Abstract
Extracellular domains of cell-surface receptors and ligands mediate cell-cell communication, adhesion, and initiation of signaling events, but most existing protein-protein “interactome” datasets lack information for extracellular interactions. We probed interactions between receptor extracellular domains, focusing on the Immunoglobulin Superfamily (IgSF), Fibronectin type-III (FnIII) and Leucine-rich repeat (LRR) families of Drosophila, a set of 202 proteins, many of which are known to be important in neuronal and developmental functions. Out of 20503 candidate protein pairs tested, we observed 106 interactions, 83 of which were previously unknown. We ‘deorphanized’ the 20-member subfamily of defective in proboscis IgSF proteins, showing that they selectively interact with an 11-member subfamily of previously uncharacterized IgSF proteins. Both subfamilies interact with a single common ‘orphan’ LRR protein. We also observed new interactions between Hedgehog and EGFR pathway components. Several of these interactions could be visualized in live-dissected embryos, demonstrating that this approach can identify physiologically relevant receptor-ligand pairs.
Introduction
In metazoans, cell surface and secreted proteins play essential roles in intercellular communication, cellular adhesion, and developmentally important signaling pathways by binding to other proteins in the extracellular milieu (Ben-Shlomo et al., 2003). Protein domain families that mediate these extracellular processes have expanded greatly during the evolution of complex multicellular organisms (Vogel and Chothia, 2006). Consequently, cell surface and secreted proteins comprise a substantial fraction of the human proteome (Almén et al., 2009; da Cunha et al., 2009; Diehn et al., 2006). Although vast amounts of protein interactome data have been generated in the last decade, extracellular and transmembrane proteins are greatly underrepresented in these data sets, due to the technical challenges that extracellular proteins present for systems biology and proteomics approaches (Wright et al., 2010). Producing extracellular molecules requires special conditions enabled by secretion, such as an oxidizing environment (for disulfide bonds) and specific post-translational modifications (predominantly glycosylation) for folding and function. Methods that target proteins to intracellular compartments, such as the nucleus in Yeast-Two Hybrid (Y2H), are unlikely to allow functional folding of most extracellular proteins. Furthermore, low-affinity interactions (i.e. KD ∼μM), which are difficult to detect, are known to dominate the extracellular interactome (van der Merwe and Barclay, 1994). Moreover, since many extracellular proteins contain transmembrane regions, affinity pull-downs and mass spectrometric methods tend to miss or mis-assign interactions as a consequence of the presence of the plasma membrane and/or detergents non-specifically subsuming hydrophobic, “sticky” and nearest neighbor interactions. For these reasons, there is a deficiency of reliable global interaction data for extracellular proteins.
The lack of reliable interaction data for transmembrane and extracellular proteins has been previously noted in large protein interactome studies and databases (Braun et al., 2009; Guruharsha et al., 2011; Miller et al., 2005). Braun et al. (2009) showed that extracellular proteins were refractory to detection by existing interactome methodologies, including Y2H. Also, a recent affinity-purification/mass spectrometry (AP-MS)-based interactome, the Drosophila Protein Interaction Mapping (DPiM) Project, underrepresents every one of the six extracellular and transmembrane protein classifications. These include cell adhesion molecules, cell junction proteins, defense/immunity proteins-IgSF family, extracellular matrix proteins, receptors, signaling molecules and transmembrane proteins. By contrast, only one intracellular category out of the remaining 21 was underrepresented (Guruharsha et al., 2011).
To address these problems, recent work has focused on the development of eukaryotic expression systems that use oligomerization to identify and assess low-affinity interactions between extracellular proteins. Clustering of ligands in various formats was found to be necessary for detecting interactions between DSCAM splice variants (Wojtowicz et al., 2007). Multimerization was also shown to enhance detection of interactions among the extracellular domains (ECD) of zebrafish Immunoglobulin Superfamily (IgSF) and leucine-rich repeat (LRR) proteins in an extracellular interaction screening screening assay (AVEXIS; Bushell et al., 2008; Söllner and Wright, 2009). Similarly, Ramani et al. (2012) has utilized a protein microarray format with multivalent protein-coated beads for a group of human IgSF, where multivalency enhanced binding signal by 10 to >150-fold.
