Abstract
Eukaryotic genomes harbor transposable elements and other repetitive sequences that must be silenced. Small RNA interference pathways play a major role in their repression. Here, we reveal another mechanism for silencing these sequences in Drosophila. Depleting the linker histone H1 in vivo leads to strong activation of these elements. H1-mediated silencing occurs in combination with the heterochromatin-specific histone H3 lysine 9 methyltransferase Su(var)3-9. H1 physically interacts with Su(var)3-9 and recruits it to chromatin in vitro, which promotes H3 methylation. We propose that H1 plays a key role in silencing by tethering Su(var)3-9 to heterochromatin. The tethering function of H1 adds to its established role as a regulator of chromatin compaction and accessibility.
Eukaryotic genomes are packaged into chromatin, which is composed of highly conserved repetitive units referred to as nucleosomes. The nucleosome consists of ~145 base pairs of DNAwrapped around an octamer of core histones, H2A, H2B, H3, and H4. Chromatin also contains the linker histone H1, which binds to the linker DNA between nucleosomes and facilitates folding of nucleosome arrays into more compact structures (1). Chromatin is organized into regions of euchromatin and more densely packed heterochromatin, which is generally silenced. The mechanisms leading to heterochromatic silencing are not well understood (2, 3).
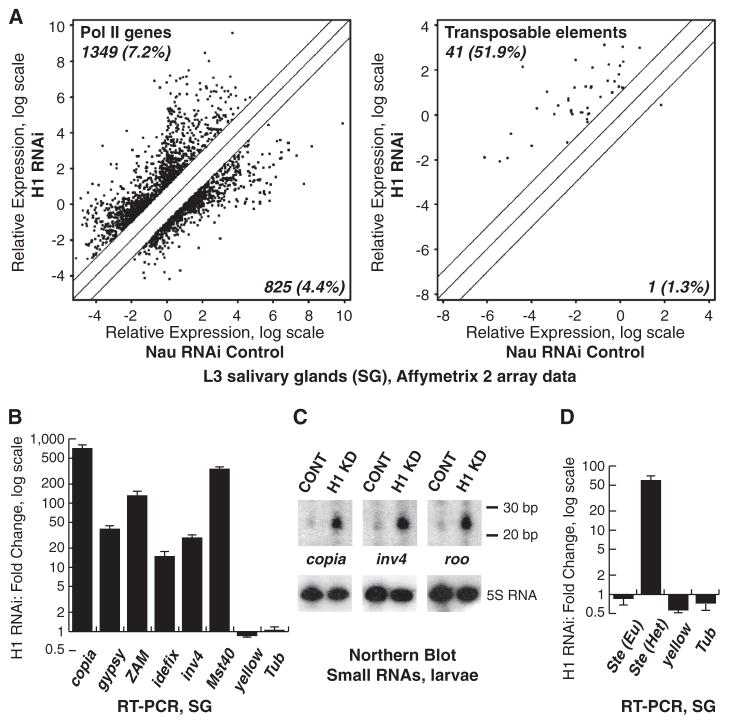
Depleting H1 in Drosophila by RNA interference (RNAi) leads to marked disruption of salivary gland (SG) polytene chromosome structure, including pericentric heterochromatin, and a decrease in nucleosome spacing (4). We compared the RNA expression profiles of SGs depleted of H1 and control Nautilus (Nau) RNAi SGs. We found only a modest difference in the mRNA profile, with only 2174 (11.5%) protein-coding genes showing a change of twofold or more (P < 0.05) (Fig. 1A). However, H1 depletion caused significant changes in the abundance of transcripts derived from transposable elements (TEs). Of the 79 annotated TE transcripts, the abundance of 42 (53.2%) changed by twofold or more (Fig. 1A and table S1), with more than 98% of changes representing increased expression. Quantitative reverse transcription polymerase chain reaction (QRT-PCR) of 11 different RNAs representing various classes of Drosophila TEs confirmed that their expression is activated from 15-fold to as much as 800-fold (Fig. 1B). Thus, the repressive function of H1 in vivo is directed particularly toward TEs.
