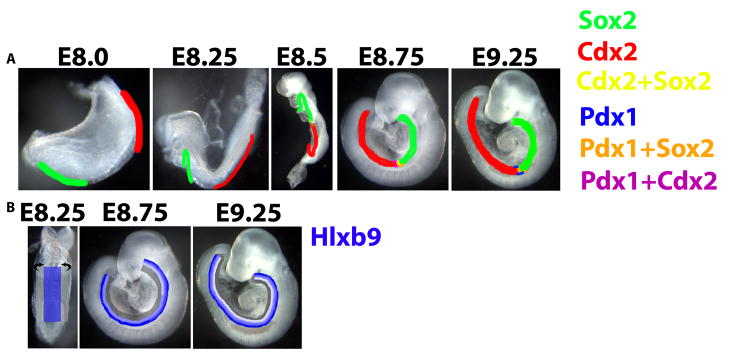
Schematization of anterior-posterior and dorsal-ventral transcriptional dynamics in the endoderm. (A) Endodermal Sox2 (green) and Cdx2 (red) expression start at the anterior and posterior ends of the endoderm, respectively. Between E8.0–E8.5, the domains of these transcription factors expand toward each other, most likely as a result of endodermal cells that do not express either factor initiating expression of Sox2 or Cdx2. At E8.5 (~11–13-somite stage), the Sox2 and Cdx2 expression domains meet, and between E8.75–E9.25, these domains overlap slightly, although by E9.25, co-expression becomes rare. Pdx1 expression begins on the ventral and slightly later the dorsal side of the endoderm at E8.5 at the border of the Cdx2 and Sox2 domains, and the initial Pdx1-expressing cells co-express either Sox2 or Cdx2. By E9.25, dorsal and ventral Pdx1-expressing cells lose expression of Sox2 and Cdx2, although weaker Pdx1 expression is still detected in Sox2-expressing cells immediately anterior to the border of Sox2 and Cdx2. (B) Endodermal Hlxb9 expression (blue) is present at E8.25 at uniform levels in the medial half of the endoderm. Through tubulogenesis, the medial endoderm stays dorsal while the Hlxb9− lateral endoderm wraps ventrally. By E8.75, the most dorsal quarter of the endoderm expresses Hlxb9 strongly, while the next quarter of the dorsal endoderm expresses Hlxb9 more weakly. A gradient of Hlxb9 expression emerges first in the anterior endoderm, which undergoes tubulogenesis earliest, and progressively more posteriorly, such that by E9.25, Hlxb9 is expressed in a gradient in the dorsal half of the endoderm throughout the anterior-posterior axis of the endoderm.
The extension of Cdx2 and Sox2 expression could occur by the migration of cells already expressing these genes, by oriented cell division of expressing cells, or by de novo expression in previously non-expressing cells. Lineage tracing of endoderm suggests that the latter possibility is the most likely. Several lineage tracing studies of mouse somite-stage endoderm have been performed by injecting dye into endodermal cells at specific locations and analyzing their descendants after further development (Lawson and Pedersen, 1987; Tremblay and Zaret, 2005; Franklin et al., 2008), and these studies do not suggest an extensive anterior-posterior migration that would be required for pre-existing Sox2 and Cdx2-expressing cells to colonize the entire endoderm. Thus, anteriorizing and posteriorizing signaling mechanisms must exist that spread expression of these two genes. The nature of these mechanisms is unclear and could consist of signaling molecules spreading toward the center of the embryo from the anterior and posterior poles or of timing-based mechanisms that allow cells that previously were exposed to morphogenic signals to begin to express these genes.
Once Cdx2 and Sox2 meet, a repressive interaction can be posited to stop the intermingling of these two expression domains. In fact, introduction of ectopic Cdx2 into Sox2-expressing foregut cells represses endogenous Sox2, while ectopic Sox2 does not repress Cdx2, suggesting that Cdx2 repression of Sox2 may cause the abrupt border between these genes. On the dorsal and ventral sides, Cdx2 also forms a border with Pdx1. While ectopic expression analysis was not performed to determine the effect of Cdx2 on Pdx1 expression, expression of Cdx4, a homolog of Cdx2, has been shown to limit pancreatic domain expansion in zebrafish (Kinkel et al., 2008). The ability of a more posteriorly expressed transcription factor to downregulate an anterior transcription factor and the lack of the reciprocal effect provides a molecular basis for a commonly invoked concept known as posterior dominance (reviewed in (McGinnis and Krumlauf, 1992; Duboule and Morata, 1994). Posterior dominance has been invoked in endoderm patterning (Kumar et al., 2003; Kimura et al., 2007), and this transcriptional repressive activity may help to explain this phenomenon.
