Summary
Large non-coding RNAs are emerging as an important component in cellular regulation. Considerable evidence indicates that these transcripts act directly as functional RNAs rather than through an encoded protein product. However, a recent study of ribosome occupancy reported that many large intergenic ncRNAs (lincRNAs) are bound by ribosomes, raising the possibility that they are translated into proteins. Here, we show that classical non-coding RNAs and 5′-UTRs show the same ribosome occupancy as lincRNAs demonstrating that ribosome occupancy alone is not sufficient to classify transcripts as coding or non-coding. Instead, we define a metric based on the known property of translation whereby translating ribosomes are released upon encountering a bona fide stop codon. We show that this metric accurately discriminates between protein-coding transcripts and all classes of known non-coding transcripts, including lincRNAs. Taken together, these results argue that the large majority of lincRNAs do not function through encoded proteins.
Introduction
The mammalian genome encodes many thousands of genes thought to encode large non-coding RNAs (Birney et al., 2007; Carninci et al., 2005; Derrien et al., 2012), including ∼3500 termed ‘large intergenic non-coding RNAs’ (lincRNAs) (Guttman et al., 2009; Guttman et al., 2010). The lincRNAs have recently been shown to play key roles in diverse biological processes and are emerging as an important new class of regulatory molecules (Guttman et al., 2011; Guttman and Rinn, 2012; Orom et al., 2010; Ulitsky et al., 2011; Wang and Chang, 2011). The lincRNAs resemble messenger RNAs (mRNAs) with respect to transcription, length, and splicing structure yet differ in that (i) they do not contain open reading frames (ORFs) that show the cross-species mutational pattern expected of evolutionarily conserved proteins (Guttman et al., 2009; Guttman et al., 2010); (ii) they do not contain ORFs that show the within-species substitution patterns expected of recently evolved proteins (Figure S1, see Methods); (iii) they tend to be highly enriched in the nucleus (Derrien et al., 2012); (iv) many physically interact with chromatin regulatory proteins to affect gene expression (Guttman et al., 2011; Guttman and Rinn, 2012; Wang and Chang, 2011); and (v) they rarely produce proteins detectable by mass-spectrometry (Banfai et al., 2012; Slavoff et al., 2013). Together, these results suggest that lincRNAs function directly as RNA molecules rather than through a translated protein product.
Recently an approach, termed ribosome profiling, was described that enables the global analysis of translation in a wide range of organisms including yeast and mammals. This method involves digestion of RNA followed by separation of 80S ribosomes based on their size and density through a sucrose cushion (Ingolia et al., 2009; Ingolia et al., 2011). The associated RNAs are then sequenced to assess the occupancy of ribosomes on RNAs. Ribosome profiling can be used to identify mRNAs associated with 80S ribosomes, distinguish ribosome-free 3′-untranslated regions (3′-UTRs) on these messages, and assess the quantitative dynamics of translation within translated regions (Ingolia et al., 2009; Ingolia et al., 2011). Global identification of the locations of 80S ribosomes can be used to identify novel translation events. Defining the full set of translation products is important for identifying regions that encode functional proteins (Stern-Ginossar et al., 2012). Yet, not all translation events lead to stable, functional polypeptides. Instead, this translation could be important for regulation of a down-stream ORF (Geballe and Morris, 1994), contribute to the antigenic potential of pathogens (Starck et al., 2008), regulate mRNA stability by inducing nonsense-mediated decay (Smith and Steitz, 1998), and may even serve as a source of proto-proteins enabling the evolution of novel proteins (Carvunis et al., 2012). Finally, some events may have no biological function at all – representing biological noise without deleterious consequence to the organism. Thus, the detection of 80S ribosomes on an RNA does not, alone, provide evidence that the RNA encodes a functional protein. For example, the host messages for a number of snoRNAs are translated and this is used to induce rapid degradation through the nonsense mediated decay pathway, thereby allowing disposal of the mature RNA after the snoRNAs have been extracted from intronic regions (Smith and Steitz, 1998). Thus, it is important to develop strategies for using the 80S footprint data provided by ribosome profiling experiments to distinguish true messengers that encode functional proteins from those that are non-coding.
Given the many lines of evidence that lincRNAs do not encode functional proteins, we examined the ribosome profiling data in greater detail. In particular, we considered whether non-coding RNAs should be expected to resemble 3′-UTRs with respect to their ribosome profiles. There is an important distinction between these two situations. When a ribosome engaged in translating an open reading frame (ORF) encounters a stop codon, it is actively disassembled and is recycled (Jackson et al., 2010; Kisselev and Buckingham, 2000) (Figure 1a); this creates a sharp drop in the ribosome occupancy in 3′-UTRs relative to the ORF. In contrast, in transcripts that have not been selected to produce a distinct protein product, it is possible that the ribosome initiates translation at multiple locations, and thus may show significant occupancy across regions within the transcript or even over the entire transcript. (In theory, ribosomes might also scan the transcript without engaging in translation; we will address this possibility elsewhere.) Thus, non-coding transcripts might show very different ribosome profiles than 3′-UTRs.
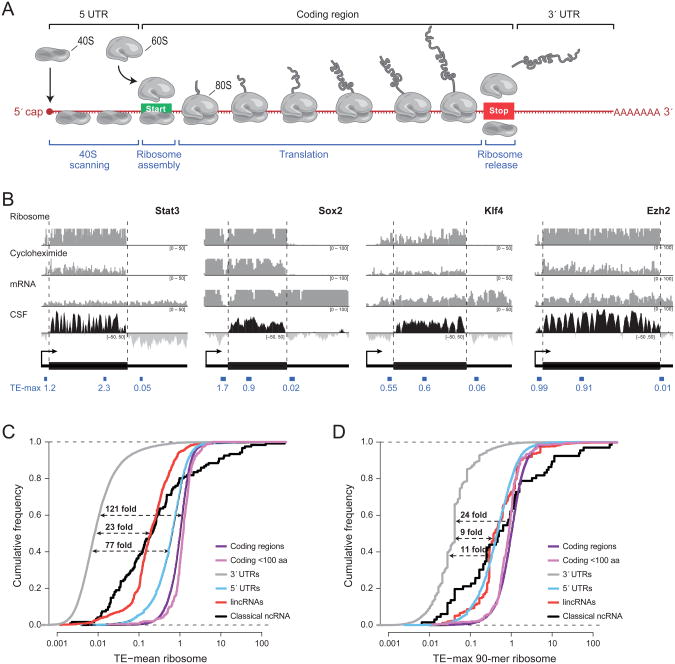
Figure 1. Properties of the translational efficiency score.