Fig. 1. Drosophila H1 represses repetitive elements.
(A) Transcript expression was examined by micro-array analyses in H1-depleted and control (Nau RNAi) SGs (4). Signal intensities are shown for transcripts in control (x axis) versus H1-depleted (y axis) samples. The diagonal lines indicate equal expression level or a twofold change. Significantly affected transcripts above or below twofold threshold are indicated by dots. (Left) Signals for protein-coding gene probes; (right) signals for probes annotated as TEs. Numbers in the top left and bottom right corners represent percentages of transcripts that are up- or down-regulated above threshold, relative to the total number of probes (18,833 protein coding genes, 79 TEs). (B) TE transcripts in SGs were analyzed by QRT-PCR. Fold changes were calculated as a ratio of signals for H1-depleted samples to those for control samples and normalized to RP49. Standard deviations are from triplicate PCR reactions for three independent experiments. (C) RNA was extracted from H1-depleted (H1 KD) and control (CONT) larvae and (top) analyzed by Northern blot with TE-specific probes. (Bottom) Hybridization with the 5S RNA probe (loading control). (D) QRT-PCR of transcripts for euchromatic (Eu) and heterochromatic (Het) copies of Ste was analyzed as in (B).
Preferential repression of TEs is also observed in normal, mitotically dividing Drosophila cells. RNA expression profiling of Kc cells depleted of H1 to ~30% of control levels (fig. S1A) showed similar effects on derepression of TE transcripts and significant overlap with those observed in SGs, as well as a similar limited effect on protein-coding genes (fig. S1B). Furthermore, measurements of transposon transcript levels by QRT-PCR in four other tissue sources (whole larvae, brains, ovaries, and testes) from H1-depleted animals showed that expression of TEs is activated from 50- to more than 500-fold (fig. S2, A and B). Therefore, H1 exerts a strong repressive effect on TE expression in a variety of cell types [see also (5)].
Repeat-associated small interfering RNA (rasiRNA) pathways are involved in the negative regulation of TEs (6, 7). In Drosophila germ cells, 24- to 28-nucleotide (nt) PIWI-interacting RNAs (piRNAs) are generated by the dicer activity of AGO3, whereas in ovarian soma piRNA-dependent silencing relies on the activity of PIWI, and in other types of somatic cells, TEs are repressed by 21-nt endo-siRNAs and AGO2 [reviewed in (8, 9)]. To test whether TE up-regulation in H1-depleted cells is due to decreased small RNA expression, we extracted total RNA from H1-depleted and control larvae and analyzed small RNAs homologous to copia, invader 4, roo, and idefix transposable elements by Northern blotting. In each case, we observed a marked increase in the abundance of the corresponding small RNAs (Fig. 1C). We also used massive parallel sequencing to quantify small RNAs in SGs and ovaries from H1-depleted and control larvae. In each tissue we found that the majority of TE-specific small RNAs (both endo-siRNAs and piRNAs) are strongly up-regulated upon H1 depletion (table S2). Thus, activation of TE expression is not due to a decrease in the concentration of the repressive small RNAs.
TE insertions in Drosophila are thought to be mostly located in heterochromatin (10, 11) and proximal heterochromatin-euchromatin transition zones (12). We hypothesized that H1 may silence TEs through its ability to regulate the activity of heterochromatin, with TEs responding differently to H1 depletion depending upon their insertion site. Stellate (Ste) (13) exhibits several features similar to TEs, with multiple tandem copies at two distinct loci, one euchromatic (Eu Ste) and the other in pericentric heterochromatin (Het Ste) (14). Ste expression is regulated by Su(Ste) encoding piRNAs that silence Ste (14, 15). Heterochromatic and euchromatic copies of Ste exhibit single-nucleotide polymorphisms that allow discrimination between transcripts originating from either locus. Depletion of H1 strongly up-regulates only Het Ste transcripts, whereas Eu Ste transcripts are not substantially affected (Fig. 1D). Although transcripts from both loci are negatively regulated by Su(Ste)-derived small RNAs, H1 specifically silences Het Ste, presumably through its role in regulation of heterochromatin function.
