Abstract
Although the potential human health impacts from exposure to engineered nanoparticles (ENPs) are uncertain, past epidemiological studies have established correlations between exposure to ambient air pollution particulates and the incidence of pneumonia and lung infections. Using amorphous silica and superparamagnetic iron oxide (SPIO) as model high production volume ENPs, we examined how macrophage activation by bacterial lipopolysaccharide (LPS) or the lung pathogen Streptococcus pneumoniae is altered by ENP pre-treatment. Neither silica nor SPIO treatment elicited direct cytotoxic or pro-inflammatory effects in bone marrow-derived macrophages. However, pre-treatment of macrophages with SPIO caused extensive reprogramming of nearly 500 genes regulated in response to LPS challenge, hallmarked by exaggerated activation of oxidative stress response pathways and suppressed activation of both pro- and anti-inflammatory pathways. Silica pre-treatment altered regulation of only 67 genes, but there was strong correlation with gene sets affected by SPIO. Macrophages exposed to SPIO displayed a phenotype suggesting an impaired ability to transition from a M1 to M2-like activation state, characterized by suppressed IL-10 induction, enhanced TNFα production, and diminished phagocytic activity toward S. pneumoniae. Studies in macrophages deficient in scavenger receptor A (SR-A) showed SR-A participates in cell uptake of both the ENPs and S. pneumonia, and co-regulates the anti-inflammatory IL-10 pathway. Thus, mechanisms for dysregulation of innate immunity exist by virtue that common receptor recognition pathways are used by some ENPs and pathogenic bacteria, although the extent of transcriptional reprogramming of macrophage function depends on the physicochemical properties of the ENP after internalization. Our results also illustrate that biological effects of ENPs may be indirectly manifested only after challenging normal cell function. Nanotoxicology screening strategies should therefore consider how exposure to these materials alters susceptibility to other environmental exposures.
Keywords: nanotoxicology, macrophage, iron oxide, amorphous silica, Streptococcus pneumoniae, lipopolysaccharide, scavenger receptor
Concern over the potential health impacts of engineered nanoparticles (ENPs) is due in part to our limited knowledge of how ENPs interact with cells and the subsequent biological pathways impacted. With the rapid emergence of a growing diversity of ENPs and increased importance placed on in vitro test systems for nanotoxicology, the appropriate selection of biological endpoints for use in hazard analysis becomes essential. Frequently however, the primary focus of nanotoxicity studies is on measures of the direct cytotoxic or pro-inflammatory effects of ENPs. This targeted approach supports the need for rapid hazard screening,1 but sheds limited insight into the much broader array of biological pathways that may be impacted by ENPs. In fact, genomic studies have shown that even ENPs which are often categorized as biologically inert can alter cellular expression of hundreds of gene products.2-3 Such effects on gene expression may not manifest with immediate and observable phenotypic changes, but could influence the ability of cells to adapt and respond to additional environmental stressors. The indirect consequences of ENP exposure on the susceptibility of biological systems to other environmental agents is a poorly understood aspect of nanotoxicology.
Although adverse human health effects of ENPs have not been clearly documented, prior epidemiological studies have established correlations between human exposure to ultrafine ambient particulates and increased mortality, particularly among elderly individuals.4-8 Escalated hospitalization rates associated with spikes in urban air pollution have been associated with increased incidence of secondary lung infections.6, 8 Indeed, a recent two-year case control study found exposure to ambient particulates with diameters less than 2.5 μm (PM2.5) was associated with increased hospitalization rates for community-acquired pneumonia.9 These findings and related studies in rodents that show exposure to air pollution particles increases susceptibility to lung infection10-12 suggest particulate exposure may be a contributing risk factor for the overall rising incidence of pneumonia observed in the United States.13 Similar studies with engineered materials are limited, but two studies have reported that exposure of mice to nanomaterials which alone cause pulmonary damage, impairs lung clearance of bacteria.14-15 Macrophage cytotoxicity, diminished phagocytic capability, and altered macrophage innate immune signaling have all been hypothesized as potential mechanisms for these interactions.14-16
The potential for biological interactions between ENP and pathogen exposures should not be unanticipated, since macrophages have evolved conserved mechanisms for recognition and removal of “non-self” entities. Our previous work3, 17-18 suggests that macrophages recognize and respond to ENPs using common core signaling modules and receptor pathways that evolved for purposes of pathogen detection. For instance, through analysis of transcriptomic studies, we identified a core transcriptional response module macrophages use to respond to both ENPs and Salmonella infection.17 More recently, we found that that cellular uptake of some anionic ENPs is mediated through the class A macrophage scavenger receptor (SR-A, CD204),18 a transmembrane glycoprotein whose natural ligands also include bacterial cell wall components and structurally altered lipoproteins.19-21 In mice, deficiency of SR-A results in impaired clearance of bacteria and increased susceptibility to infection.22-23 SR-A may also cooperate with toll receptors to regulate inflammatory signaling,24 although conflicting reports exist. Some studies report SR-A confers resistance to lipopolysaccharide (LPS)-induced lethality in mice, 23, 25-26 whereas others report SR-A exacerbates LPS-mediated inflammation and endotoxic shock.27-28
In this study, we examined how the phenotype and innate immune signaling response of macrophages challenged with LPS or bacterial pathogen is impacted by previous exposure to ENPs. We hypothesize that prior exposure to ENPs would alter the functional phenotype of macrophages and their ability to phagocytose bacteria, and that transcriptomic profiles can be used to identify these phenotypic consequences. Silica and superparamagentic iron oxide (SPIO) were chosen as experimental ENPs because they represent materials produced in high volumes and we have found these ENPs gain entry into macrophages through the SR-A phagocytic pathway.18, 29 Our study was also motivated in part by early work that demonstrated that poorly soluble particles accumulate in macrophages after inhalation, and can be retained in the lung for prolonged periods.30-31 Our results illustrate that exposure to ENPs which have no cytotoxic or proinflammatory effects alone can significantly disrupt macrophage gene regulation, activation, and phagocytic activity toward pathogens. The similarities in the pathways macrophages use to recognize and internalize ENPs and pathogenic bacteria can create potential mechanisms where by ENPs dysregulate cellular innate immune responses.
Results and Discussion
Nanoparticles and Characterization
Fluorescent amorphous silica and superparamagnetic iron oxide nanoparticles (SPIO), both generally considered low toxicity materials, were chosen as experimental proxies for a broader group of poorly soluble low toxicity ENPs which have widespread and high volume use in occupational settings. The physicochemical properties of the ENPs when suspended in medium used for cellular studies are provided in Supporting Information, Table S1. Transmission electron microscopy showed the SPIO particles had a primary diameter of 13 nm, compared to 50 nm for silica. Zeta potentiometry conducted at 100 μg/ml concentrations showed SPIO and silica particles had similar net zeta potentials in complete cell culture medium of -21.9 and -23.5 mV, respectively. Dynamic light scattering analysis indicated the SPIO particles agglomerated to a greater extent than silica particles in water (453 nm, 87 nm, respectively). When SPIO particles were suspended in medium supplemented with serum proteins however, their average hydrodynamic diameter was reduced to 274 nm. Both particle types were found to be free of endotoxin contamination (<0.01 EU/ml). Cytotoxicity studies were also conducted in bone marrow-derived macrophages following 24 hr exposure to the ENPs. ELISA analysis of cellular LDH enzyme leakage showed that neither SPIO nor silica ENPs caused significant cytotoxicity, even up to concentrations of 200 μg/ml (see Supporting Information, Figure S1).
Impact of ENPs and LPS on Macrophage Gene Regulation
To investigate how the phenotype of macrophages that have engulfed ENPs differs from untreated macrophages, we conducted Affymetrix microarray studies to identify the gene regulatory pathways affected by the ENPs. Mouse bone marrow-derived macrophages were used as an experimental model since, similar to alveolar macrophages,22 these cells express high levels of class A scavenger receptors including SR-A and MARCO, and high cell yields needed for large scale transcriptomics are readily obtained. To mimic potential occupational exposure scenarios, the experimental design involved pretreatment of macrophages with the ENPs (25 μg/ml, 24 hr), followed by removal of residual ENPs and acute challenge with the TLR4 ligand and surrogate bacterial stimulus, lipopolysachharide (LPS, 10 ng/ml, 4 hr). The 4 hr challenge time was chosen based on preliminary studies which showed many of the pro-inflammatory gene expression responses peak between 2-6 hr after LPS treatment (not shown). Microarray analysis revealed a total of 1029 and 67 genes that were differentially expressed (p<0.05, 1.5-fold change) in macrophages treated with SPIO or silica alone, respectively, compared to controls (Table 1), demonstrating that SPIO exposure had a much greater impact on basal transcription in macrophages at these exposure concentrations. Consistent with previous reports,32-34 LPS challenge alone produced a robust transcriptional response, with 5027 genes differentially expressed compared to untreated controls (p<0.05, 1.5-fold change) (Table 1). Using MetaCore, we determined the biological processes that were significantly overrepresented by the gene expression profiles for each treatment group (see Supporting Table S2). The most significant biological processes up-regulated by LPS in macrophage cells were associated with inflammation and immune response, including interferon signaling, innate inflammatory response, lymphocyte proliferation, phagocytosis and IL-10, TCR (T-cell receptor), TREM1 (triggering cells expressed on myeloid cells 1), MIF (macrophage migration inhibitory factor), IFN-gamma and IL-6 signaling. Other processes significantly (p<0.01) down-regulated by LPS exposure included those associated with DNA damage, cell cycle, WNT signaling, circadian rhythm and the lactosylceramide pathway. These biological processes are similar to those previously described for LPS treatment.33-34 Overall, the activation of inflammatory and immune response processes was unique to the LPS treatment group. No enrichment of these processes was observed for genes that were up-regulated by either the SPIO or silica pretreatments. In addition, common gene markers associated with pro-inflammatory activation of macrophages, such as TNFα, IL-12α, and IL-6, were not significantly altered by either SPIO or silica pretreatments alone.