(a) An overview of mRNA translation. (b) Examples of ribosome profiling data over four mRNAs: Stat3, Sox2, Klf4, and Ezh2. The first three rows show, respectively, the sequencing coverage in counts (y-axis) of the ribosome-associated fraction, ribosome-associated fraction after treatment with cycloheximide, and polyA-selected total RNA per nucleotide (x-axis) on the associated transcript. The fourth row shows the codon substitution frequency (CSF) score across the mRNA which indicates the degree to which the sequence shows the evolutionary conservation pattern expected in protein-coding regions. Black corresponds to conserved coding potential (CSF>0) and light grey to lack of conserved coding potential (CSF<0). Dashed lines correspond to the boundaries of the coding region of the mRNA and the location and score of the max 90-mer translational efficiency (TE) score is shown for the 5′-UTR, 3′-UTR (thin black boxes), and coding region (thick black boxes). (c) Cumulative distribution of the average TE score across coding regions (purple line), small coding regions (magenta line), 3′-UTRs (gray line), 5′-UTRs (blue line), classical ncRNAs (black line), and lincRNAs (red line). The dashed lines show the median separation relative to 3′-UTRs for 5′-UTRs (bottom), lincRNAs and classical ncRNAs (middle line), and coding regions (top line). (d) Cumulative distribution of the TE computed using the max 90-mer window across the same classes. See also Figure S1.
Here, we study the pattern of ribosome occupancy on transcripts to evaluate whether lincRNAs show evidence of encoding functional proteins. In particular, we analyze the existing ribosome profiling data across other non-coding regions, including 5′-UTRs and classical non-coding RNAs (such as small nuclear and nucleolar RNAs, microRNAs precursors, and large ncRNAs such as telomerase RNA and RNase P) all of which are well-established to be non-coding(Eddy, 2001). We find that known non-coding controls also show ribosome profiles that differ from those of the 3′-UTRs of protein-coding genes. Moreover, 5′-UTRs resemble coding regions of protein-coding genes more closely than do lincRNAs.
To systematically analyze the pattern of ribosome occupancy across different classes of RNAs, we developed a metric that we term the ribosome release score (RRS), which identifies functional protein-coding transcripts with greater sensitivity by detecting the termination of translation at the end of an open reading frame (Jackson et al., 2010; Kisselev and Buckingham, 2000). We show that the RRS sharply distinguishes between the class of well-established protein-coding regions and other classes such as 5′-UTRs and classical ncRNAs, and that the class of lincRNAs closely resembles the other non-coding classes with respect to this metric. Because 5′-UTRs and classical ncRNAs do not encode functional proteins, the similar pattern of ribosome occupancy on the lincRNAs indicates that they too are unlikely, as a class, to produce functional proteins.
Results
We began by defining a variety of control sets whose coding status is well-accepted: (i) the translated regions, 5′-UTRs, and 3′-UTRs of 10,050 known protein-coding transcripts (“genes encoding typical-size proteins”), (ii) the translated regions of 639 known protein-coding transcripts encoding a protein with <100 amino acids (“genes encoding small proteins”), and (iii) 130 well-established non-coding RNAs that are well expressed in mouse ES cells (referred to as “classical non-coding RNAs”), including small nuclear RNAs, small nucleolar RNAs, microRNA precursors, and large ncRNAs, such as telomerase RNA, RNase P, Neat1, and Neat2/Malat1 (see Methods).
Relative to these control sets, we analyzed a set of 167 expressed lincRNAs whose RNA levels were well-represented in the Ingolia et al. dataset (see Methods). We previously defined these lincRNA transcripts based on a chromatin signature of active transcription (Guttman et al., 2009). This lincRNA set excludes a small group of 11 genes that have short open reading frames (ORFs) that show a pattern of evolutionary conservation characteristic of protein-coding genes (Guttman et al., 2009; Lin et al., 2011) and thus may encode a small protein; we refer to this set as the CSF+ set (see Methods). In addition, this lincRNA set excludes a small group of 7 genes that have non-conserved long ORFs but have been annotated as putative protein-coding genes based on homology to other proteins (see Methods).
We analyzed the previously published ribosome profiling data in mouse ES cells (Ingolia et al., 2011) for each class of RNAs. We note that the absolute level of ribosome occupancy (as assayed by the ribosome profiling method) is not a useful metric, because it is strongly correlated with RNA levels for both coding and non-coding transcripts (r=0.89). To overcome this problem, a relative metric was proposed called the “translational efficiency” (TE) (Ingolia et al., 2011). For each gene, this relative metric is defined as the number of sequencing reads observed among ribosome-associated RNA divided by the number of reads in total poly-A+ RNA. The TE score can be computed as either (i) the average value (TE-mean) across an entire feature (transcript or region) or (ii) the maximum value (TE-max) in windows of a specified size within a feature. Ingolia et al. based their conclusions on the TE-max metric with a window size of 90 bases, in order to account for potential translation in short coding regions within a longer non-coding transcript. Accordingly, we used the same metric.
We first confirmed the previous observations that (i) coding regions differ sharply from 3′-UTRs (Figure 1b, c, d) and (ii) the lincRNAs resemble coding regions more closely than 3′-UTRs (Figure 1c,d, 2a). It was these observations that raised the possibility that most lincRNAs are translated into proteins.