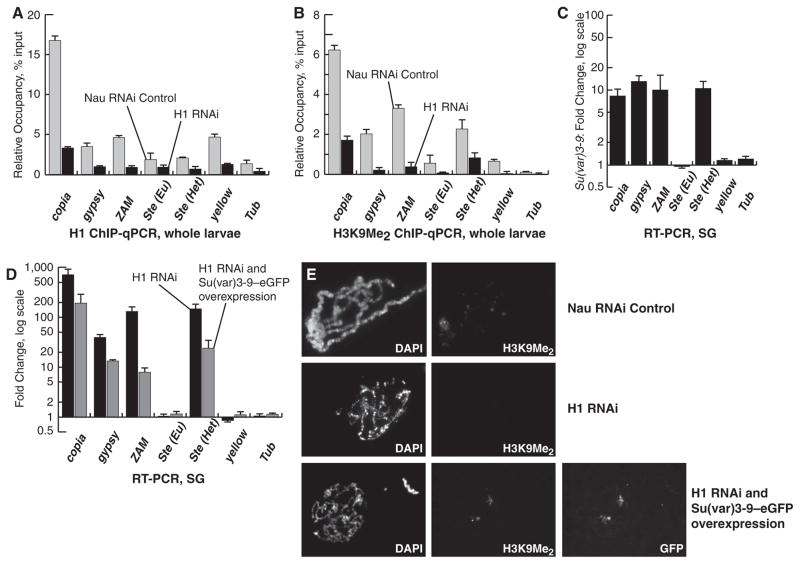
H1 depletion causes a reduction in dimethylation of histone H3 lysine 9 (H3K9Me2) (4). Quantitative chromatin immunoprecipitation (QChIP) in H1-depleted and control larvae at the regulatory regions of several TEs—including copia, gypsy, and ZAM—and at Het Ste, revealed a marked decline in the presence of H3K9Me2 accompanying the loss of H1 (Fig. 2, A and B). Although H1 depletion also leads to reduced H3K9Me2 at Eu Ste and other euchromatic loci like yellow (Fig. 2B), only heterochromatic loci are derepressed (Fig. 1, B and D), consistent with the existence of additional, H3 methylation–independent silencing mechanisms outside of heterochromatin. H3K9Me2 modification is catalyzed primarily by the histone methyltransferase (HMT) Su(var)3-9 (16). Su(var)3-9 null mutation also leads to strong up-regulation of TEs and Het Ste but not Eu Ste (Fig. 2C), which suggests that H1 and Su(var)3-9 may function in concert. Although H3K9Me2 is severely reduced in Su(var)3-9 mutant larvae, H1 occupancy at repetitive sequences and its distribution in polytene chromosomes is not substantially affected (fig. S2, C to E). These results suggest that H1 acts upstream of Su(var)3-9 to regulate hetero-chromatin identity. If H1 and Su(var)3-9 cooperate to silence TEs, then overexpression of Su(var)3-9 might ameliorate the effects of H1 depletion. Indeed, overexpression of Su(var)3-9 partially reverses the activation of TE and Het Ste expression accompanying H1 depletion in SGs (Fig. 2D) and larvae (fig. S2F). In addition, Su(var)3-9 overexpression partially restores the decreased viability of flies caused by H1 depletion (table S3). Conversely, Su(var)3-9 mutation strongly enhances the lethality caused by even moderate H1 depletion (table S3). Furthermore, Su(var)3-9 overexpression in the H1-depleted SG reinstates the H3K9Me2 mark in pericentric heterochromatin (Fig. 2E). However, it does not rescue the global defects in the morphology of polytene chromosomes, which suggests that the combined regulation of chromatin structure by H1 and Su(var)3-9 is directed specifically toward heterochromatin.