Table 1.
Summary of differentially expressed genes in mouse bone marrow-derived macrophages following exposure to SPIO, silica or LPS.
| Pretreatment | Challenge | DE genesa vs. Control | DE genesa vs. LPS (GTA)b |
|---|---|---|---|
| None | None | -- | -- |
| Silica (25 μg/ml) | None | 67 | -- |
| SPIO (25 μg/ml) | None | 1029 | -- |
| None | LPS (10 ng/ml) | 5027 | -- |
| Silica | LPS | 4954 | 44 (15) |
| SPIO | LPS | 5483 | 1044 (499) |
|
| |||
| Union | 6831 | 1058 (503) | |
Differential expression (DE) criteria of p<0.05 and 1.5-fold change compared to control or LPS alone. Significance was calculated by ANOVA with unequal variance and Tukey's post-hoc test (p<0.05) and false discovery rate calculation.
Greater-than-average (GTA) responses were identified by transforming the data to account for the magnitude of change induced by the ENP alone prior to statistical comparison with LPS. These genes show a magnitude of change in the combined pretreatment/challenge groups that is greater than would be predicted by the sum of the individual treatments.
Among the 1029 genes differentially expressed in macrophages after SPIO treatment, 515 genes (50%) were up-regulated and 514 genes were down-regulated, indicating that specific biological processes were impacted by exposure and that the ENPs did not simply cause a general repression of transcription (Figure 1A). Although silica treatment altered expression of fewer genes (67), over 65% of these genes were also significantly altered by SPIO treatment and the remaining genes followed a similar expression trend as found for SPIO. To identify the potential biological consequences of ENP exposure in macrophages, we calculated the enrichment of biological and metabolic pathways separately for the genes that were up- or down-regulated by SPIO or silica treatment, as an indicator of whether these processes were enhanced or suppressed after treatment. A table of all significant processes (p<0.01) associated with ENP treatment is shown in Figure 1C. Most of the processes significantly affected by silica treatment are also significant for SPIO, supporting the overlap observed in gene regulation between the treatments, and include processes for cell adhesion, chemotaxis and blood coagulation. Processes that were uniquely up-regulated by SPIO but not silica include those associated with hypoxia and oxidative stress and several lipid metabolic pathways. Processes down-regulated by SPIO were instead associated with cell cycle, cell adhesion, inflammation and immune response. In fact, several of the inflammation-related processes that were significantly up-regulated by LPS were down-regulated by SPIO pretreatment, including interferon signaling, MIF signaling, leukocyte chemotaxis and Th17-derived cytokine response (Figure 1C and Supporting Table S2). In particular, the decrease in the interferon signaling process by SPIO was due to significant down-regulation of interferon stimulatory genes and regulatory factors (Isg15, Isg54, Ifi44, Irf7, Gbp2), IL-1 beta, IL-21 receptor and several chemokines (Ccl12 and Ccl7), which were stimulated by LPS. These data suggest that SPIO treatment in macrophage cells leads to decreased cell proliferation and interferon-dependent activation and stimulates lipid biosynthesis and oxidative stress response signaling. The enhanced oxidative stress response is mediated through transcriptional up-regulation of antioxidant enzymes such as catalase, thioredoxin reductase, peroxiredoxin-6, glutatamate, and glutathione-s-transferases A3 (Gsta3) and Mu2 (Gstm2).
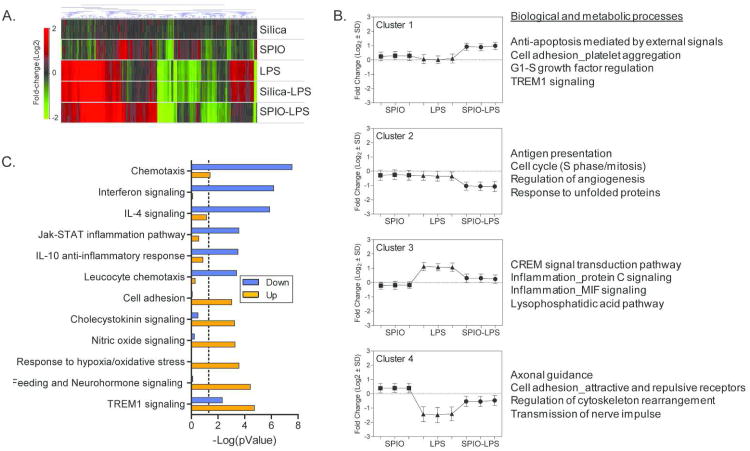
Figure 1. Effects of Silica and SPIO ENPs on Macrophage Gene Regulation.
Microarray analysis identified a total of 1052 genes that were differentially expressed (p<0.05, 1.5-fold change) between ENP treatments and controls. (A) Bidirectional hierarchical clustering of all differentially regulated genes. Values are log2 fold-change compared to controls. Green, red and black represent down-regulation, up-regulation or no change in gene expression, respectively. (B) Venn diagram comparing differentially regulated genes between silica and SPIO treatment groups. (C) Table of all significantly enriched (p<0.01) biological and metabolic processes from MetaCore (GeneGo) for genes that were up-regulated or down-regulated by SPIO or silica compared to untreated controls.
ENP Pretreatment Selectively Shifts the LPS-Regulated Inflammatory Responses
We next examined the subset of genes whose regulation in response to LPS challenge was specifically modulated by ENP pretreatment. Among the total genes whose expression was altered in the ENP-LPS treatment groups (eg., ENP pretreatment followed by LPS challenge), we identified 1044 (SPIO-LPS) and 44 (silica-LPS) genes for that were statistically different (p<0.05) when compared to LPS treatment alone. Furthermore, approximately 50% of the genes (499/1044) regulated by SPIO-LPS (Table 1, Figure 2) showed response patterns that would not have been predicted by examination of transcriptional responses to either agent alone, which we define as “greater than additive” behavior. In comparison with SPIO, pretreatment of cells with silica resulted in very few changes in LPS-mediated gene regulation; however, the data were significantly correlated (p<0.0001, Spearman r = 0.707) with over 68% of the genes differentially expressed in the silica-LPS group overlapping with the SPIO-LPS group, again suggesting similar mechanisms by which ENP exposures modulate LPS-mediated signaling. Hierarchical clustering of the data (Figure 2A) illustrates the dramatic shift in the transcriptional response to LPS in SPIO-pretreated macrophages. Genes that followed interesting trends suggesting co-regulation of LPS signaling by SPIO were further identified using K-means clustering (for full list, see Supporting Table S3). In some cases, the induction or suppression of genes by LPS was completely blocked in macrophages pretreated with SPIO (Figure 2B clusters 3 and 4, respectively). For example, transcriptional signaling through the CREM (cAMP response element modulator) pathway, the lysophosphatidic acid pathway, and several inflammatory response pathways induced by LPS alone were repressed by pre-treatment with SPIO, despite the fact that the expression of genes in these pathways was not significantly affected by SPIO treatment alone (Figure 2B, cluster 3). Genes uniquely regulated (both up and down) in the combined treatments were also identified and suggest unique regulation of apoptotic signaling and antigen presentation processes by the combined SPIO-LPS treatment group that was not observed for either treatment alone (Figure 2B, clusters 1 and 2).
Figure 2. Pretreatment with SPIO Shifts the LPS Activation Profile of Macrophages.