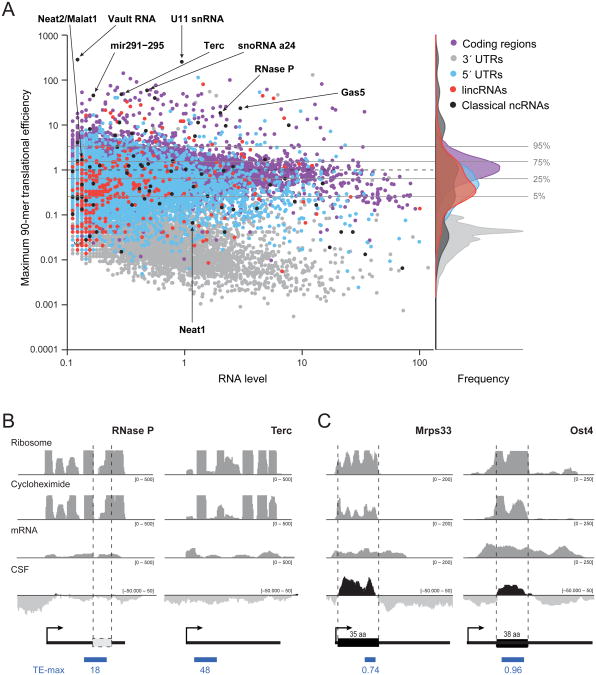
Figure 2. Translational efficiency of the maximum 90-mer fails to separate translated and non-translated RNAs.
(a) Scatter plot of RNA expression (log scale, x-axis) compared to the TE of the maximum 90-mer (log scale, y-axis) for coding regions (purple dots), 3′-UTRs (gray dots), 5′-UTRs (blue dots), classical ncRNAs (black dots), and lincRNAs (red dots). Horizontal lines correspond to the indicated percentiles of the TE-max score for protein-coding regions. The overlaid density distributions of the TE-max scores for each feature are shown. (b) Two examples of classical ncRNAs that have very high translational efficiency scores: RNase P and the telomerase RNA (Terc). The four rows (ribosome, cycloheximide, mRNA and CSF) are as described in legend of Figure 1. Beneath is an ideogram of the RNA, the location of a potential ORF (white box), and score of the maximum 90-mer (blue box). (c) Examples of two small coding genes encoding 35- and 38-amino acid peptides. See also Figure S2.
Interestingly, we found that the same observations applied to the non-coding controls – that is, the classical non-coding RNAs and 5′-UTRs. As with the lincRNAs, these non-coding controls also more closely resemble protein-coding regions than 3′-UTRs with respect to either the TE-mean (Figure 1c) or the TE-max metric (Figure 1d, 2a). Indeed, ∼48% of these non-translated controls have translational efficiency scores that exceed the 95th percentile of the scores for 3′-UTRs (Figure 1c,d).
5′-UTRs
One possible explanation for the high TE scores for 5′-UTRs is that it results solely from the translation of a small number of upstream ORFs (uORFs). To explore this possibility, we first excluded regions within the 5′-UTR that contained ORFs beginning with a canonical start codon (AUG). After removing these uORFs, we still observed a striking difference between 5′ UTRs and 3′ UTRs using both the TE-mean and TE-max (Figure S1). We further excluded the regions surrounding all ORFs beginning with a near-cognate start codon (CUG, GUG, or UUG), which together with canonical ORFs account for roughly 75% of proposed uORFs (Fritsch et al., 2012; Ingolia et al., 2011; Lee et al., 2012). After removing these near-cognate uORFs, we still observed a similar difference compared to 3′-UTRs (Figure S1). While some of the observed ribosome occupancy in 5′-UTRs is surely due to uORFs (a topic which we discuss elsewhere), the results indicate that the observed ribosome occupancy cannot be explained solely by sharply defined uORFs with cognate or near-cognate start codons.
Classical ncRNAs
We considered the possibility that the ribosome occupancy measure may be inflated for the classical ncRNAs, because many lack polyA tails. This is potentially relevant because the “translational efficiency” defined by Ingolia et al. is based on normalization to polyA-selected mRNA levels whereas the “ribosome-associated fraction” involved no polyA-selection (Ingolia et al., 2011). To control for polyA status, we generated a control RNA-Seq expression dataset that was not polyA-selected (see Methods). Importantly, when normalizing by the non-polyA selected RNA levels, we observe a nearly identical result – with the lincRNAs closely resembling the classical ncRNAs, and both being well separated from the 3′-UTRs (Figure S2). While this consideration affects abundance estimates for certain non-adenylated RNAs, the high observed ribosome occupancy is not simply due to the lack of poly-A tails on the classical ncRNAs.
Notably, some of the strongest translational efficiency scores (>99th percentile of 3′-UTRs in both polyA and non-polyA normalized samples) are observed for very well-characterized ncRNAs, including the telomerase RNA, RNase P, small nuclear RNAs, small nucleolar RNAs, vault RNA, miRNAs, and other RNAs which have been clearly demonstrated to function as RNA molecules (Bartel, 2004; Eddy, 2001; Shippen-Lentz and Blackburn, 1990; Stark et al., 1978) (Figure 2, Figure S2). This observation raised the possibility that some background RNA fragments, not protected by a translating 80S ribosome, contribute to the apparent translation of these non-coding RNAs. Indeed, such background could arise in transcripts that are highly structured and embedded in ribonucleoprotein complexes. A strategy for identifying true 80S ribosome footprints will be presented elsewhere.
Yet, even after removing likely non-80S ribosomal background reads, the translational efficiency score still does not distinguish between protein-coding genes and the non-coding controls (Figure S2). Accordingly, the “translational efficiency” score per se does not reliably indicate whether a class of transcripts – such as lincRNAs – are translated into functional proteins. The fact that the TE-max metric is not significantly better at separating these classes than the TE-mean implies that the observed results are not due to small patches within these regions.
Ribosome release separates classes of coding and non-coding RNAs
Because the translational efficiency score does not distinguish between the classes of coding and non-coding RNAs, we explored whether we could develop a metric that does distinguish between these classes.
We first attempted to define a metric based on translational start sites. Specifically, we used ribosome-profiling data generated after treatment with harringtonine, a drug that binds 60S ribosomes and has been reported to block initiation (Ingolia et al., 2011). As such data are expected to show enrichment at sites of translation initiation, we tested whether we could distinguish between the annotated start codons in protein-coding genes and randomly chosen start codons in classical non-coding RNAs. We observed little difference in enrichment in the former set relative to the latter set. This was true regardless of whether we studied the maximum enrichment seen over any start codon or the enrichment seen at the start codon of the ORF with the highest ribosome occupancy (see Methods, Figure S2). Because enrichment in the presence of harringtonine in this dataset did not distinguish between known protein-coding RNAs and classical non-coding RNAs, we could not use these data to study the translational status of lincRNAs.