In the present study, we utilize a high-throughput oligomerization-based methodology for detecting extracellular interactions between individually expressed recombinant ECDs in Drosophila melanogaster, which offers the advantage of rapid and robust functional tools to assess function. We applied these methods to nearly all the members of the IgSF, FnIII, and LRR families of the Drosophila melanogaster extracellular proteome. We expressed 202 proteins, and evaluated a total of 20503 unique pairwise interactions. We found 106 protein pairs that displayed detectable interactions, of which 83 (78%) are previously unknown. We confirmed several of these interactions using quantitative biophysical methods, and demonstrated that previous large-scale interactomes had failed to detect these interactions. We elucidated new interactions amongst known signaling pathways, and discovered that a 20-member IgSF subfamily of unknown function, the Dprs, interacts with an 11-member subfamily, also of unknown function. We demonstrated that these protein-protein interactions can be visualized in vivo by using oligomerized fusion proteins to stain live-dissected Drosophila embryos. We found that Dprs and their binding partners label specific subsets of cells within the central nervous system (CNS). For one Dpr-ligand pair, we used loss-of-function (LOF) and gain-of-function (GOF) genetics to demonstrate that ligand-receptor interactions discovered in vitro also occur in live embryos. Collectively, this study provides a framework with which to identify bona fide receptor-ligand partners, which can then be functionally defined in vivo during development, using genetic methodologies. The eventual extension of this approach to the entire Drosophila extracellular proteome will facilitate an understanding of the mechanisms through which these classes of proteins influence development and function.
Results
The choice of domain families for the interactome
The IgSF is the most highly represented extracellular protein domain in humans (∼0.3% of human protein-coding genes). Among all large protein domain families, the numbers of IgSF domains encoded in a genome correlate the most with organismal complexity (Vogel and Chothia, 2006). IgSF proteins are essential for intercellular communication during development of organ systems (Williams and Barclay, 1988). In the nervous system, they are required for cell migration, axon guidance, synaptogenesis, and synaptic plasticity (Yamagata et al., 2003). In the immune system, IgSF proteins are involved in many cell communication, migration and signaling events, and in molecular recognition of self vs. nonself (Pålsson-McDermott and O'Neill, 2007). IgSF proteins are known to engage other IgSF proteins in homophilic and heterophilic complexes that mediate distinct functionalities, such as formation of adhesion structures (e.g., Özkan et al., 2013), but as yet there has been no experimental cataloging of all IgSF protein interactions from any species. FnIII domains are structurally and functionally related to the IgSF fold, as both are composed of beta-sheet sandwiches and they often coexist within the same protein.
LRR proteins constitute another prominent extracellular family heavily utilized in the nervous and innate immune systems (Dolan et al., 2007). They have been found to be receptors for developmentally important signals in both Drosophila and vertebrates (Dolan et al., 2007; de Wit et al., 2011), and are involved in innate immune response (Valanne et al., 2011). In the Drosophila nervous system, LRRs mediate cell recognition events required for target selection by motor neurons and olfactory neurons (Hong et al., 2009; Kurusu et al., 2008). Since the pervasive nature of IgSF, FnIII and LRR proteins, we chose to focus our interactome on these three domain types. Furthermore, a large number of these genes are not annotated, the majority of them have not been studied, and only a small percentage have known protein interactors, so efforts to deorphanize them could fill this void and likely yield interactions that are functionally important.