Among 1058 genes found to be differentially expressed in the ENP-LPS treatment groups compared to control, a subset of genes (503) showed a greater than average gene expression response (p<0.05) which would not be predicted compared to ENP or LPS exposures alone; 499 genes of these genes were regulated by SPIO-LPS and 15 genes by silica-LPS treatment groups. (A) Heatmap of hierarchical clustering of all 503 genes. Values represent log2 fold-change for all treatments compared to control. Green, red and black indicate down-regulation, up-regulation or no change in gene expression, respectively. (B) K-means clustering to identify processes uniquely activated or repressed by SPIO pre-treatment. Data for four selected clusters are shown, representing 46% (227/499) of the genes regulated by SPIO-LPS treatment. Graphs show average log2 fold-change expression ± s.d. for all genes in each cluster for SPIO (squares), LPS (triangles) and SPIO-LPS (circles) treatments. The top four most significantly enriched (p<0.05) biological or metabolic processes (MetaCore) are provided for each gene cluster. A full list of the genes in each cluster is included in Supplemental Table S2. (C) Graph of significantly enriched (p<0.001) biological processes (MetaCore) for genes up or down regulated by the SPIO-LPS treatment compared to LPS. The dashed line indicates a significance level of p<0.05.
Further evaluation of the 499 genes that were statistically different between SPIO-LPS and LPS alone (“greater-than-additive gene set”) identified several LPS-regulated processes that were enhanced by SPIO pretreatment, including oxidative/nitrative stress response, cholesystokinin signaling and cell adhesion (Figure 2C, orange bars). In contrast, the expression of genes associated with inflammation-related processes, including leukocyte chemotaxis, interferon signaling and Jak-Stat inflammation signaling, were generally suppressed by SPIO pretreatment (Figure 2C, blue bars). The only inflammatory signaling process category found to be significantly overrepresented in gene datasets that were both up- and down-regulated was the TREM signaling process. In part, this is due to overlap among the genes represented in the TREM process and genes represented in the IL-10 and JAK-STAT process categories, such as IL-10 (downregulated), TNF and IL-12 (both upregulated).
SR-A Dependent Disruption of IL-10 Negative Feedback Signaling
Approximately half (49%) of the genes that displayed expression patterns which suggest co-regulation (greater than additive response) between SPIO pretreatment and LPS challenge belong to inflammatory-related processes (Figure 3 and Supporting Table S4). The transcriptional response of macrophages to LPS has been widely investigated, and involves not only stimulation of proinflammatory cytokines, but parallel induction of negative feedback pathways and anti-inflammatory pathways which function to self-regulate inflammation. Over time these feedback pathways reprogram the cells to an endotoxin tolerant state.34-35 Although the transcriptional signature of endotoxin tolerance varies in different cellular contexts, consistent features of this phenotype include a decreased production of proinflammatory cytokines (eg., TNFα, IL-12), stimulation of anti-inflammatory cytokine pathways (eg., IL-10, SOCS3) and increased phagocytic activity following LPS exposure.34-35 Endotoxin tolerance was recently proposed to represent a distinct state of alternative (M2) macrophage activation,34 which is also characterized by high IL-10, low pro-inflammatory (TNFα, IL-12) cytokine production and enhanced phagocytic capability.36 In this context, many of the transcriptional responses to LPS that were modulated in macrophages pretreated with SPIO suggest that a major effect of ENP exposure is the selective impairment of regulatory pathways involved in transition to an endotoxin tolerant or M2-like state. For instance, whereas LPS-induced expression of anti-inflammatory genes (IL-10, SOCS1, SOCS3) was inhibited, the induction of TNFα and IL-12 mRNA was augmented in SPIO-pretreated cells compared to LPS alone (Figure 3). Furthermore, the induction of several cell surface receptors and complement components involved in macrophage phagocytic activity following LPS challenge was diminished by SPIO pretreatment (Figure 3).
Figure 3. Identification of Potential Co-Regulated Gene Pathways.
Heatmap schematic of select genes associated with the LPS inflammatory response that displayed ‘greater-than-additive’ behavior after pretreatment with SPIO. Genes that were part of the inflammatory response comprised 49% of the total genes in the SPIO-LPS greater-than-additive dataset recognized for analysis in Ingenuity Pathway Analysis (Ingenuity Systems). Values in the heatmap represent z-scores calculated across the treatment groups with blue coloring indicating relative down-regulation and yellow representing relative up-regulation. A full list of the inflammatory response genes is available in Supporting Information, Table S4.
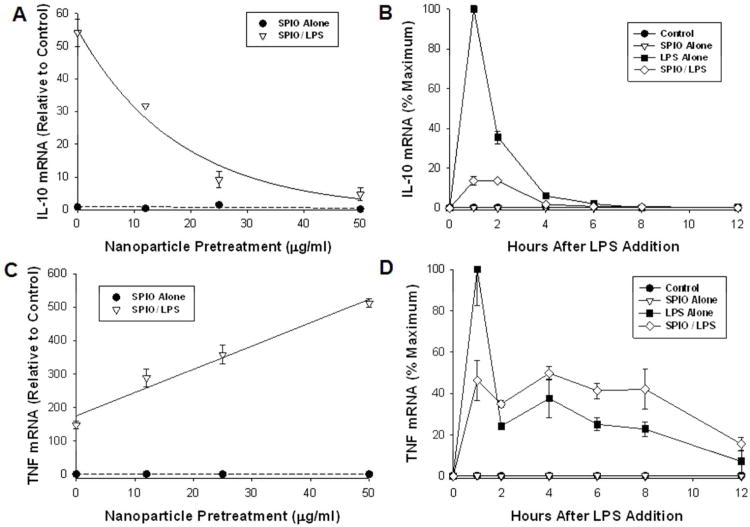
To our knowledge, the suppression of anti-inflammatory pathways by ENP exposure has not been previously reported. Our microarray results are consistent with the hypothesis that some ENPs may disrupt innate immune function through dysregulation of normal feedback control mechanisms. In particular, IL-10 functions as a potent negative feedback inhibitor of transcription of TNFα and other TH1 cytokines.37-38 It is noteworthy that the SR-A gene (MSR1) was previously identified as part of a candidate susceptibility locus involved in differential regulation of plasma IL-10 levels among various mouse strains in response to endotoxin.39 We therefore conducted orthogonal experiments to corroborate the microarray findings and to investigate whether SR-A is involved in regulation of the IL-10 pathway by ENPs. Quantitative RT-PCR analysis of independent biological samples confirmed the microarray results that pretreatment of macrophages with SPIO caused a dose-dependent suppression of IL-10 induction following LPS challenge (Figure 4A). Time course studies also demonstrated that the suppression of IL-10 was not simply due to a delay in transcription of IL-10, since peak IL-10 mRNA levels occurred at similar times after LPS challenge in control and SPIO-pretreated cells (Figure 4B). Consistent with the role of IL-10 as a feedback inhibitor of TNF transcription,40 we also found that the induction of TNF mRNA by LPS (measured at 4 hr) was augmented in a dose-dependent manner by SPIO pretreatment (Figure 4C). Although peak TNF mRNA induction (1 hr) was diminished in SPIO-pretreated cells, there was a slower decay of TNF mRNA levels in SPIO-pretreated cell comparing to cells receiving LPS alone, resulting in overall elevated TNF mRNA from 2 - 8 hrs after LPS stimulation (Figure 4D).
Figure 4. SPIO Pretreatment Disrupts the IL-10/TNFα Feedback Loop.
Wildtype macrophages were pretreated with the indicated concentrations of SPIO for 24 hr, then challenged with LPS (10 ng/ml). Quantitative RT-PCR analysis was used to measure the relative levels of IL-10 and TNFα mRNA expression either 2 hr (A, C) or at the times indicated (B, D) after LPS challenge. Results are mean + s.d. of at least three biological replicates.
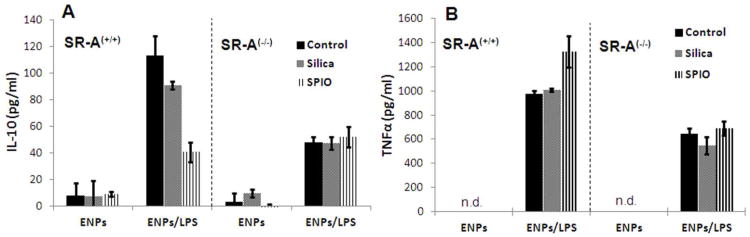
We confirmed the mRNA results by ELISA analyses of secreted IL-10 and TNFα protein. In macrophages derived from wildtype (SR-A+/+) mice, pretreatment with either SPIO or silica ENPs also inhibited the secretion of IL-10 protein, although greater inhibition occurred with SPIO (Figure 5). In macrophages derived from SR-A(-/-) mice however, LPS-stimulated IL-10 protein secretion was reduced to less than half that observed in wildtype macrophages (without ENP pretreatments), and no further inhibition of IL-10 secretion was caused by ENP pretreatment. In concurrence with the mRNA results, we found that levels of secreted TNF protein after LPS challenge were also augmented in wildtype macrophages by pretreatment with SPIO (Figure 5). In contrast, lower levels of TNF were secreted in response to LPS in SR-A deficient macrophages, and the enhanced TNF secretion caused by SPIO pretreatment observed in wildtype macrophages was also abolished. Pretreatment with silica ENPs was not sufficient to stimulate LPS-mediated TNF secretion in wildtype macrophages. This result may reflect the fact that IL-10 protein is exceedingly potent at suppressing TNF synthesis37 and the amorphous silica used in these studies was less effective than SPIO in blocking IL-10 induction. Thus, while pretreatment with silica caused a similar SR-A dependent trend in IL-10 regulation as SPIO, the smaller reduction in IL-10 protein levels caused by silica may not be sufficient to alter its effectiveness as a feedback inhibitor of TNF production.