We next attempted to define a metric based on translational stop sites. Because translating ribosomes are known to be released upon encountering a stop codon (Jackson et al., 2010; Kisselev and Buckingham, 2000), we reasoned that translation of a discrete (non-overlapping) open reading frame should be associated with a sharp decrease in ribosome occupancy between a protein-coding region and the subsequent 3′-UTR. Indeed, such a striking pattern occurs in known protein-coding transcripts (Ingolia et al., 2011). We thus searched for such a decrease in the other classes of non-coding transcripts (see Figures 1b, 2c). To do this, we defined putative coding regions within a transcript as all regions contained within an ORF– that is, a region in any reading frame that begins with a start codon and ends with the next in-frame stop codon. We defined the corresponding putative 3′-UTRs as a region beginning immediately downstream of the ORF and ending at the first subsequent start codon (in any reading frame); we truncated the putative 3′-UTR at the first subsequent start codon to allow for the possibility of poly-cistronic transcripts.
We defined the ribosome release score (RRS) to be the ratio between the total number of reads that are contained within the putative coding region and the total number of reads contained within the putative 3′-UTR, normalized by the respective lengths of these regions. We then normalized by the ratio of reads in the mRNA coverage between the two regions, which has the effect of correcting for any erroneous annotations of the 3′-UTRs (see Methods).
We found that the RRS does an excellent job of distinguishing between the class of known protein-coding genes and the class of non-coding control transcripts (Figure 3a). The median RRS for the protein-coding genes is ∼112, meaning that ribosome occupancy is ∼112-fold higher in the coding region preceding the stop codon compared to the region immediately after the stop codon (Figure 3a). By contrast, the RRS across all ORFs within the classical ncRNAs, 5′-UTRs and 3′-UTRs has a median of 0.81, reflecting roughly equal coverage before and after the stop codon (Figure 3a).
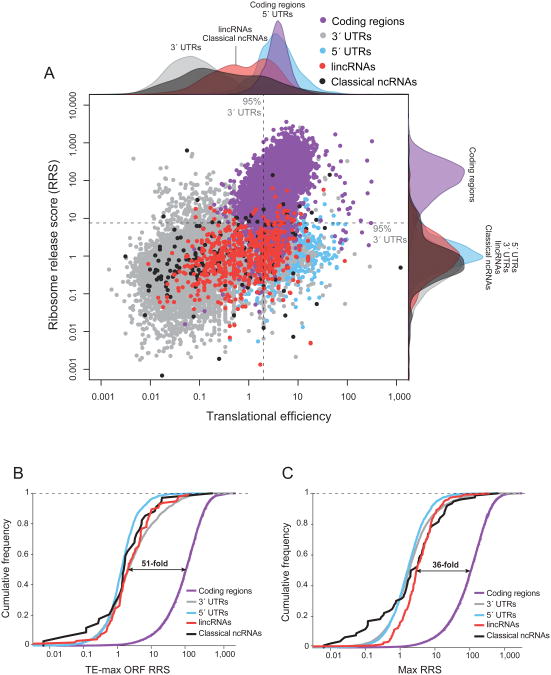
Figure 3. Ribosome release score separates translated and non-translated RNAs.
(a) Scatter plot of the TE-mean score for each ORF (log scale, x-axis) compared to its ribosome release score (log scale, y-axis) for coding genes (purple), 5′-UTRs (blue), 3′-UTRs (gray), classical ncRNAs (black), and lincRNAs (red). For known coding regions, we show the annotated ORF and for all other features we computed all possible ORFs (see Methods). The TE-mean score reflects the mean over each ORF. The dashed lines represent the 95th percentile of 3′-UTR values. Along each axis, all points are summarized using an overlaid density plot. (b) Cumulative density distribution of the RRS for the putative ORF with the highest ribosome occupancy (see Methods) for protein-coding regions (purple), 3′-UTRs (gray), 5′-UTRs (blue), classical ncRNAs (black), and lincRNAs (red). The dashed line indicates the fold difference between the median score for lincRNAs and protein-coding regions. (c) A cumulative density distribution of the maximum RRS over any ORF within a transcript (see Methods). See also Figure S3.
We then examined the RRS for lincRNAs. Importantly, the median RRS for lincRNA ORFs is ∼1, which is similar to the other non-coding regions and very different than for protein-coding regions (Figure 3a, Figure 4a,d,e).
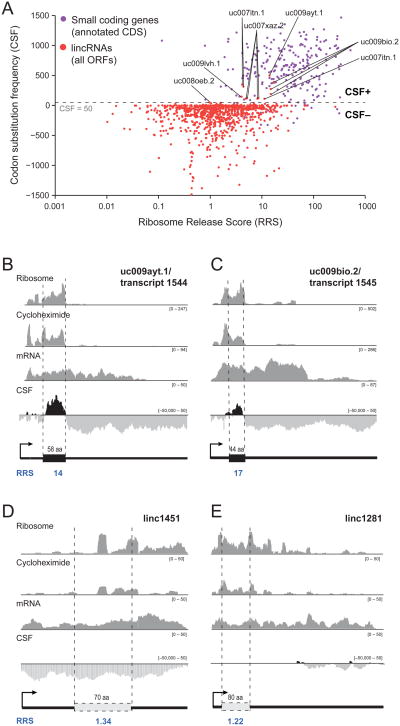
Figure 4. Ribosome release separates lincRNAs from small coding genes.