Curation of Gene List and Cloning for Overexpression
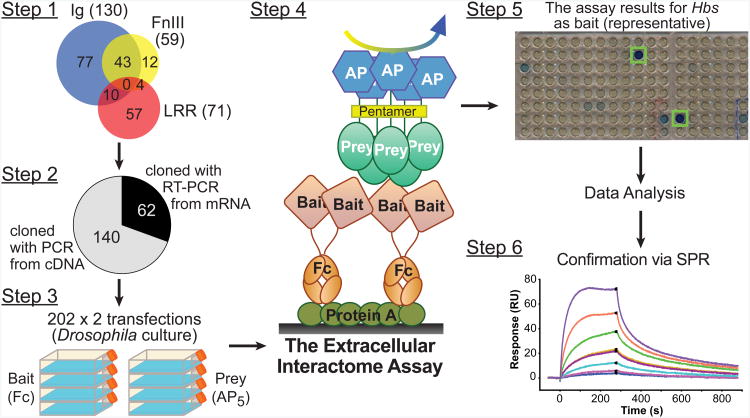
Previous efforts have been made to identify and annotate extracellular IgSF and LRR family proteins of D. melanogaster (Dolan et al., 2007; Vogel et al., 2003). We further searched the fly genome for proteins containing extracellular IgSF, FnIII and LRR domains (Extended Experimental Protocols: Bioinformatics, and Table S1), and identified 130 IgSF, 71 LRR, and 59 FnIII-containing extracellular and transmembrane proteins, counting one splice variant per gene, resulting in 203 non-redundant genes (Figure 1, step 1). We determined the boundaries of mature extracellular regions, “extracellular domains (ECD),” of these proteins by predicting signal peptides and, for membrane-bound proteins, transmembrane helices or glycosylphosphatidylinositol (GPI)-linkage sites. ECDs were cloned into our Drosophila expression vectors (Figure S1A) using conventional PCR if there were existing cDNAs, or with RT-PCR from primary RNA samples (Figure 1, step 2). Out of the 203 candidates, we were successful in cloning all but eight of these (96%) into expression vectors. For three proteins, we examined multiple splice variants; we also included three additional proteins of interest, for a total of 202 clones. Our collection is now available to the scientific community via the Berkeley Drosophila Genome Project (http://www.fruitfly.org/).
Figure 1. Schematic representation of the Extracellular Interactome.
Schematic representation of the Extracellular Interactome in the following steps: (1) Annotation of all proteins containing extracellular IgSF, FnIII and LRR domains (see also Table S1), (2) Cloning of the ECDs of target genes with PCR from cDNA or RT-PCR from mRNA into Drosophila expression plasmids, (3) Expression of all cloned ECDs in bait and prey formats in Drosophila cell culture, (4) Application of the Extracellular Interactome Assay, (5) Collection and analysis of the assay results, and (6) Confirmation of hits via biophysical methods, such as Surface Plasmon Resonance. See Figure S1 for a detailed explanation of tags used for bait and prey.
The Extracellular Interactome Assay (ECIA)
To create a system that would accurately assess interactions between extracellular proteins, we devised a high-throughput assay comprised of the following components (Figures 1 and S1B): (1) Bait and prey are expressed and secreted from cultured Drosophila Schneider 2 (S2) cells (Figure 1, step 3); (2) Bait are fused with human dimeric Fc, which allows for easy capture from conditioned media on staphylococcal Protein A-coated plates (Figure 1, step 4, Figure S1B); (3) Prey proteins are oligomerized into pentamers by fusing the ECDs to a pentameric helical region of rat COMP (cartilage oligomeric matrix protein) (Bushell et al., 2008; Holler et al., 2000; Voulgaraki et al., 2005). This avidity enhancement is a powerful means of capturing low affinity interactions and also recovering interacting pairs when the prey is only expressed at low protein concentrations (Figure 1, step 4, Figure S1B); and (4) Prey proteins are fused with human placental alkaline phosphatase (AP) for facile enzymatic detection using colorigenic substrates (Figure 1 steps 4 and 5, Figure S1B). The sensitivity in detection is amplified by the pentamerization of the AP tag.