Fig. 2. H1 represses repetitive elements in conjunction with Su(var)3-9.
(A) The occupancy of H1 in larval chromatin was measured by qChIP. The ordinate indicates the amounts of qChIP DNA samples relative to input DNA. All experiments were performed in triplicate. Error bars, standard deviation. (B) The occupancy of the H3K9Me2 was measured by qChIP and presented as in (A). (C) QRT-PCR assays were performed in homozygous Su(var)3-9[6] and wild-type SGs. The data were analyzed as in Fig. 1B. (D) RNA was prepared from SGs from control, H1-depleted, and H1-depleted UAS:Su(var)3-9–eGFP larvae. QRT-PCR assays were performed as in (C). Black bars, H1-depleted SGs; gray bars, H1-depleted UAS:Su(var)3-9-eGFP SGs. (E) SGs from control (top), H1-depleted (middle), and H1-depleted UAS:Su(var)3-9–eGFP (bottom) larvae were dissected, and polytene spreads were stained with 4′,6′-diamidino-2-phenylindole (DAPI) and the indicated antibodies.
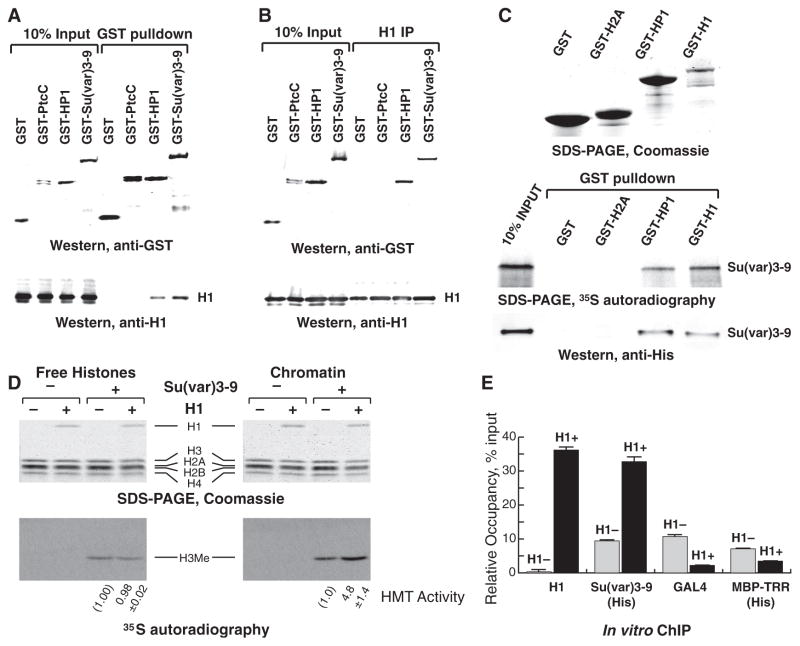
We next asked if H1 and Su(var)3-9 physically interact. A glutathione S-transferase (GST) fusion of Su(var)3-9 was expressed in Drosophila S2 cells, and immunoprecipitation (IP) of nuclear extracts with GST-specific antibody, followed by immunoblotting for H1, showed that endogenous H1 and GST-Su(var)3-9 associate (Fig. 3A). Drosophila H1 also interacts with heterochromatin protein 1 (HP1) (Fig. 3A), which parallels previous observations with their mammalian counterparts (17–19). Reciprocal IP with H1 antiserum confirmed the interaction of endogenous H1 with GST-Su(var)3-9 and GST-HP1 (Fig. 3B). Recombinant GST-H1 purified from bacteria also directly interacts with 35S-labeled Su(var)3-9 translated in vitro and with purified recombinant Su(var)3-9–His6 (Fig. 3C). The absence of binding between Su(var)3-9 and another histone protein (GST-H2A) shows that interaction of Su(var)3-9 with H1 is not due to the high net positive charge of H1 or to a bridging artifact owing to contaminating nucleic acids.