Figure 5. SR-A Dependent Regulation of the IL-10 Feedback Pathway.
Macrophages isolated from either wildtype or SR-A deficient mice were pretreated with SPIO or silica ENPs (25 μg/ml, 24 hr), and then challenged with LPS. At 6 hr following addition of LPS, the amount of secreted IL-10 or TNF protein was determined by ELISA analyses. N.D. indicates not detectable.
Cellular Reprogramming of Macrophage Phagocytic Activity
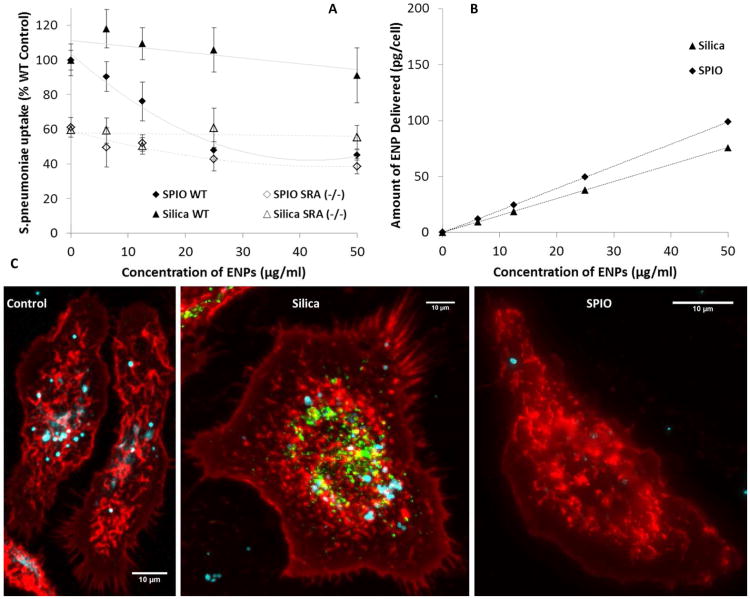
It is well documented that secreted IL-10 stimulates macrophage phagocytic activity, opposing the inhibitory effects of exogenous TNFα on bacterial phagocytosis.41–42 We conducted additional experiments to determine whether these altered patterns of gene regulation were associated with functional changes in the ability of the cells to recognize and phagocytize pathogens, and to investigate whether SR-A was involved in regulating these processes. An experimental protocol similar to the microarray studies was used, where macrophages isolated from wildtype or SR-A deficient mice were pre-exposed to either silica or SPIO, and after removal of ENPs were challenged for 2 hr with Streptococcus pneumoniae, a model pathogen and the leading cause of community-acquired pneumonia.9 Flow cytometry analysis showed that pretreatment of wildtype macrophages with SPIO caused an ENP dose-dependent suppression of bacterial phagocytosis (Figure 6A). Maximal inhibition of phagocytosis was achieved in cells pretreated with ∼25 μg/ml SPIO, corresponding to ∼50% of the activity observed in control cells without ENP pretreatment. In contrast, pretreatment with up to 100 μg/ml silica ENPs for 24 hr did not significantly reduce the number of total phagocytosed bacteria (Figure 6A). We also observed a 40% reduction in bacterial uptake in untreated cells derived from SR-A(-/-) mice compared to wildtype controls, indicating that loss of SR-A expression alone was sufficient to reduce phagocytic activity. These results confirm a previous report that SR-A is involved in macrophage recognition of S. pneumonia.22 Although SPIO pretreatment caused a strong dose-dependent reduction in bacterial uptake in wildtype macrophages, this inhibitory effect was significantly dampened in SR-A deficient macrophages. At the highest SPIO pretreatment concentrations tested (25-50 μg/ml), the number of intracellular bacteria were statistically identical in wildtype and SR-A-deficient macrophages, and was only slightly less than that caused by SR-A deficiency alone, suggesting the majority of the effects of SPIO on pathogen phagocytosis involve SR-A. It is noteworthy that we have also observed similar inhibitory effects of SPIO pretreatment on bacterial phagocytosis using Salmonella typhimurium (data not shown), indicating phagocytic pathways involving both gram negative and gram positive pathogens are impacted. Furthermore, in separate studies using the RAW 264.7 macrophage cell line, we observed a similar inhibition of bacterial phagocytosis in SPIO pretreated cells based on direct quantitation of intracellular bacteria counts by fluorescent microscopy analysis (Supporting Figure S2).
Figure 6. Effects of ENP Pretreatment on Macrophage Phagocytosis of S. pneumoniae.
A) Macrophages from wildtype or SR-A deficient mice were pretreated with the indicated concentrations of SPIO or silica ENPs for 24 hrs, followed by 2 hr challenge with S. pneumonia (MOI=10). Flow cytometry was used to measure bacterial uptake and is expressed as a percent of uptake relative to wildtype controls. B) Total SPIO and silica doses delivered to cell monolayer in culture. Values were obtained using the ISDD model.43 C) Confocal microscopy images showing macrophages (control, silica pretreated, or SPIO pretreated) after 2hr challenge with fluorescent labeled S. pneumoniae. Cell membrane is labeled with a red membrane dye and bacteria are labeled with blue dye. Fluorescent silica ENPs are labeled green. The results illustrate that macrophages that have internalized silica ENPs maintain the ability to phagocytose bacteria, whereas phagocytic activity is significantly diminished in SPIO-pretreated cells.
The differential effects of silica and SPIO on macrophage phagocytic activity correlate well with the differential impacts of these particles on macrophage gene regulation. Although both particle types are within the size limits for uptake by endocytosis, we considered the possibility that either different rates of gravitational settling in culture medium and/or poor cellular internalization of silica particles may explain the differences in their biological potency. However, calculations of the amount of silica and SPIO particles deposited on the cells using the ISDD dosimetry model43 showed that the doses of particles delivered in culture were similar for both ENP types (Figure 6B). Confocal microscopy also verified that silica ENPs were effectively taken up by macrophages, and cells which had high levels of internalized silica ENPs remained capable of phagocytizing bacteria (Figure 6C). Furthermore, using flow cytometry and magnetic particle detection29 to quantify uptake of silica and SPIO ENPs in macrophages derived from either wildtype or SR-A deficient mice, we confirmed our previous findings18, 29 and recent reports44-45 that show SR-A facilitates cellular uptake of both ENP types (see Supporting Figure S3). Thus, the differential impacts on bacterial phagocytosis observed for silica and SPIO are not due to preferential uptake of SPIO versus silica, but likely involve specific physicochemical properties of the ENPs that are manifested after cellular internalization by the SR-A pathway.
Conclusions and Perspective
The concept that exposure to engineered nanomaterials may modulate the susceptibility of biological systems to other environmental agents has received little attention. Our results illustrate that potential mechanisms for dysregulation of innate immunity exist by virtue that common receptor recognition pathways are used by some ENPs and pathogenic bacteria. Our results confirm studies from our and other groups that anionic ENPs and S. pneumonia are both internalized in macrophages in an SR-A dependent manner.18, 22, 29 We further demonstrate that both silica and SPIO ENPs modulate TLR4-mediated gene regulation, though the extent of cross-regulation of gene expression is quite different for the two ENP types. The transcriptional reprogramming induced by SPIO in particular resulted in a phenotype that resembles an impaired ability of the macrophage to transition normally from an M1 to a M2-like activation state, characterized by reduced phagocytic capacity toward pathogens. It is noteworthy that the effects of SPIO are not simply a result of an overall downregulation of gene expression, but involve similar numbers of upregulated and downregulated genes. Furthermore, reductions in phagocytic activity were observed in macrophages at the lowest concentrations of SPIO tested (6.25 -12.5 μg/ml). Based on measurements using magnetic particle detection, we estimate the cellular dose of SPIO from these concentrations is far below levels that cause “macrophage overload”, which is thought to occur at particle loads corresponding to ∼6% of the macrophage cell volume.46 The potential that endotoxin contamination of the ENPs is responsible for the observed effects is also unlikely, since both ENPs test negative for endotoxin. Furthermore, treatment of macrophages with the ENPs alone had no effect on TNFα expression, a highly sensitive indicator of endotoxin contamination. Thus, the altered LPS activation profile of macrophages previously exposed to SPIO, and to lesser extent silica, appears to reflect a specific reprogramming of cell function rather than a general suppression of macrophage function. We postulate that this phenotypic shift is in part a consequence of disruption of negative feedback pathways normally responsible for resolution of inflammation and stimulation of phagocytic processes needed for tissue repair.