Slightly before Sox2 and Cdx2 meet, at the 6–8-somite stage, the first detectable hepatopancreatic regional marker, Onecut1, begins to be expressed in an anterior-posterior stripe spanning the Sox2 and Cdx2 domains. Onecut1 expression spans the entire dorsal-ventral axis, whereas Prox1 and Pdx1, which initiate expression at the 9–11-somite stage, are initially expressed first ventrally and then dorsally by the 13–15-somite stage but not in the medial endoderm (although by E9.0, Pdx1 is expressed weakly in medial cells that will become the posterior stomach and duodenum), and Hnf4a is expressed solely ventrally. The notochord is known to play a vital role in induction of Pdx1 in the dorsal endoderm (Kim et al., 1997; Hebrok et al., 1998), and the notochord becomes separated from the endoderm in the pancreatic region by the dorsal aorta between the 12–15-somite stage (Yoshitomi and Zaret, 2004). Thus, Pdx1 can first be detected in the dorsal endoderm immediately after the notochord loses contact with the endoderm. This delay may be caused by the time taken for notochord-induced transcription of Pdx1 to be evidenced as protein expression and highlights the precise coordination of morphogenesis necessary for organogenesis, as the displacement of the notochord by the dorsal aorta is necessary to induce Ptf1a expression (Lammert et al., 2001; Yoshitomi and Zaret, 2004).
At their inception, Onecut1, Prox1, Pdx1, and Hnf4a do not share borders, and even at E9.5, Pdx1 and Prox1 do not share an anterior border in the pancreatic domain. The presence of more expression domains than organs in the hepatopancreatic region suggests that multiple signaling mechanisms could be responsible for the initiation of expression of these genes. These additional hepatopancreatic domains could be vital for the formation of distinct structures within this region such as the pancreatic and bile ducts.
While anterior-posterior heterogeneity can be easily tied to the formation of organs at different positions along the anterior-posterior axis, there are fewer clues to the significance of the dorsal-ventral heterogeneity of Hlxb9. Embryos deficient in Hlxb9 have defects in dorsal but not ventral pancreas formation (Harrison et al., 1999; Li et al., 1999), suggesting a role for Hlxb9 in facilitating patterning of dorsal endoderm in the pre-pancreatic region. However, Hlxb9 is expressed throughout the dorsal endoderm, so it is feasible that its absence causes more subtle defects in dorsal-ventral patterning in other regions of the endoderm. The separation of the dorsal esophagus from the ventral trachea and the dorsal curvature and cell type disparity in the stomach are two events that could receive patterning input from Hlxb9.
While the functional significance of Hlxb9 in dorsal-ventral endoderm patterning is murky, its expression pattern clearly suggests that Hlxb9 is interpreting one or multiple dorsal-ventrally asymmetrical cues. Hlxb9 expression is expressed in the medial half of the endoderm by E8.0, and by E8.75, when the medial endoderm has become the dorsal endoderm due to ventral folding of the lateral endoderm during tubulogenesis, its expression level declines in the most ventral Hlxb9-expressing cells (summarized in Figure 7b). Subsequently, slightly increased expression in the most dorsal Hlxb9-expressing cells and graded declines more ventrally create a linear gradient of expression in which each cell has less Hlxb9 than its dorsal neighbor. The linearity and slope of this gradient are within range of the values obtained for Drosophila Bicoid, whose concentration decreases ~7-fold between the anterior pole and the midpoint at a similar concentration/cell slope (Gregor et al., 2007). The data additionally suggest that all cells expressing Hlxb9 at E9.5 are descended from E8.5 Hlxb9-positive cells, barring significant cell movement, because the dorsal 50% of the endoderm consistently expresses Hlxb9.
Thus, the endoderm appears to be receiving graded morphogenic input along the dorsal-ventral axis, the nature of which is uncertain. Hlxb9 could directly or indirectly be positively regulated by signals deriving from medially/dorsally located tissues such as the notochord or negatively regulated by signals from the lateral plate mesoderm, or inputs could be combinatorial. Identifying these signals will be crucial to understanding endoderm organogenesis. Additionally, as Hlxb9 was the only transcription factor found to have dorsal-ventral polarity, it will be important to flush out the other transcription factors involved in dorsal-ventral patterning. One enticing candidate is Hhex, against which we have not found a working commercial antibody but which seems to be preferentially expressed in early ventral endoderm and is necessary for proper formation of the ventral pancreas and liver (Thomas et al., 1998; Martinez Barbera et al., 2000).