(a) A scatter plot of the RRS (log scale, x-axis) versus the CSF (y-axis) is plotted for each ORF of the lincRNAs (red points) and known small peptides (purple points). The dashed line corresponds to a CSF score of 50, the cutoff used to define a CSF+ set (CSF≥50) and CSF- set (CSF<50) (see Methods). (b) An example of a representative CSF+ transcript encoding a likely 58 amino acid protein with an RRS of 14. The four rows (ribosome, cycloheximide, mRNA and CSF) are as described in legend of Figure 1. The RRS score is noted in blue beneath the ideogram. (c) Another representative CSF+ transcript encoding a likely 44 amino acid protein with an RRS of 17. (d) A representative CSF- transcript, linc1451. The putative ORF (white) is defined as the ORF with the highest ribosome occupancy and has an RRS of 1.34. (e) Another representative CSF-transcript, linc1281. The putative ORF (white) has an RRS of 1.22. See also Figure S4.
To account for the possibility that there may be a single ORF buried within a long non-coding transcript, we repeated the analysis by analyzing only the ORF with the highest ribosome occupancy (as defined by the TE score). We still observed a striking difference (∼51-fold) between the class of protein-coding regions (median RRS ∼112) and both the classical ncRNAs and lincRNAs sets (median RRS 2.24 and 2.09 respectively) (Figure 3b).
In addition, even when looking only at the ORF with the highest RRS (RRS-max), we still observe a dramatic separation between protein-coding RNAs and the non-coding RNAs, with a ∼36-fold separation between the medians of coding regions and lincRNAs (Figure 3c). Notably, this separation is larger than the 24-fold separation observed between the medians of coding regions and 3′-UTRs using the TE-max metric (see Figure 1d).
Importantly, we note that the ability of the RRS metric to separate the classes of protein-coding regions and non-coding RNAs is robust to the precise implementation of the metric. For example, the RRS described above counts all reads overlapping the putative coding region. This provides a robust estimate of the ribosome occupancy and allows for the detection of tiny coding regions, including those that are smaller than the fragments (∼30nts) generated in the ribosome profiling assay. Indeed, the only limitation for detection using this approach is the size of the 3′-UTR, which would have to be larger than the fragment length generated in the ribosome profiling assay. A more conservative approach would include only reads that are fully contained within the putative coding region, which increases confidence in the assignment of the ribosome reads but would prevent detection of tiny coding regions. Using this alternative implementation, we obtain similar separations between the classes of known protein-coding regions and all non-coding RNAs (Figure S3). Similarly, the RRS described above used a trimmed 3′-UTR to account for possible polycistronic transcripts. If we alternatively define the putative 3′-UTR as the entire region following a stop codon, we observe similar separations (Figure S4).
While the RRS metric does an excellent job of distinguishing between the class of protein-coding genes and the class of non-coding RNAs, it does not provide a perfect classifier for individual transcripts within these sets because there is overlap between the distributions for coding and classical non-coding transcripts (Figure 3a). The known protein-coding transcripts with low RRS are primarily for transcripts with short 3′-UTRs (less than the fragment length), where estimation of the 3′-UTR counts are less reliable. Other cases occur when the 3′-UTRs have higher read coverage possibly due to ribosome read-through, overlapping translation of antisense transcription, or non-ribosomal contamination. As such, conclusions about coding potential of any individual transcript based on the RRS alone should be taken with care.
Importantly, the RRS is intended as a relative metric to compare different classes of transcripts. As such, the absolute level of the RRS may not be informative since it will be impacted by various features of a particular dataset. Similarly, the RRS is not intended to measure the relative proportion of heterogeneous translation states, where some proportion of a given transcript may be translated and the remainder non-translated; rather, it is meant to provide predominant translational features of a class of transcripts (e.g. mRNAs are predominately protein-coding and lincRNAs are predominantly non-coding). Furthermore, the RRS is not designed to identify specific translated regions within a transcript containing multiple overlapping or nearby translated regions. (We note that overlapping translation can act as a regulatory mechanism (Lu et al., 2004) or even produce alternative functional proteins (e.g. Ink/ARF (Sharpless and DePinho, 1999)).)
Taken together, the RRS shows strikingly different properties than the TE score. The TE score distinguishes the class of 3′-UTRs from all other RNA classes, while the RRS score distinguishes protein-coding regions from all other RNA classes. The TE score indicates that lincRNAs are bound by ribosomes to the same extent as 5′-UTRs or classical ncRNAs, while the RRS indicates that lincRNAs, like 3′-UTRs, 5′-UTRs and classical ncRNAs, rarely show the known signatures of translational termination that are characteristic of protein-coding regions.
Examination of specific cases
To further study the utility of the RRS, we revisited the previously published lincRNA collections (Guttman et al., 2011; Guttman et al., 2010). In 7 cases, transcripts that contain long ORFs (>100 amino acids) but lacked any conservation of their open reading frames had been subsequently reannotated as putative protein-coding genes based on their homology to other proteins (Pruitt et al., 2012). (These include four RNAs associated with pluripotency (Guttman et al., 2011).) Because these transcripts had been removed from our lincRNA collections, they provided useful test cases for the RRS method. While the translational efficiency scores for these 7 transcripts resemble those for lincRNAs and coding regions, the RRS scores clearly distinguish the 7 transcripts from lincRNAs and match those for coding regions; this provides independent evidence that these 7 transcripts are likely to be translated. (We note that our lincRNA catalogue contains a handful of additional transcripts that contain long ORFs, such as the Xist ncRNA, but show no evidence of coding potential based on evolutionary conservation or homology to known proteins; these cases have low RRS.)
In addition to these 7 transcripts, 12 additional lincRNAs showed high RRS but low CSF. These 12 transcripts also fail to show other detectable signatures of translation such as protein homology; they may also represent noise within the RRS distribution as a comparable proportion of the classical ncRNA transcripts would be similarly mischaracterized based on the RRS at this threshold (Figure 3a). This is consistent with the observation above that RRS is not a perfect classifier for individual transcripts. In contrast, methods such as CSF demonstrate sharper discriminative power between coding and non-coding transcripts (Guttman et al., 2009; Guttman et al., 2010) and are therefore more reliable on a per-transcript basis, but they fail to detect proteins that are not conserved (Lin et al., 2011).