The Fc-tagged bait proteins are captured by Protein A-coated plates directly from conditioned S2 media, and the AP-tagged proteins are directly assayed in conditioned media; therefore, the Extracellular Interactome Assay (ECIA) does not require any sample purification or buffer exchange, and is amenable to multiplexing and high-throughput measurements. Starting from initiation of cell culture work to the detection of interactions, the assay can be completed within five days (Figure 1, steps 3-5). The most labor-intensive aspects of the Extracellular Interactome are the cloning of genes from mRNA (Figure 1, step 2) and protein expression by transient transfection in eukaryotic cell culture (Figure 1, step 3). However, the labor burden for these steps increases only linearly with the number of proteins tested, allowing for many interactions to be tested without advanced robotic instrumentation. Yet, the ECIA is scalable, and could be automated for true high throughput, enabling this methodology to be expanded to very large collections, such as an entire proteome (i.e. millions of possible pairs).
The Extracellular Interaction Network of Drosophila IgSF, LRR and FnIII proteins
We performed the ECIA initially for a subset of 125 proteins, covering nearly all IgSF. Upon demonstrating feasibility, we expanded it to all the extracellular members of the IgSF, FnIII and LRR protein families. The two ECIA results closely matched, confirming the reproducibility of the assay (Figures S2A and S2B, see Extended Experimental Protocols: ECIA Data Internal Consistency and Reproducibility for detailed analysis,). For the analysis of our data, we utilized simple statistics, mainly Z-scores (see Extended Experimental Protocols: Interactome Data Analysis). All detected interactions observed were further verified by repeating the assay in both bait/prey orientations with fresh batches of proteins. We also performed titrations of the bait and prey samples in six selected interactions, which could be fit using concentration-dependent, single-site binding models. We observed that the strength of the binding signal was determined by the interaction partner present in lower abundance, (Figures S1C-G).
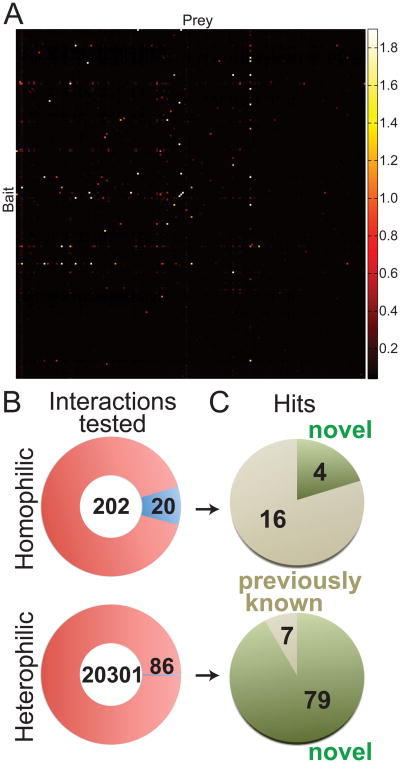
The ECIA dataset comprises a pairwise matrix of 42025 experiments (Figure 2A), corresponding to 20503 unique pairwise interactions (Figure S2B). Each heterophilic interaction was tested twice, with bait and prey in reverse orientations, and these results were in good agreement (see Extended Experimental Protocols: ECIA Data Internal Consistency and Reproducibility). The experimental matrix included testing of 202 potential homophilic pairs. Of the observed 106 unique interactions, 20 were homophilic (Figure 2B, Table S2). Based on the Drosophila protein interaction database DroID (Giot et al., 2003; Guruharsha et al., 2011; Murali et al., 2011) and an exhaustive literature search, 79 (92%) of the heterophilic and four (20%) of the homophilic interactions have not been previously reported (Figure 2C).
Figure 2. Interactions detected by our Extracellular Interactome.
(A) The matrix of data for 202×202 pairwise interactions measured using the Extracellular Interactome Assay (ECIA). Each row represents a single bait, while each column represents a single prey protein. The color scale maps to Absorbance values at 650 nm. See Figure S2 for the processed data matrix, and Table S2 for list of interactions detected.
(B) Number of interactions observed (blue slices) out of all unique homophilic and heterophilic interactions tested (red circles).
(C) Observed interactions are further classified as novel (green) vs. previously known (beige).