This work has provided a large volume of information about the genes that underlie organogenesis and has provided detailed characterization of the dynamics of anterior-posterior and dorsal-ventral patterning that guides the process. Linking these expression patterns and dynamics to causal mechanisms will be the next challenge. Meeting this challenge will likely be essential if one is to achieve efficient directed differentiation of embryonic stem cells to endoderm organ progenitors.
Supplementary Material
Graphs plotting ratio of mean nuclear intensity of Hlxb9 to Foxa2 (Y-axis) in endodermal cells at E8.75. Cells are plotted by their position along the dorsal-ventral axis (X-axis) from dorsal to medial. Equation and R2 value are shown for each graph. (A–D) Dorsal half (A, C) and ventral half (B, D) of the Hlxb9-expressing posterior (A–B) or anterior (C–D) endoderm. Panels A–B are sub-regions of Figure 6F and panels C–D are sub-regions of Figure 6G, split to demonstrate differences in the dorsal and ventral halves of the Hlxb9 expression domain.
Expression of these transcription factors is >3-fold higher in at least one organ than in other organs. Expression is indicated as absent (−), present (+), present at low levels (lo), or present at high levels (hi). Eso=esophagus, Liv=liver, Panc=pancreas, Int=intestine.
Expression of these cell surface proteins is >3-fold higher in one organ than in all other organs, and expression is not detected significantly above background in other organs.
Acknowledgments
D.A.M. is an investigator of the Howard Hughes Medical Institute. R.I.S. was supported by a National Science Foundation Graduate Research Fellowship and the Sternlicht Fellowship. This work was supported by the N.I.H. The authors would like to thank Brian Tilton, Anastasie Kweudjeu, George Kenty, and the Molecular Genetics Core Facility at Children’s Hospital Boston for technical assistance, Christopher Wright for reagents, and William Anderson for review of the manuscript. The G8.8, F6A11, and 74.5A5 antibodies were obtained from the Developmental Studies Hybridoma Bank.
References
- Ang SL, Wierda A, Wong D, Stevens KA, Cascio S, Rossant J, Zaret KS. The formation and maintenance of the definitive endoderm lineage in the mouse: involvement of HNF3/forkhead proteins. Development. 1993;119:1301–1315. doi: 10.1242/dev.119.4.1301. [DOI] [PubMed] [Google Scholar]
- Beck F, Erler T, Russell A, James R. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn. 1995;204:219–227. doi: 10.1002/aja.1002040302. [DOI] [PubMed] [Google Scholar]
- Beck F, Tata F, Chawengsaksophak K. Homeobox genes and gut development. Bioessays. 2000;22:431–441. doi: 10.1002/(SICI)1521-1878(200005)22:5<431::AID-BIES5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Burke Z, Oliver G. Prox1 is an early specific marker for the developing liver and pancreas in the mammalian foregut endoderm. Mech Dev. 2002;118:147–155. doi: 10.1016/s0925-4773(02)00240-x. [DOI] [PubMed] [Google Scholar]
- Burlison JS, Long Q, Fujitani Y, Wright CV, Magnuson MA. Pdx-1 and Ptf1a concurrently determine fate specification of pancreatic multipotent progenitor cells. Dev Biol. 2008;316:74–86. doi: 10.1016/j.ydbio.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dathan N, Parlato R, Rosica A, De Felice M, Di Lauro R. Distribution of the titf2/foxe1 gene product is consistent with an important role in the development of foregut endoderm, palate, and hair. Dev Dyn. 2002;224:450–456. doi: 10.1002/dvdy.10118. [DOI] [PubMed] [Google Scholar]
- Dessimoz J, Opoka R, Kordich JJ, Grapin-Botton A, Wells JM. FGF signaling is necessary for establishing gut tube domains along the anterior-posterior axis in vivo. Mech Dev. 2006;123:42–55. doi: 10.1016/j.mod.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Deutsch G, Jung J, Zheng M, Lora J, Zaret KS. A bipotential precursor population for pancreas and liver within the embryonic endoderm. Development. 2001;128:871–881. doi: 10.1242/dev.128.6.871. [DOI] [PubMed] [Google Scholar]
- Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Franklin V, Khoo PL, Bildsoe H, Wong N, Lewis S, Tam PP. Regionalisation of the endoderm progenitors and morphogenesis of the gut portals of the mouse embryo. Mech Dev. 2008;125:587–600. doi: 10.1016/j.mod.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A, Majithia AR, Melton DA. Key events of pancreas formation are triggered in gut endoderm by ectopic expression of pancreatic regulatory genes. Genes Dev. 2001;15:444–454. doi: 10.1101/gad.846001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W. Probing the limits to positional information. Cell. 2007;130:153–164. doi: 10.1016/j.cell.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison KA, Thaler J, Pfaff SL, Gu H, Kehrl JH. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet. 1999;23:71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- Hebrok M, Kim SK, Melton DA. Notochord repression of endodermal Sonic hedgehog permits pancreas development. Genes Dev. 1998;12:1705–1713. doi: 10.1101/gad.12.11.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Jung J, Zheng M, Goldfarb M, Zaret KS. Initiation of mammalian liver development from endoderm by fibroblast growth factors. Science. 1999;284:1998–2003. doi: 10.1126/science.284.5422.1998. [DOI] [PubMed] [Google Scholar]
- Kim SK, Hebrok M, Li E, Oh SP, Schrewe H, Harmon EB, Lee JS, Melton DA. Activin receptor patterning of foregut organogenesis. Genes Dev. 2000;14:1866–1871. [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Hebrok M, Melton DA. Notochord to endoderm signaling is required for pancreas development. Development. 1997;124:4243–4252. doi: 10.1242/dev.124.21.4243. [DOI] [PubMed] [Google Scholar]
- Kimura W, Yasugi S, Fukuda K. Regional specification of the endoderm in the early chick embryo. Dev Growth Differ. 2007;49:365–372. doi: 10.1111/j.1440-169X.2007.00933.x. [DOI] [PubMed] [Google Scholar]
- Kinkel MD, Eames SC, Alonzo MR, Prince VE. Cdx4 is required in the endoderm to localize the pancreas and limit beta-cell number. Development. 2008;135:919–929. doi: 10.1242/dev.010660. [DOI] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Jordan N, Melton D, Grapin-Botton A. Signals from lateral plate mesoderm instruct endoderm toward a pancreatic fate. Dev Biol. 2003;259:109–122. doi: 10.1016/s0012-1606(03)00183-0. [DOI] [PubMed] [Google Scholar]
- Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294:564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- Landry C, Clotman F, Hioki T, Oda H, Picard JJ, Lemaigre FP, Rousseau GG. HNF-6 is expressed in endoderm derivatives and nervous system of the mouse embryo and participates to the cross-regulatory network of liver-enriched transcription factors. Dev Biol. 1997;192:247–257. doi: 10.1006/dbio.1997.8757. [DOI] [PubMed] [Google Scholar]
- Lawson KA, Pedersen RA. Cell fate, morphogenetic movement and population kinetics of embryonic endoderm at the time of germ layer formation in the mouse. Development. 1987;101:627–652. doi: 10.1242/dev.101.3.627. [DOI] [PubMed] [Google Scholar]
- Lazzaro D, Price M, de Felice M, Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- Li H, Arber S, Jessell TM, Edlund H. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat Genet. 1999;23:67–70. doi: 10.1038/12669. [DOI] [PubMed] [Google Scholar]
- Manley NR, Capecchi MR. The role of Hoxa-3 in mouse thymus and thyroid development. Development. 1995;121:1989–2003. doi: 10.1242/dev.121.7.1989. [DOI] [PubMed] [Google Scholar]
- Martinez Barbera JP, Clements M, Thomas P, Rodriguez T, Meloy D, Kioussis D, Beddington RS. The homeobox gene Hex is required in definitive endodermal tissues for normal forebrain, liver and thyroid formation. Development. 2000;127:2433–2445. doi: 10.1242/dev.127.11.2433. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Yoshitomi H, Rossant J, Zaret KS. Liver organogenesis promoted by endothelial cells prior to vascular function. Science. 2001;294:559–563. doi: 10.1126/science.1063889. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R. Homeoboxgenes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- Molotkov A, Molotkova N, Duester G. Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev Dyn. 2005;232:950–957. doi: 10.1002/dvdy.20256. [DOI] [PubMed] [Google Scholar]
- Monaghan AP, Kaestner KH, Grau E, Schutz G. Post implantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- Nierhoff D, Ogawa A, Oertel M, Chen YQ, Shafritz DA. Purification and characterization of mouse fetal liver epithelial cells with high in vivo repopulation capacity. Hepatology. 2005;42:130–139. doi: 10.1002/hep.20735. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Pan FC, Chen Y, Bayha E, Pieler T. Retinoic acid-mediated patterning of the pre-pancreatic endoderm in Xenopus operates via direct and indirect mechanisms. Mech Dev. 2007;124:518–531. doi: 10.1016/j.mod.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Pierreux CE, Poll AV, Jacquemin P, Lemaigre FP, Rousseau GG. Gene transfer into mouse prepancreatic endoderm by whole embryo electroporation. JOP. 2005;6:128–135. [PubMed] [Google Scholar]
- Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausa F, Samadani U, Ye H, Lim L, Fletcher CF, Jenkins NA, Copeland NG, Costa RH. The cut-homeodomain transcriptional activator HNF-6 is coexpressed with its target gene HNF-3 beta in the developing murine liver and pancreas. Dev Biol. 1997;192:228–246. doi: 10.1006/dbio.1997.8744. [DOI] [PubMed] [Google Scholar]
- Rossi JM, Dunn NR, Hogan BL, Zaret KS. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Prezioso VR, Darnell JE, Jessell TM. Sequential expression of HNF-3 beta and HNF-3 alpha by embryonic organizing centers: the dorsal lip/node, notochord and floor plate. Mech Dev. 1993;44:91–108. doi: 10.1016/0925-4773(93)90060-b. [DOI] [PubMed] [Google Scholar]
- Serls AE, Doherty S, Parvatiyar P, Wells JM, Deutsch GH. Different thresholds of fibroblast growth factors pattern the ventral foregut into liver and lung. Development. 2005;132:35–47. doi: 10.1242/dev.01570. [DOI] [PubMed] [Google Scholar]
- Sherwood RI, Jitianu C, Cleaver O, Shaywitz DA, Lamenzo JO, Chen AE, Golub TR, Melton DA. Prospective isolation and global gene expression analysis of definitive and visceral endoderm. Dev Biol. 2007;304:541–555. doi: 10.1016/j.ydbio.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Shin D, Shin CH, Tucker J, Ober EA, Rentzsch F, Poss KD, Hammerschmidt M, Mullins MC, Stainier DY. Bmp and Fgf signaling are essential for liver specification in zebrafish. Development. 2007;134:2041–2050. doi: 10.1242/dev.000281. [DOI] [PubMed] [Google Scholar]
- Shiojiri N. Enzymo-and immunocytochemical analyses of the differentiation of liver cells in the prenatal mouse. J Embryol Exp Morphol. 1981;62:139–152. [PubMed] [Google Scholar]
- Stafford D, Prince VE. Retinoic acid signaling is required for a critical early step in zebrafish pancreatic development. Curr Biol. 2002;12:1215–1220. doi: 10.1016/s0960-9822(02)00929-6. [DOI] [PubMed] [Google Scholar]
- Thomas PQ, Brown A, Beddington RS. Hex: a homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development. 1998;125:85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- Tiso N, Filippi A, Pauls S, Bortolussi M, Argenton F. BMP signalling regulates anteroposterior endoderm patterning in zebrafish. Mech Dev. 2002;118:29–37. doi: 10.1016/s0925-4773(02)00252-6. [DOI] [PubMed] [Google Scholar]
- Tremblay KD, Zaret KS. Distinct populations of endoderm cells converge to generate the embryonic liver bud and ventral foregut tissues. Dev Biol. 2005;280:87–99. doi: 10.1016/j.ydbio.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Wang J, Kilic G, Aydin M, Burke Z, Oliver G, Sosa-Pineda B. Prox1 activity controls pancreas morphogenesis and participates in the production of “secondary transition” pancreatic endocrine cells. Dev Biol. 2005;286:182–194. doi: 10.1016/j.ydbio.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Wells JM, Melton DA. Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development. 2000;127:1563–1572. doi: 10.1242/dev.127.8.1563. [DOI] [PubMed] [Google Scholar]
- Yoshitomi H, Zaret KS. Endothelial cell interactions initiate dorsal pancreas development by selectively inducing the transcription factor Ptf1a. Development. 2004;131:807–817. doi: 10.1242/dev.00960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Graphs plotting ratio of mean nuclear intensity of Hlxb9 to Foxa2 (Y-axis) in endodermal cells at E8.75. Cells are plotted by their position along the dorsal-ventral axis (X-axis) from dorsal to medial. Equation and R2 value are shown for each graph. (A–D) Dorsal half (A, C) and ventral half (B, D) of the Hlxb9-expressing posterior (A–B) or anterior (C–D) endoderm. Panels A–B are sub-regions of Figure 6F and panels C–D are sub-regions of Figure 6G, split to demonstrate differences in the dorsal and ventral halves of the Hlxb9 expression domain.
Expression of these transcription factors is >3-fold higher in at least one organ than in other organs. Expression is indicated as absent (−), present (+), present at low levels (lo), or present at high levels (hi). Eso=esophagus, Liv=liver, Panc=pancreas, Int=intestine.
Expression of these cell surface proteins is >3-fold higher in one organ than in all other organs, and expression is not detected significantly above background in other organs.