In addition to a general analysis, earlier work noted two specific lincRNAs whose ribosomal profiles suggest the presence of a discrete translated region over an ORF (see Figure 5b,d in Ingolia et al.). These two examples both show TE scores similar to other lincRNAs but have very high RRS, consistent with those observed for other small coding regions. Notably, both of these examples belong to the CSF+ set noted above – that is, the small set of 11 chromatin-defined genes that show the pattern of evolutionary conservation characteristic of protein-coding genes and thus may be translated (Figure 4a,b,c). These two examples are thus among a handful of cases previously noted as likely to encode proteins (Guttman et al., 2009; Guttman et al., 2010). Notably, the genes in the CSF+ set have higher RRS scores (comparable to that seen for genes known to encode small proteins) – supporting the notion that many indeed are translated into functional proteins (Figure 4a). Importantly, we note that demonstrating that these or other transcripts encode novel proteins will require more direct experiments including identification of the encoded protein product in vivo.
These results support the notion that detailed analysis of ribosome release can aid in identifying transcripts that are translated into functional proteins and separating them from the numerous transcripts likely to function directly as RNAs.
Discussion
The observation that lincRNAs have higher ribosome occupancy than 3′-UTRs raised questions about the biological function of lincRNAs. Here, we show that a more complete analysis of the ribosome profiling data argues that most do not function by encoding small proteins. First, the ribosome occupancy observed on lincRNAs is comparable to that seen on other known non-coding RNAs: this implies that ribosome occupancy per se is not a reliable indicator of whether a transcript functions by encoding a protein. Second, the ribosomal-profiling data indicates that most lincRNAs, like other well-known non-coding RNAs, do not show the sharp decrease in ribosome occupancy following a stop codon that is consistently seen for known protein-coding genes (including those encoding small proteins). Taken together, these findings clearly demonstrate that lincRNAs are likely to function directly as RNA molecules rather than through encoded protein products.
If 5′-UTRs, lincRNAs and other known non-coding RNAs do not act through encoded proteins, why do they exhibit any ribosome occupancy at all? There are several possible explanations.
One possibility is that the fragments obtained in the ribosome profiling experiments result from protection of RNA by non-ribosomal RNA-protein complexes or other sources of non-ribosomal contamination. Indeed, some of the observed RNA fragments likely do not represent true 80S ribosomal footprints in that there are many fragments observed from classical non-coding RNAs that are predominantly localized in the nucleus (such as telomerase RNA, RNase P, snRNAs, and others). Consistent with this notion, the fragments derived from both cytoplasmic and nuclear classical non-coding RNAs show a different size distribution than that observed for protein-coding genes. In contrast, the distributions for 5′-UTRs and lincRNAs resemble the distributions observed for protein-coding genes. However, it is not possible to determine whether individual fragments arise from the ribosome based solely on size, and at least some of the reads on lincRNAs may also be non-ribosomal contamination. Future work including the affinity purification of 80S ribosomes and mass-spectrometry of small peptides will help address this topic as well as provide more accurate determination of the translational status of individual transcripts.
The RRS metric is well-suited to distinguish real translation from non-ribosomal contamination since it is robust to potential protection by non-ribosomal proteins as such protection should show no bias for the presence of a stop codon. Furthermore, as the RRS involves comparing putative translated regions within a transcript, it is robust to other potential sources of bias that lead to higher levels of contaminating non-ribosomal reads on specific classes of RNA, such as that observed on the classical ncRNAs.
An attractive possibility is that the presence of ribosomes on some ncRNAs may reflect the default engagement of the translation machinery with any capped transcripts present in the cytosol. In the absence of selective pressure for the use of a specific start codon, ribosomes may initiate translation at heterogeneous sites, with a bias towards the beginning of the transcript. Additionally, ribosomes that encounter short open reading frames may be capable of reinitiating translation after encountering stop codons. This may lead to the translation of multiple, overlapping reading frames, biased towards the 5′ end of the transcript. Such engagement may produce 80S footprints, but without the translation of a predominant functional reading frame and the sharp decrease in ribosome occupancy at its stop codon. The RRS metric is well suited to distinguish this behavior and thus provides a valuable metric to prioritize candidates for more in-depth characterization using labor intensive approaches such as mass spectrometry and or epitope tagging that have been used to evaluate whether translation events observed in ribosome profiling experiments contribute to a cells proteome (Banfai et al., 2012; Slavoff et al., 2013; Stern-Ginossar et al., 2012).
If classical ncRNAs and lincRNAs are bound by ribosomes, even though they lack canonical features of translation, this raises fascinating questions similar to those raised by pervasive transcription which occurs even in regions of the genome that do not produce stable RNA products (Birney et al., 2007; Carninci et al., 2005; Kapranov et al., 2007). Translation can have functional implications beyond the production of a protein product by affecting the stability, structure, and localization of an RNA. It can also reflect noise with no direct functional role that can be coopted over the course of evolution in order to create new coding sequence. More work will be needed to determine the biological implications of these diverse effects and their impact on the biogenesis and functions of non-coding RNAs.
Experimental Procedures
Filtering and alignment of sequencing reads
We downloaded the data from the Gene Expression Omnibus (GEO) using accession number GSE30839 (Ingolia et al., 2011). Fastq files were split into two sets based on whether the libraries were generated by polyA tailing or linker ligation. Samples generated by polyA tailing were directly aligned. For the remaining samples, the fastq files were filtered by first clipping all reads for the adaptor sequences using the fastx toolkit version 0.013. We then aligned all reads to the genome by using Tophat version 1.4.1 including a transcriptome reference defined using our ab initio transcriptome reconstruction of mouse embryonic stem cells (Guttman et al., 2010) along with all RefSeq- and UCSC-defined transcripts.
We note that all analyses presented of translational efficiency and ribosome release use the non-cycloheximide treated ribosome profiling data for consistency with the previous analysis (Ingolia et al., 2011). However, we note that the results are comparable even when using cycloheximide treated ribosome profiling data.