The physicochemical basis for the differences in biological potency between silica and SPIO ENPs is not yet clear, but these differences are nonetheless informative from a mechanistic perspective. The large differences in the number of macrophage gene regulation changes caused by SPIO and silica illustrate that the physicochemical properties of the ENPs play important roles in modulating macrophage activation, as expected. This study (Figure S3) and our previous work18, 29 show that the uptake of both particle types is facilitated by SR-A to similar extents, and we have shown that fluorescent-labeled silica ENPs co-localize with SR-A in macrophages.47 In this respect, our data suggest the primary role of SR-A is in mediating uptake and intracellular dose of the ENPs, rather than participating in a direct signaling function. On the other hand, correlations among the specific gene regulatory changes induced by silica and SPIO suggest some overlapping mechanisms between these ENPs, and our results also suggest engagement of SR-A by ENPs may be sufficient to modulate specific TLR4 regulated pathways, such as the IL-10 pathway. Although SPIO was more effective than the silica ENPs used in this study in suppressing IL-10 expression in response to LPS challenge, both responses were dependent on SR-A. The finding that macrophages from SR-A knockout mice show significantly reduced IL-10 induction in response to LPS also provides strong support for a co-regulatory role of SR-A and TLR4 in this pathway.
Although the IL-10 pathway is only a subset of the gene regulatory pathways we identified to be affected by ENP exposure, disruption of this negative feedback loop provides a potential contributing mechanism whereby nanomaterial exposure may inhibit bacterial clearance and exacerbate LPS-mediated inflammation in vivo.14-15, 48 Consistent with the inhibition of IL-10 and enhanced TNF expression we observed in primary macrophages, mice deficient in IL-10 show an exaggerated response to endotoxin associated with high levels of TNF production.49 The reduced phagocytic activity of macrophages exposed to SPIO is also consistent with the opposing regulatory roles of exogenous IL-10 and TNFα on phagocytosis, and the role of IL-10 in regulating expression of the Fcγ phagocytosis receptor,41-42, 49 which we also found was significantly reduced in macrophage pretreated with SPIO.
Recent work suggests SR-A binds SPIO through charge interactions between anionic groups on the ENP surface and lysine-rich regions of the receptor collagen-like (CL) domain.45 Our experiments were conducted in medium supplemented with serum proteins, which clearly adsorb and modify the ENP surface chemistry. Nonetheless, zeta potentiometry showed the ENPs in our study maintain a net negative potential in culture medium, consistent with the average net negative charge of serum proteins. We cannot exclude the possibility that differential ENP surface chemistries and corresponding profiles of adsorbed proteins selectively modulate cellular responses to SPIO or silica. However, it is interesting to hypothesize that the SR-A pathway may have evolved to recognize general characteristics of the ‘protein corona’ (eg., net charge) in order to promote clearance of a broad range of particle types and surface chemistries. For instance, in the lung environment, surfactant proteins expected to comprise the major adsorbed corona proteins have also been shown to enhance cell surface levels of SR-A and promote uptake of iron oxide nanoparticles by alveolar macrophages.50-51 Future studies which determine whether ENPs made “stealth” to scavenger receptor recognition by cationic or polymeric surface modifications cause similar phenotypic and gene regulatory effects in macrophages would be informative.
Overall, our results suggest several contributing mechanisms by which ENP exposure may modulate pathogen recognition by macrophages (Figure 7). For instance, availability of membrane receptors needed for pathogen recognition may be limited at the transcriptional level as well as by general membrane turnover mediated by ENP-stimulated endocytosis. We expect that the endocytic internalization of SR-A following ENP binding18 would both reduce the amount of cell surface SR-A available to interact with bacteria cell wall components and promote the amount of intracellular SR-A available to interact with selected toll receptor signaling complexes.52 While this mechanism of “ligand hijacking” may explain some of the observed effects on LPS activation patterns, additional processes initiated intracellularly after SR-A dependent uptake of ENPs must also play a prominant role in determining the impact of ENP exposure on macrophage activation. For instance, unlike silica, SPIO caused upregulation of multiple antioxidant gene pathways in macrophages. The importance of oxidative stress as a predictive measure of nanoparticle toxicity has been previously discussed by Nel and colleagues.53 Although the low level of oxidative stress caused by SPIO does not result in cytotoxicity, subcytotoxic levels of intracellular reactive oxygen species (ROS) may impair cytoskeletal actin assembly and macrophage phagocytic activity, and are hypothesized to be a contributing risk factor for pneumonia associated with acute respiratory distress.54 It was also reported that ROS inhibits the endocytosis of SR-A,55 which could also contribute to the reduced phagocytosis of S. pneumoniae observed following SPIO exposure in our study. Thus, the fact that macrophages use common pathways in the recognition of ENPs and pathogens creates several plausible mechanisms for interaction among these agents. The impacts each of these potential mechanisms has on susceptibility to lung infection in vivo need to be further quantified in vivo. However, previous studies which demonstrated pre-exposure of mice by inhalation to copper nanoparticles15 or carbon nanotubes14 results in reduced bacterial lung clearance suggest common macrophage-mediated processes observed in our study likely occur in vivo. Given additional epidemiological evidence of linkages between particulate exposure and increasing incidence of pneumonia,9 further investigations are warranted to sort these mechanisms out and to determine the ENP exposure levels where these effects become important. This complexity also highlights the importance of recognizing that the potential health hazards following exposure to ENPs may be indirectly mediated, and nanotoxicology screening strategies must consider whether these emerging materials alter susceptibility to other environmental exposures.
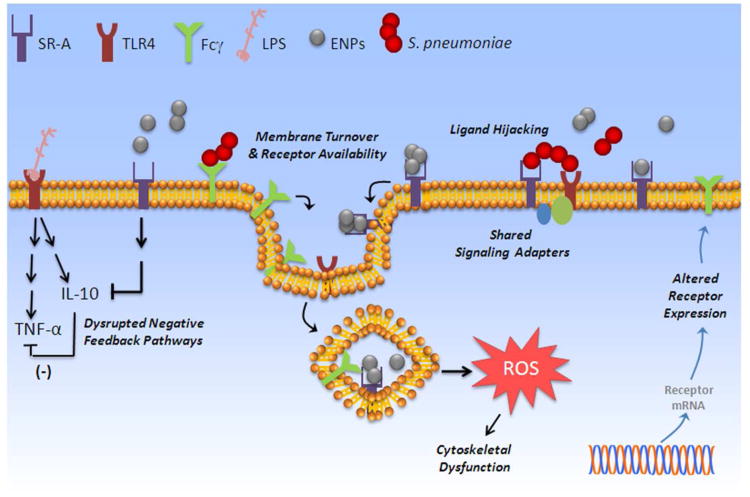
Figure 7. Potential Mechanisms of Interaction Between ENP and Bacterial Pathways in Macrophages.
Exposure to ENPs may impact normal macrophage phenotype and functional recognition of pathogenic bacteria through multiple mechanisms. Potential contributing mechanisms identified include changes in transcriptional levels of key regulators of macrophage activation, altered membrane availability of cell surface phagocytic receptors (scavenger receptors, toll receptors, Fc receptors) due to ENP-stimulated endocytosis, as well as compensatory changes in cytoskeletal function and phagocytic activity associated with low levels of oxidative stress.
Materials and Methods
Nanomaterials and Nanomaterial Characterization
Superparamagnetic iron oxide (SPIO) nanoparticles (Fe3O4) were prepared by chemical co-precipitation at 23°C, as previously described.29 Briefly, 0.2M FeCl3 (anhydrate, EM Science) and 0.1 M FeSO4·7H2O (J.T. Baker) were mixed in 100 ml H2O under nitrogen, followed by injecting 10ml of 29 weight % NH3·H2O into the mixture with vigorous stirring. A black color change upon addition of NH3·H2O implied formation of iron hydroxide in the solution. After the solution was dehydrated with overnight stirring, SPIO colloidal nanoparticles were collected by precipitation with a neodymium magnet (Magnetics, Inc.) and washed three times with deionized water (>18 megaohm). All reactions were conducted under nitrogen atmosphere or in closed containers to protect samples from oxidation. Independent batches of SPIO particles synthesized by the same methodology were used for the microarray studies and bacterial phagocytosis assays. Fluorescent-labeled amorphous silica nanospheres (50 nm) were obtained from Corpuscular Inc. (Cold Spring, USA). Endotoxin was tested using Toxinsensor Chromogenic LAL (GenScript, L00350) at a nanoparticle concentration of 100 μg/ml. All ENPs tested below detection limits (< 0.01 EU/ml). Dynamic light scattering (DLS) and zeta potentiometry measurements were conducted at a particle concentration of 100 μg/ml using a BI 90 particle sizer (Brookhaven Instruments Corp., Holtsville, NY). The particles were initially dispersed in fetal bovine serum (FBS), sonicated, and the final volume was brought up to the required nanoparticle concentration. DLS values for water-dispersed particles were generated with vortex mixed samples. The DLS and zeta potential measurements reported are the average of at least 5 replicates. The cytotoxic potential of the ENPs was determined by measuring release of intracellular lactate dehydrogenase using the CytoTox-ONE membrane integrity assay (Promega, G7890) per manufacturer's recommendation.