Fig. 3. H1 physically interacts with and recruits Su(var)3-9 to chromatin in vitro.
(A) GST fusion proteins were ectopically expressed in S2 cells and immunoprecipitated. The input and IP material was analyzed by immunoblotting with (top) GST- and (bottom) H1-specific antibodies. PtC, the C-terminal tail of Hedgehog receptor Patched (25) (negative control). (B) Reciprocal IP experiments with H1-specific antibody were performed as in (A). (C) Su(var)3-9 was expressed and 35S-labeled by in vitro translation in reticulocyte lysates or purified as a 6His-tagged protein from bacteria. GST fusion proteins were expressed in E. coli and incubated with Su(var)3-9. The pulled-down material was examined by SDS–polyacrylamide electrophoresis (SDS-PAGE) and Coomassie staining (top), autoradiography (middle), or 6His-specific antibody immunoblotting (bottom). (D) Free histones (left) or reconstituted chromatin (right) with and without H1 were incubated with radioactive S-adenosylmethionine (SAM) in the presence or absence of recombinant Su(var)3-9–His6. H3 methylation was examined by autoradiography (bottom) and corrected for H3 loading (top, Coomassie). H3 methylation was quantified in two independent experiments; the average and standard deviation are shown at the bottom. (E) In vitro reconstituted chromatin with (H1+, black bars) or without H1 (H1–, gray bars) was incubated with Su(var)3-9–His6, GAL4-VP16, or MBP-TRR–His6, crosslinked, and analyzed by in vitro QChIP. The ordinate indicates the amounts of qChIP DNA samples relative to input DNA. All experiments were performed in triplicate. Error bars, standard deviation.
To better understand the functional significance of this physical interaction, we studied Su(var)3-9 binding and its HMT activity toward chromatin reconstituted in vitro with and without H1. Recombinant Su(var)3-9–His6 (fig. S3A) was assayed for HMT activity on free histones and oligonucleosomal templates (fig. S3, B and C). Su(var)3-9 can methylate histone H3 both when H3 is in the context of nucleosomes and in its dissociated native form (Fig. 3D). However, whereas the presence of H1 did not affect Su(var)3-9 activity toward H3 in solution, it strongly stimulated methylation of H3 assembled in chromatin (Fig. 3D). Thus, H1 does not stimulate the intrinsic enzymatic activity of Su(var)3-9, rather it promotes H3 methylation within the chromatin substrate. Furthermore, in vitro ChIP demonstrated a greater magnitude of Su(var)3-9 association with the H1-containing oligonucleosome substrate versus that with the H1-free substrate (Fig. 3E). In control ChIP experiments, the presence of H1 did not promote but, rather, strongly inhibited the occupancy of purified recombinant fusion of yeast GAL4 and herpes simplex virus VP16 proteins (GAL4-VP16), which can bind GAL4 sites present in the template, and MBP-TRR-His6, a fusion protein of Trithorax-related, H3K4-specific HMT (Fig. 3E, fig. S3A). These results indicate that H1 can specifically recruit Su(var)3-9 to chromatin where it methylates histone H3 in nucleosomes.
Our results indicate that Drosophila H1 and Su(var)3-9 work together in repressing the transcriptional activity of TEs and TE-like sequences in heterochromatin. Su(var)3-9 physically associates with H1 and is recruited to H1-containing chromatin, where it mediates H3K9 methylation. Considering the previously observed interactions between Su(var)3-9, HP1 and H3K9Me2/3 [re-viewed in (20)], H1 and HP1 (Fig. 3, A and B) (17–19, 21) and the physical interaction and joint activities of H1 and Su(var)3-9 reported here, we propose that these known heterochromatin effectors and components additionally require linker histone H1 for the establishment of heterochromatin identity and for repression of its genetic activity.