Defining a set of classical non-coding RNAs
We defined a set of classical non-coding RNAs. We first manually curated all non-coding RNAs included in the RefSeq catalog. Specifically, we retained ncRNAs that have been well-characterized as non-coding, including snRNAs, snoRNAs, miRNA precursors, snoRNA host genes, and large ncRNAs with publications demonstrating a role as an RNA (such as Neat1, Neat2/Malat1, and Gas5). We then added all small nuclear RNAs and miRNA precursors downloaded from the functional RNA database (Kin et al., 2007). Finally, we added a more extensive collection of literature curated large ncRNAs, taken from the functional RNA database (Kin et al., 2007). We mapped all RNAs to the genome and filtered those that overlapped a known protein-coding gene. We excluded all tiny RNAs <50nts because their short size (less than the length of a sequencing read) made it hard to estimate coverage. We merged these different sets and collapsed all overlapping transcripts into a single merged transcript. We excluded all translation-associated RNAs including tRNAs and ribosomal RNAs.
Defining a set of lincRNAs
We started with all chromatin-defined lincRNAs with transcripts that had been reconstructed from RNA-Seq data (lincRNA V3). The lincRNA set was filtered for all transcripts that contained a high CSF score or overlapped a known protein-coding transcript in the latest version of RefSeq (Pruitt et al., 2012) as previously described (Guttman et al., 2009; Guttman et al., 2010). As the data set of Ingolia et al. contains significantly lower coverage (by ∼5-fold) than the data used to reconstruct the lincRNA transcripts, we sought to analyze only transcripts with adequate coverage. Accordingly, we calculated the expression levels of each lincRNA from the data set of Ingolia et al. and only those lincRNAs with significant expression relative to the randomized genomic average (p<0.01). This yielded a set of 167 lincRNAs.
Defining ORFs
Using the orientation of each transcript, we defined all three possible frames and identified all possible start codons. For each start codon, we then searched for an in-frame stop codon. All regions defined by a start- and stop-codon pair were defined as a putative ORF.
Excluding upstream ORFs (uORFs)
To exclude the possibility that the high observed ribosome occupancy observed in 5′-UTRs was due to the presence of upstream ORFs, we defined all ORFs based on the presence of an AUG followed by an in-frame stop codon within the 5′-UTR regions. We then created a modified transcript which contained all regions except the putative ORFs and calculated the translational efficiency for the remaining regions. To exclude all possible near-cognate ORFs, we defined ORFs based on the presence of an AUG, CUG, GUG, or UUG start codon followed by an in-frame stop codon. We then computed translational efficiency over the remaining regions.
Defining a CSF+ set of transcripts
Using the 167 lincRNA genes, we computed all possible ORFs and calculated the codon substitution frequency (CSF) score for each ORF using the PhyloCSF package (Lin et al., 2011). We utilized a PhyloCSF cutoff of 50 to generate a CSF- set (CSF<50) and a CSF+ set (CSF>50). We have previously shown that a CSF cutoff of 50 accurately separates known protein-coding genes from known non-coding sequences (Guttman et al., 2010). Using this cutoff, we identified 11 chromatin-defined genes that exceed this cutoff (CSF+); based on their evolutionary conservation pattern, these genes are likely to encode proteins.
Computing Translational Efficiency score
We computed the translational efficiency (TE) score by counting the number of reads overlapping a feature in the ribosome fraction and dividing by the number of reads in the total RNA sample. For each feature, we counted only reads that were fully contained within the feature (thus avoiding counting reads that overlap multiple features). As the TE score is not a reliable estimator at low expression levels, we computed a TE score only for those features that had significant mRNA expression above a randomized genomic background (p<0.01). We placed no restrictions on the significance level for the ribosome coverage.
Defining the maximum TE score
We scanned a window of 90-nts across each transcript, with windows starting and ending fully within the transcript or feature. We then calculated the TE score, excluding windows with insufficient mRNA expression. We counted only reads that were fully contained in each window. To account for the difference in coverage level between ribosome and mRNA experiments, we normalized the score by dividing by the median TE-max of coding genes. For consistency with the methods utilized previously by Ingolia et al., we also computed the TE-max by identifying the 90-mer window with the largest number of positions covered by a ribosome read. In this case, ribosome reads were assigned to a nucleotide position as previously described (Ingolia et al., 2011). Briefly, we assigned reads to positions from the 5′-end of the fragment based on the following rules. For reads between (i) 29-30 nucleotides in length, we assigned the 15th position, (ii) 31-33 nucleotides in length, we assigned the 16th position, and (iii) 34-35 nucleotides we assigned the 17th position. We excluded all reads with a size <29 nucleotides and >35 nucleotides from all calculations because these are thought to represent non-ribosomal contamination. If multiple windows contained the same number of covered positions, we chose the 5′ most window from the transcript start site as the maximum window as previously described. We then computed the TE score for this defined window across all features. We identified comparable separations across classes using both of these measures (Figure S2).
Computing ORFs with the highest ribosome occupancy
We defined putative ORFs to be any region starting with an AUG start codon and ending with an in-frame stop codon. We computed the TE score for each ORF in the feature and retained the ORF with the highest TE score as the ORF containing the highest ribosome occupancy. We similarly defined all non-ORFs as regions that occurred between a stop codon and the next start codon (in any frame). To score the TE-max and TE-mean for ORF and non-ORF regions, we defined all ORF and non-ORFs within a transcript. We then computed the TE-max or TE-mean using only the regions contained within any of the ORF or non-ORF regions, respectively.
Generating a non-polyA selected RNA-Seq dataset
To generate a control mRNA dataset that was not limited to transcripts that were poly-adenylated, we generated a non-polyA selected RNA-Seq dataset using ribosomal depletion. We isolated RNA from wild-type mouse ES cells. RNA was fragmented using 1× Fragmentation buffer (Ambion # AM8740) by heating at 70C for 15 minutes. RNA was then dephosphorylated using Alkaline Phosphatase (Thermo # EF0651). RNA was ligated using the partial Illumina adaptor sequence with a 5′-phosphate and 3′-dideoxyC (AGATCGGAAGAGCGTCGTGTA) using T4 RNA ligase 1 (NEB # M0204). Ribosomal RNA was depleted using 120-mer biotinylated probes complementary to the 28S, 18S, and 5.8S ribosomal RNA sequences. The hybrids were captured with streptavidin magnetic beads (Invitrogen # 650-01) and removed. The remaining RNA was reverse transcribed using a primer complimentary to the linker (TACACGACGCTCTTCCGAT) using AffinityScript RT (Agilent # 200436) at 55C for 45 minutes. The cDNA was ligated with a second partial Illumina adaptor sequence with a 5′-phosphate and 3′-dideoxyC (AGATCGGAAGAGCACACGTCT) using T4 RNA ligase 1. A library was then generated using PCR containing primers with the entire Illumina adaptor sequences using Phusion polymerase (NEB # M0531) for 14 cycles. Libraries were sequenced on the Illumina HiSeq using 44 base single end reads.