Macrophage Isolation and Treatment
Wildtype and SR-A deficient mice bred on a C57/BL6 background (originally obtained from Jackson Laboratory) were maintained at the University of Washington and shipped to PNNL for use in isolation of bone marrow derived macrophages (BMDMs). All procedures were approved by the PNNL Institutional Animal Care and Use Committee. Primary bone marrow cells were isolated from 4-6 week old mice following euthanasia by CO2 asphyxiation. Cells were flushed from isolated femurs and cultured at a concentration of 6×106 cells in 100 cm2 dishes with 10 mL RPMI 1640 supplemented with 10% FBS and 20% conditioned medium obtained from L929 cell cultures. Every two days nonadherent cells were washed off and fresh medium was added. Conditioned medium was obtained from L929 cells cultured in RPMI supplemented with L-glutamine, Pen-Strep, 10% FBS. For cell treatments, 1.5×106 adherent cells were re-plated in 6-well plates in RPMI 1640 supplemented with L-glutamine, penicillin/streptomycin, and 10% FBS. Just prior to exposure, the nanoparticles were sonicated (Fisher Scientific, Model 100) in FBS and brought up with media to the required concentration and volume. Complete media without ENPs was used as a control. After 24 hrs of ENP exposure, media was aspirated, cells were washed, and 2 ml fresh media with or without 10 ng/ml LPS (Escherichia coli O111:B4, LIST Biological Laboratories Inc.) was added.
Bacterial Phagocytosis
Streptococcus pneumoniae (ATCC 6303) was obtained from American Type Culture Collection (VA, USA). The bacteria were initially grown overnight on blood agar plates in an incubator maintained at 37°C and 5% CO2. A bacterial colony was collected using an inoculating loop and cultured with shaking in 4ml of Todd Hewitt broth supplemented with 0.5% Yeast Extract (BD Difco, USA) maintained at 37°C until an OD600 of 0.3-0.4 was reached. To label the bacteria, 100 μl of 0.1 mg/ml Alexa Fluor® 647 Succinimidyl Ester (Invitrogen) was added to 109 heat-inactivated bacteria in phosphate buffer (pH 8) and incubated for 1 hour. Excess dye was removed by centrifugation and the bacteria were resuspended in phosphate buffered saline (PBS). For exposure, 1:10 ratio of bacteria to macrophages (MOI) was determined and incubated in FBS for 30 minutes before resuspending in RPMI-1640. Cells pretreated with ENPs were washed three times with PBS and exposed to fresh medium containing bacteria. To facilitate uniform cell exposure to the bacteria at t = 0 hr, the plates were centrifuged at 1000g for 10 min and uptake was allowed to occur for 2 hrs.
Flow Cytometry and Microscopy Analysis
Following exposure to fluorescent bacteria or LPS for the required time, cells were washed with phosphate-buffered saline (PBS), harvested by mild trypsinization and scraping, centrifuged at 1000 g for 5 min and resuspended in PBS. The cell-associated ENPs, bacteria or LPS were analyzed using a FACSCalibur (BD Biosciences, San Jose, CA). All experiments were performed using triplicate samples and at least 10,000 cells per treatment, and the mean fluorescence was determined using FCS Express software (De Novo Software, Los Angeles, CA). For microscopy analysis, cells were grown on glass slides and after exposure to ENPs and bacteria for the required experimental time, were fixed using 3% paraformaldehyde washed and mounted using ProLong® Gold (Invitrogen, Carlsbad, CA). Confocal images were obtained using a Zeiss LSM 710 upright confocal microscope (Carl Zeiss, USA) with a 40× water immersion objective. Image analysis was performed on reconstructed z-stack images using Volocity 3D Image Analysis software (Perkin-Elmer, Waltham, MA).
ELISA and RT-PCR Analysis
Protein levels in cell culture supernatant were measured by enzyme-linked immunosorbent assay (ELISA, R&D Systems) per manufacturer's instructions. RNA was isolated from the 6-well plates using RNeasy Mini Kit (Qiagen, 74104) protocol with a final elution volume of 30μl RNAse free H20. cDNA was generated using QuantiTect Reverse Transcription (Qiagen, 205310), per manufacturer instructions. Real time PCR amplification was performed using a final volume of 20μl containing 3μl CDNA, 2μl Primer Mix (5μM each forward and reverse, Eurofins MWG Operon), 5μl RNAse free H2O and 10μl POWER SYBR Green (Applied Biosystems) on a ABI StepOnePlus thermocycler (Applied Biosystems). The amplification protocol was according to manufacturer's recommendation and included an initial denaturing step at 90°C for 10min, followed by 45 cycles with denaturation at 95°C for 15 sec and an annealing/elongation step of 60°C for 1min. Melting curve analyses were performed in each run. Relative expression was determined using the ΔΔCT method with samples normalized to the expression level of the cyclophilin A (CPHA) transcript. The primers used included (5′- 3′): CPHA sense: GAGCTGTTTGCAGACAAAGTTC, CPHA antisense: CCCTGGCACATGAATCCTGG; IL10 sense: CAGAGCCACATGCTCCTAGA, IL10 antisense: GTCCAGCTGGTCCTTTGTTT; TNFα sense, TCTTCTCATTCCTGCTTGTGG, and TNFα antisense, GGTCTGGGCCATAGAACTGA.
Microarray Analysis
Whole genome microarray analysis was performed using Affymetrix Mouse Genome 430A 2.0 chips (Affymetrix, Santa Clara, CA, USA; 22,690 probesets). Total RNA was collected using the RNeasy Kit (Qiagen, Valencia, CA). RNA integrity and purity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Complementary DNA was synthesized from 3 μg of total RNA in the presence of an oligo-dT primer containing a T7 RNA polymerase promoter, and an in vitro transcription reaction was performed in the presence of a mixture of biotin-labeled ribonucleotides to produce biotinylated cRNA from the cDNA template, according to manufacturer's protocols (Affymetrix One-Cycle Target Labeling Kit). Biotin-labeled cRNA (15 μg) was fragmented to a size range between 50-200 bases for array hybridization. After hybridization, the arrays were washed, stained with streptavidin-phycoerythrin, and scanned at a resolution of 2.5 microns using an Affymetrix GeneChip Scanner 3000. Quality control parameters were assessed throughout the experimental process to measure the efficiency of transcription, integrity of hybridization, and consistency of qualitative calls. The synthesis of the cDNA and cRNA, and the fragmentation of cRNA were assessed using the Agilent 2100 Bioanalyzer. Spike-in control transcripts also were monitored to verify hybridization integrity.
Raw intensity data were quantile normalized by Robust Multi-Array Analysis summarization and analyzed by ANOVA unequal variance with Tukey's post-hoc and 5% false discovery rate (FDR) calculation using GeneSpring v.11 (Silicon Genetics, Redwood City, CA). Criteria for differential expression were an absolute fold change of 1.5 and a FDR adjusted p-value < 0.05 for comparison to control or LPS treatment groups. To identify gene subsets whose expression profiles indicate potential co-regulation between ENP pretreatment and LPS challenge groups, data were transformed to ENP minus treated controls prior to statistical analysis to subtract the individual effect of ENP pretreatment from the comparison. Thus, genes identified as significantly different from the resulting comparison of the ENP-LPS group to the LPS group alone are defined as displaying greater than additive behavior. Genes that passed the differential expression criteria with an FDR adjusted p-value of <0.05 for ENP-LPS combined treatments compared to the LPS treatment alone were included for downstream analysis. Raw and normalized Affymetrix data files are available online through Gene Expression Omnibus accession GSE 44294.
Bioinformatic Analysis
Unsupervised bidirectional hierarchical clustering of microarray data was performed using Euclidean distance metric and centroid linkage clustering to group treatments and gene expression patterns by similarity. K-means clusters of genes were calculated by Euclidean distance for 14 clusters over 13 iterations. The clustering algorithms, heat map visualizations and centroid calculations were performed with Multi-Experiment Viewer software based on log2 expression ratio values or Z-scores. Z-scores, which were used for clustering and heatmap visualizations, were calculated on gene expression data for the ENP-LPS and LPS treatment groups by subtracting the mean and dividing by standard deviation across the dataset. Functional enrichment statistics were determined with Metacore (GeneGo, St. Joseph, MI) to identify the most significant biological and metabolic processes affected by silica, SPIO and LPS. The statistical scores in MetaCore are calculated separately for up-regulated and down-regulated genes within a treatment group using a hypergeometric distribution where the p-value represents the probability of a particular mapping arising by chance for experimental data compared to the background, which included all genes on the Affymetrix platform. Ingenuity Pathway Analysis (IPA; Ingenuity Systems) was used to determine the significant regulation of inflammatory response pathways with prediction of pro- and anti-inflammatory responses after exposure to ENPs and LPS. Inflammation pathways that were significantly enriched in our dataset were calculated by right-tailed Fisher exact test (p<0.05) as a measure of the likelihood that the association between a set of genes and a related function is significantly greater than random association compared to all genes on the Affymetrix platform.