H1 is thought to be nearly ubiquitous in the genome, but several studies report its consistently higher abundance in heterochromatin (22–24). We propose that higher concentrations of H1, equal in stoichiometry to nucleosomes, along extended chromatin domains may be essential to achieve its optimal function as a repressor, whereas substoichiometric or local deposition may only allow for a limited ability to repress genetic activity in euchromatin (Fig. 1A, left). In the future, it will be interesting to compare H1 abundance in various parts of the genome by physical fractionation of chromatin and to study the effects of H1 on activity of other chromatin-modifying enzymes.
Supplementary Material
Acknowledgments
We are grateful to S. Elgin, A. Imhof, T. Kornberg, and G. Reuter for fly stocks and DNA constructs; J. Kadonaga, M. Khuong, and T. Kusch for purified recombinant GAL4-VP16 and MBP-TRR; A. Lusser for critical reading of the manuscript; and E. Vershilova for technical assistance. This work was supported by grants from the NIH to D.V.F. (GM074233) and A.I.S. (GM093190 and CA079057). A.I.S. also receives support from National Cancer Institute, NIH, Cancer Center Grant 2P30CA13330. S.N.W. was supported by 5T32GM0728. Data have been deposited in the GEO database and assigned Series accession no. GSE44148.
Footnotes
References and Notes
- 1.Wolffe AP. Chromatin: Structure and Function. Academic Press; San Diego: 1995. [Google Scholar]
- 2.Elgin SC, Grewal SI. Curr Biol. 2003;13:R895. doi: 10.1016/j.cub.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Eissenberg JC, Reuter G. Int Rev Cell Mol Biol. 2009;273:1. doi: 10.1016/S1937-6448(08)01801-7. [DOI] [PubMed] [Google Scholar]
- 4.Lu X, et al. Genes Dev. 2009;23:452. doi: 10.1101/gad.1749309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vujatovic O, et al. Nucleic Acids Res. 2012;40:5402. doi: 10.1093/nar/gks224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khurana JS, Theurkauf W. J Cell Biol. 2010;191:905. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malone CD, Hannon GJ. Cell. 2009;136:656. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czech B, Hannon GJ. Nat Rev Genet. 2011;12:19. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghildiyal M, Zamore PD. Nat Rev Genet. 2009;10:94. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergman CM, Quesneville H, Anxolabéhère D, Ashburner M. Genome Biol. 2006;7:R112. doi: 10.1186/gb-2006-7-11-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimitri P. Genetica. 1997;100:85. [PubMed] [Google Scholar]
- 12.Kaminker JS, et al. Genome Biol. 2002;3:RESEARCH0084. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ. Genetics. 1990;124:303. doi: 10.1093/genetics/124.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotelnikov RN, et al. Nucleic Acids Res. 2009;37:3254. doi: 10.1093/nar/gkp167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravin AA, et al. Curr Biol. 2001;11:1017. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 16.Jenuwein T. Trends Cell Biol. 2001;11:266. doi: 10.1016/s0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- 17.Daujat S, Zeissler U, Waldmann T, Happel N, Schneider R. J Biol Chem. 2005;280:38090. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen AL, et al. Mol Cell. 2001;7:729. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- 19.Trojer P, et al. J Biol Chem. 2009;284:8395. doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebert A, Lein S, Schotta G, Reuter G. Chromosome Res. 2006;14:377. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 21.Hale TK, Contreras A, Morrison AJ, Herrera RE. Mol Cell. 2006;22:693. doi: 10.1016/j.molcel.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 22.Grigoryev SA, Spirin KS, Krasheninnikov IA. Nucleic Acids Res. 1990;18:7397. doi: 10.1093/nar/18.24.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamakaka RT, Thomas JO. EMBO J. 1990;9:3997. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weintraub H. Cell. 1984;38:17. doi: 10.1016/0092-8674(84)90522-1. [DOI] [PubMed] [Google Scholar]
- 25.Lu X, Liu S, Kornberg TB. Genes Dev. 2006;20:2539. doi: 10.1101/gad.1461306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.