Scoring putative start codons using harringtonine data
The four harringtonine datasets representing varying treatment times (90s, 120s, 150s and 180s) were analyzed separately. We defined the enrichment over a start codon as follows. First, we defined a start codon using the site of the AUG start codon. We then took all reads overlapping the start codon and defined a “peak” as the total genomic span covered by reads overlapping the start codon. We then counted all reads within the peak and divided by the length of the peak. This score was then divided by the total number of reads over the transcript divided by the length of the transcript. This score was taken as the enrichment of harringtonine values over a start codon compared to the rest of the transcript. For each known coding region, we used the annotated start codon to compute the enrichment of harringtonine overlapping the start codon. For non-coding transcripts, we defined putative ORFs and used the AUG site of each putative ORF as the putative start codon for analysis. We calculated the harringtonine enrichment for non-coding transcript either by using the putative ORF with the highest TE score or the ORF with the highest harringtonine enrichment.
Defining the Ribosome Release Score
For each ORF, we computed the number of ribosome reads fully contained in the ORF divided by the number of ribosome reads fully contained in the 3′-UTR, normalizing each count by the length of the respective region to define a preliminary estimate of ribosome release strength. To account for drops in ribosome coverage due to misannotated features or alternative termination rather than a true stop codon, we also compute the same ratio on RNA reads. The RNA ratio would be expected to show a similar drop in coverage under these circumstances but not for real stop codons. The RRS is defined as the ratio of the two normalized ratios (ultimately eliminating the need to normalize by region length):
As this score is normalized within a transcript, it accounts for differences in expression levels of the transcript itself.
For known protein-coding genes, we used the annotated coding regions and 3′-UTRs to calculate the RRS. For non-coding transcripts, we computed the RRS by first defining all possible ORFs. For each putative ORF, we defined the “3′-UTR” as the region between the stop codon and the next possible start codon (in any frame). The RRS score was defined only for ORFs with a significant expression level above randomized genomic background (p < 0.01). The RRS was not defined for an ORF if there were no fully contained reads in the ribosome fractions overlapping both the ORF and its 3′UTR. Because of this, we also excluded all ORFs and 3′-UTRs whose length was less than the size of the sequencing read length. Finally, the RRS was not defined for non-coding transcripts (such as telomerase RNA) that contain no ORFs. This led to 89 lincRNAs and 79 classical ncRNAs for which the RRS was defined.
While the RRS described above increases confidence that the ribosome reads originate from the coding region, it prevents detection of tiny coding regions, smaller than the read length used in the ribosome profiling assay. While in theory, this is more conservative, since it will miss tiny ORFs, it is possible that these ORFs are the actual translated coding regions and therefore the RRS is quite low. Because of these limitations, we also implemented an alternative version which did not require that the ribosome reads be fully contained within the ORF. In this case, we reasoned that any read overlapping the coding region would arise from a ribosome overlapping the coding region and counted these as ribosome reads in the coding region. We similarly computed the 3′-UTR scores as any read overlapping the 3′-UTR (excluding reads that also overlap the coding region which will artificially inflate its estimate). Using this RRS measure, we observed comparable separations between coding and all non-coding RNAs (Figure 3, Figure S3).
Importantly, we note that even when including the entire 3′-UTR, rather than truncating it by the presence of the next ORF, we obtain a comparable separation to that observed using the truncated 3′-UTRs. This demonstrates that the RRS is a robust metric for determining translational status.
Supplementary Material
Highlights.
Ribosome occupancy levels of lincRNAs are similar to classical ncRNAs and 5′-UTRs
Ribosome occupancy does not distinguish between protein-coding and non-coding RNAs
Protein-coding RNAs show ribosome release at stop codons, known ncRNAs do not
lincRNAs do not show evidence of ribosome release for any open reading frame
Acknowledgments
We thank Manuel Garber, and John Rinn for helpful discussions and thoughtful comments on the manuscript, Moran Cabili and Jesse Engreitz for critical reading and suggestions on the manuscript, Alex Shishkin for generating the non-polyA selected RNA-Seq libraries, Shari Grossman for help with population-level dN/dS calculations and Leslie Gaffney for assistance with figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Jr, Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012;22:1646–1657. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. Proto-genes and de novo gene birth. Nature. 2012;487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Non-coding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Fritsch C, Herrmann A, Nothnagel M, Szafranski K, Huse K, Schumann F, Schreiber S, Platzer M, Krawczak M, Hampe J, et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome Res. 2012;22:2208–2218. doi: 10.1101/gr.139568.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geballe AP, Morris DR. Initiation codons within 5′-leaders of mRNAs as regulators of translation. Trends Biochem Sci. 1994;19:159–164. doi: 10.1016/0968-0004(94)90277-1. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kin T, Yamada K, Terai G, Okida H, Yoshinari Y, Ono Y, Kojima A, Kimura Y, Komori T, Asai K. fRNAdb: a platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic Acids Res. 2007;35:D145–148. doi: 10.1093/nar/gkl837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev LL, Buckingham RH. Translational termination comes of age. Trends Biochem Sci. 2000;25:561–566. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- Lee S, Liu B, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A. 2012;109:E2424–2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247:546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM, Steitz JA. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5′-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897–6909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Ow Y, Jiang V, Tokuyama M, Rivera M, Qi X, Roberts RW, Shastri N. A distinct translation initiation mechanism generates cryptic peptides for immune surveillance. PLoS One. 2008;3:e3460. doi: 10.1371/journal.pone.0003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark BC, Kole R, Bowman EJ, Altman S. Ribonuclease P: an enzyme with an essential RNA component. Proc Natl Acad Sci U S A. 1978;75:3717–3721. doi: 10.1073/pnas.75.8.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.