Supplementary Material
Acknowledgments
The authors thank Drs. Norman Karin and Cosmin Mihai for technical assistance. Support for this research was provided by the National Institute of Environmental Health Sciences through grants U19 ES019544 and RO1 ES016212.
Footnotes
Supporting Information Available: Additional data describing nanoparticle characterization, gene expression summary results, cytotoxicity analysis, and phagocytic activity are provided as Supporting Information. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Oberdorster G, Maynard A, Donaldson K, Castranova V, Fitzpatrick J, Ausman K, Carter J, Karn B, Kreyling W, Lai D, et al. Principles for Characterizing the Potential Human Health Effects from Exposure to Nanomaterials: Elements of a Screening Strategy. Part Fibre Toxicol. 2005;2:8. doi: 10.1186/1743-8977-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perkins TN, Shukla A, Peeters PM, Steinbacher JL, Landry CC, Lathrop SA, Steele C, Reynaert NL, Wouters EF, Mossman BT. Differences in Gene Expression and Cytokine Production by Crystalline Vs. Amorphous Silica in Human Lung Epithelial Cells. Part Fibre Toxicol. 2012;9:6. doi: 10.1186/1743-8977-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waters KM, Masiello LM, Zangar RC, Tarasevich BJ, Karin NJ, Quesenberry RD, Bandyopadhyay S, Teeguarden JG, Pounds JG, Thrall BD. Macrophage Responses to Silica Nanoparticles Are Highly Conserved across Particle Sizes. Toxicol Sci. 2009;107:553–569. doi: 10.1093/toxsci/kfn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dockery DW, Pope CA, 3rd, Xu X, Spengler JD, Ware JH, Fay ME, Ferris BG, Jr, Speizer FE. An Association between Air Pollution and Mortality in Six U.S. Cities. N Engl J Med. 1993;329:1753–1759. doi: 10.1056/NEJM199312093292401. [DOI] [PubMed] [Google Scholar]
- 5.Schwartz J. Air Pollution and Hospital Admissions for the Elderly in Detroit, Michigan. Am J Respir Crit Care Med. 1994;150:648–655. doi: 10.1164/ajrccm.150.3.8087333. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz J. Air Pollution and Hospital Admissions for Heart Disease in Eight U.S. Counties. Epidemiology. 1999;10:17–22. [PubMed] [Google Scholar]
- 7.Pope CA, 3rd, Burnett RT, Thun MJ, Calle EE, Krewski D, Ito K, Thurston GD. Lung Cancer, Cardiopulmonary Mortality, and Long-Term Exposure to Fine Particulate Air Pollution. JAMA. 2002;287:1132–1141. doi: 10.1001/jama.287.9.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz J. What Are People Dying of on High Air Pollution Days? Environ Res. 1994;64:26–35. doi: 10.1006/enrs.1994.1004. [DOI] [PubMed] [Google Scholar]
- 9.Neupane B, Jerrett M, Burnett RT, Marrie T, Arain A, Loeb M. Long-Term Exposure to Ambient Air Pollution and Risk of Hospitalization with Community-Acquired Pneumonia in Older Adults. Am J Respir Crit Care Med. 2010;181:47–53. doi: 10.1164/rccm.200901-0160OC. [DOI] [PubMed] [Google Scholar]
- 10.Antonini JM, Roberts JR, Jernigan MR, Yang HM, Ma JY, Clarke RW. Residual Oil Fly Ash Increases the Susceptibility to Infection and Severely Damages the Lungs after Pulmonary Challenge with a Bacterial Pathogen. Toxicol Sci. 2002;70:110–119. doi: 10.1093/toxsci/70.1.110. [DOI] [PubMed] [Google Scholar]
- 11.Zhou H, Kobzik L. Effect of Concentrated Ambient Particles on Macrophage Phagocytosis and Killing of Streptococcus Pneumoniae. Am J Respir Cell Mol Biol. 2007;36:460–465. doi: 10.1165/rcmb.2006-0293OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang HM, Antonini JM, Barger MW, Butterworth L, Roberts BR, Ma JK, Castranova V, Ma JY. Diesel Exhaust Particles Suppress Macrophage Function and Slow the Pulmonary Clearance of Listeria Monocytogenes in Rats. Environ Health Perspect. 2001;109:515–521. doi: 10.1289/ehp.01109515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanobetti A, Woodhead M. Air Pollution and Pneumonia: The “Old Man” Has a New “Friend”. Am J Respir Crit Care Med. 2010;181:5–6. doi: 10.1164/rccm.200909-1445ED. [DOI] [PubMed] [Google Scholar]
- 14.Shvedova AA, Fabisiak JP, Kisin ER, Murray AR, Roberts JR, Tyurina YY, Antonini JM, Feng WH, Kommineni C, Reynolds J, et al. Sequential Exposure to Carbon Nanotubes and Bacteria Enhances Pulmonary Inflammation and Infectivity. Am J Respir Cell Mol Biol. 2008;38:579–590. doi: 10.1165/rcmb.2007-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JS, Adamcakova-Dodd A, O'Shaughnessy PT, Grassian VH, Thorne PS. Effects of Copper Nanoparticle Exposure on Host Defense in a Murine Pulmonary Infection Model. Part Fibre Toxicol. 2011;8:29. doi: 10.1186/1743-8977-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braydich-Stolle LK, Speshock JL, Castle A, Smith M, Murdock RC, Hussain SM. Nanosized Aluminum Altered Immune Function. ACS Nano. 2010;4:3661–3670. doi: 10.1021/nn9016789. [DOI] [PubMed] [Google Scholar]
- 17.McDermott JE, Archuleta M, Thrall BD, Adkins JN, Waters KM. Controlling the Response: Predictive Modeling of a Highly Central, Pathogen-Targeted Core Response Module in Macrophage Activation. PLoS One. 2011;6:e14673. doi: 10.1371/journal.pone.0014673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orr G, Chrisler WB, Kassens KJ, Tan R, Tarasevich BJ, Markillie LM, Zangar RC, Thrall BD. Cellular Recognition and Trafficking of Anionic Nanoparticles by Macrophage Scavenger Receptor A. Nanotoxicology. 2011;5:296–311. doi: 10.3109/17435390.2010.513836. [DOI] [PubMed] [Google Scholar]
- 19.Greaves DR, Gordon S. Thematic Review Series: The Immune System and Atherogenesis. Recent Insights into the Biology of Macrophage Scavenger Receptors. J Lipid Res. 2005;46:11–20. doi: 10.1194/jlr.R400011-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Dunne DW, Resnick D, Greenberg J, Krieger M, Joiner KA. The Type I Macrophage Scavenger Receptor Binds to Gram-Positive Bacteria and Recognizes Lipoteichoic Acid. Proc Natl Acad Sci U S A. 1994;91:1863–1867. doi: 10.1073/pnas.91.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Areschoug T, Gordon S. Scavenger Receptors: Role in Innate Immunity and Microbial Pathogenesis. Cell Microbiol. 2009;11:1160–1169. doi: 10.1111/j.1462-5822.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 22.Arredouani MS, Yang Z, Imrich A, Ning Y, Qin G, Kobzik L. The Macrophage Scavenger Receptor Sr-AI/II and Lung Defense against Pneumococci and Particles. Am J Respir Cell Mol Biol. 2006;35:474–8. doi: 10.1165/rcmb.2006-0128OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki H, Kurihara Y, Takeya M, Kamada N, Kataoka M, Jishage K, Ueda O, Sakaguchi H, Higashi T, Suzuki T, et al. A Role for Macrophage Scavenger Receptors in Atherosclerosis and Susceptibility to Infection. Nature. 1997;386:292–296. doi: 10.1038/386292a0. [DOI] [PubMed] [Google Scholar]
- 24.Seimon TA, Obstfeld A, Moore KJ, Golenbock DT, Tabas I. Combinatorial Pattern Recognition Receptor Signaling Alters the Balance of Life and Death in Macrophages. Proc Natl Acad Sci U S A. 2006;103:19794–19799. doi: 10.1073/pnas.0609671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haworth R, Platt N, Keshav S, Hughes D, Darley E, Suzuki H, Kurihara Y, Kodama T, Gordon S. The Macrophage Scavenger Receptor Type A Is Expressed by Activated Macrophages and Protects the Host against Lethal Endotoxic Shock. J Exp Med. 1997;186:1431–1439. doi: 10.1084/jem.186.9.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohnishi K, Komohara Y, Fujiwara Y, Takemura K, Lei X, Nakagawa T, Sakashita N, Takeya M. Suppression of TLR4-Mediated Inflammatory Response by Macrophage Class A Scavenger Receptor (Cd204) Biochem Biophys Res Commun. 2011;411:516–522. doi: 10.1016/j.bbrc.2011.06.161. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi Y, Miyaji C, Watanabe H, Umezu H, Hasegawa G, Abo T, Arakawa M, Kamata N, Suzuki H, Kodama T, et al. Role of Macrophage Scavenger Receptor in Endotoxin Shock. J Pathol. 2000;192:263–272. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH692>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Wermeling F, Sundqvist J, Jonsson AB, Tryggvason K, Pikkarainen T, Karlsson MC. A Regulatory Role for Macrophage Class A Scavenger Receptors in TLR4-Mediated LPS Responses. Eur J Immunol. 2010;40:1451–1460. doi: 10.1002/eji.200939891. [DOI] [PubMed] [Google Scholar]
- 29.Minard KR, Littke MH, Wang W, Xiong Y, Teeguarden JG, Thrall BD. Magnetic Particle Detection (MPD) for In-Vitro Dosimetry. Biosens Bioelectron. 2012;43:88–93. doi: 10.1016/j.bios.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehnert BE. Pulmonary and Thoracic Macrophage Subpopulations and Clearance of Particles from the Lung. Environ Health Perspect. 1992;97:17–46. doi: 10.1289/ehp.929717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lay JC, Bennett WD, Kim CS, Devlin RB, Bromberg PA. Retention and Intracellular Distribution of Instilled Iron Oxide Particles in Human Alveolar Macrophages. Am J Respir Cell Mol Biol. 1998;18:687–695. doi: 10.1165/ajrcmb.18.5.2948. [DOI] [PubMed] [Google Scholar]
- 32.Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC, Kennedy K, Hai T, Bolouri H, Aderem A. Systems Biology Approaches Identify ATF3 as a Negative Regulator of Toll-Like Receptor 4. Nature. 2006;441:173–178. doi: 10.1038/nature04768. [DOI] [PubMed] [Google Scholar]
- 33.Schroder K, Irvine KM, Taylor MS, Bokil NJ, Le Cao KA, Masterman KA, Labzin LI, Semple CA, Kapetanovic R, Fairbairn L, et al. Conservation and Divergence in Toll-Like Receptor 4-Regulated Gene Expression in Primary Human Versus Mouse Macrophages. Proc Natl Acad Sci U S A. 2012;109:5925–5926. doi: 10.1073/pnas.1110156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pena OM, Pistolic J, Raj D, Fjell CD, Hancock RE. Endotoxin Tolerance Represents a Distinctive State of Alternative Polarization (M2) in Human Mononuclear Cells. J Immunol. 2011;186:7243–7254. doi: 10.4049/jimmunol.1001952. [DOI] [PubMed] [Google Scholar]
- 35.Biswas SK, Lopez-Collazo E. Endotoxin Tolerance: New Mechanisms, Molecules and Clinical Significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 36.Mosser DM, Edwards JP. Exploring the Full Spectrum of Macrophage Activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogdan C, Vodovotz Y, Nathan C. Macrophage Deactivation by Interleukin 10. J Exp Med. 1991;174:1549–1555. doi: 10.1084/jem.174.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping Gene Expression in Activated and Resting Primary Macrophages by IL-10. J Immunol. 2002;169:2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- 39.Fulton WB, Reeves RH, Takeya M, De Maio A. A Quantitative Trait Loci Analysis to Map Genes Involved in Lipopolysaccharide-Induced Inflammatory Response: Identification of Macrophage Scavenger Receptor 1 as a Candidate Gene. J Immunol. 2006;176:3767–3773. doi: 10.4049/jimmunol.176.6.3767. [DOI] [PubMed] [Google Scholar]
- 40.Murray PJ. The Primary Mechanism of the IL-10-Regulated Antiinflammatory Response is to Selectively Inhibit Transcription. Proc Natl Acad Sci U S A. 2005;102:8686–8691. doi: 10.1073/pnas.0500419102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capsoni F, Minonzio F, Ongari AM, Carbonelli V, Galli A, Zanussi C. IL-10 Up-regulates Human Monocyte Phagocytosis in the Presence of IL-4 and IFN-gamma. J Leukoc Biol. 1995;58:351–358. doi: 10.1002/jlb.58.3.351. [DOI] [PubMed] [Google Scholar]
- 42.Michlewska S, Dransfield I, Megson IL, Rossi AG. Macrophage Phagocytosis of Apoptotic Neutrophils Is Critically Regulated by the Opposing Actions of Pro-Inflammatory and Anti-Inflammatory Agents: Key Role for TNF-alpha. FASEB J. 2009;23:844–854. doi: 10.1096/fj.08-121228. [DOI] [PubMed] [Google Scholar]
- 43.Hinderliter PM, Minard KR, Orr G, Chrisler WB, Thrall BD, Pounds JG, Teeguarden JG. ISDD: A Computational Model of Particle Sedimentation, Diffusion and Target Cell Dosimetry for in Vitro Toxicity Studies. Part Fibre Toxicol. 2010;7:36. doi: 10.1186/1743-8977-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chao Y, Makale M, Karmali PP, Sharikov Y, Tsigelny I, Merkulov S, Kesari S, Wrasidlo W, Ruoslahti E, Simberg D. Recognition of Dextran-Superparamagnetic Iron Oxide Nanoparticle Conjugates (Feridex) Via Macrophage Scavenger Receptor Charged Domains. Bioconjug Chem. 2012;23:1003–1009. doi: 10.1021/bc200685a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chao Y, Karmali PP, Mukthavaram R, Kesari S, Kouznetsova VL, Tsigelny IF, Simberg D. Direct Recognition of Superparamagnetic Nanocrystals by Macrophage Scavenger Receptor SR-AI. ACS Nano. 2013;7:4289–4298. doi: 10.1021/nn400769e. [DOI] [PubMed] [Google Scholar]
- 46.Morrow PE. Possible Mechanisms to Explain Dust Overloading of the Lungs. Fundam Appl Toxicol. 1988;10:369–384. doi: 10.1016/0272-0590(88)90284-9. [DOI] [PubMed] [Google Scholar]
- 47.Orr JS, Puglisi MJ, Ellacott KL, Lumeng CN, Wasserman DH, Hasty AH. Toll-Like Receptor 4 Deficiency Promotes the Alternative Activation of Adipose Tissue Macrophages. Diabetes. 2012;61:2718–2727. doi: 10.2337/db11-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue K, Takano H, Yanagisawa R, Hirano S, Sakurai M, Shimada A, Yoshikawa T. Effects of Airway Exposure to Nanoparticles on Lung Inflammation Induced by Bacterial Endotoxin in Mice. Environ Health Perspect. 2006;114:1325–1330. doi: 10.1289/ehp.8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the Interleukin-10 Receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 50.Kuronuma K, Sano H, Kato K, Kudo K, Hyakushima N, Yokota S, Takahashi H, Fujii N, Suzuki H, Kodama T, et al. Pulmonary Surfactant Protein A Augments the Phagocytosis of Streptococcus pneumoniae by Alveolar Macrophages through a Casein Kinase 2-Dependent Increase of Cell Surface Localization of Scavenger Receptor A. J Biol Chem. 2004;279:21421–21430. doi: 10.1074/jbc.M312490200. [DOI] [PubMed] [Google Scholar]
- 51.Ruge CA, Schaefer UF, Herrmann J, Kirch J, Canadas O, Echaide M, Perez-Gil J, Casals C, Muller R, Lehr CM. The Interplay of Lung Surfactant Proteins and Lipids Assimilates the Macrophage Clearance of Nanoparticles. PLoS One. 2012;7:e40775. doi: 10.1371/journal.pone.0040775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu X, Yi H, Guo C, Zuo D, Wang Y, Kim HL, Subjeck JR, Wang XY. Pattern Recognition Scavenger Receptor Cd204 Attenuates Toll-Like Receptor 4-Induced Nf-KappaB Activation by Directly Inhibiting Ubiquitination of Tumor Necrosis Factor (TNF) Receptor-Associated Factor 6. J Biol Chem. 2011;286:18795–18806. doi: 10.1074/jbc.M111.224345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nel A, Xia T, Madler L, Li N. Toxic Potential of Materials at the Nanolevel. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 54.O'Reilly PJ, Hickman-Davis JM, Davis IC, Matalon S. Hyperoxia Impairs Antibacterial Function of Macrophages through Effects on Actin. Am J Respir Cell Mol Biol. 2003;28:443–450. doi: 10.1165/rcmb.2002-0153OC. [DOI] [PubMed] [Google Scholar]
- 55.Aguilar-Gaytan R, Mas-Oliva J. Oxidative Stress Impairs Endocytosis of the Scavenger Receptor Class A. Biochem Biophys Res Commun. 2003;305:510–517. doi: 10.1016/s0006-291x(03)00796-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.