We analyzed published interactome datasets for the protein families included in our study (Yu et al., 2008). In total, there are 285 reported interactions for extracellular IgSF, FnIII and LRR proteins in the three large-scale experimental interactome datasets present in DroID. None of our 106 interactions were in the DroID-based list (Table S3). In fact, only one of the 285 DroID interactions has been reported independently (Table S4), and a majority of DroID interactions are physically unlikely, since the putative interactors are neither cell-surface nor extracellular proteins, but include many nuclear and intra-organelle proteins (Table S3). In contrast, 22 (21%) of our interaction hits were previously described in the literature. These results demonstrate that Y2H and AP-MS-based interactome studies fail to detect extracellular interactions, and a specialized interactome increases the yield and reliability.
The Four Subfamilies of IgSF As Observed in the Extracellular Interactome
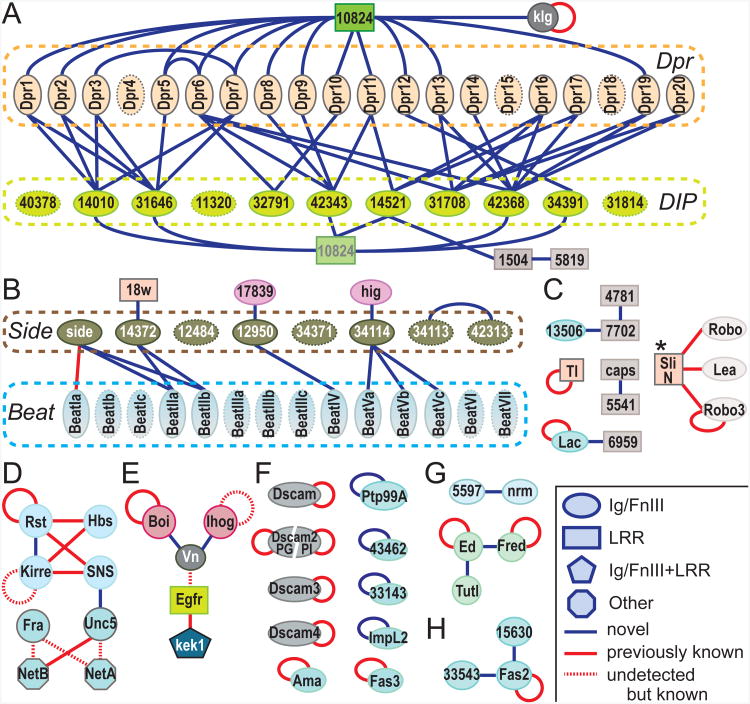
The pairwise interactions we detected are shown in Figure 3, highlighting four subfamilies and their interactions within the Drosophila IgSF (Figures 3A and 3B, Figure S3A-D). Two of these subfamilies, the “Beats” (after the protein Beaten Path) with 14 members, and Dpr homologs (after Defective in Proboscis Extension Response) with 20 members (Nakamura et al., 2002), were previously grouped by virtue of their respective sequence similarities only. For the Beat subfamily (Figure S3A), based on the recent finding that Beaten Path Ia engages another IgSF protein, Sidestep (also known as Side; Siebert et al., 2009), it was proposed that another seven proteins with related sequences to Sidestep might interact with Beats, but this was not experimentally demonstrated (Aberle, 2009; Zinn, 2009). The ECIA revealed these physical interactions from an unbiased screen, thus establishing a new network of Beat/Side interactions, and a new subfamily of Beat-interacting proteins that we term the “Sides” (Figures 3B, S3B).
Figure 3. The Extracellular Interactome of the Drosophila IgSF, FnIII and LRR.
The Extracellular Interactome of the Drosophila IgSF, FnIII and LRR, with the four IgSF subfamilies (Dpr, DIP, Side and Beat) delineated. See the inset for the classification of protein domain families and interactions. Uncharacterized gene names in the CGxxxx(x) format are shortened by removing the prefix CG. Homophilic interactions of boi, fred and robo3 were considered previously known based on the similar properties of their paralogs, ihog, ed and lea (robo2). * indicates interactions of the N-terminal fragment of Slit. The subsections highlight separate classes of proteins and interactions: (A) Dprs and the Dpr-DIP interaction network, (B) Beat and Side families, (C) proteins involved in IgSF–LRR interactions, (D) homologs of SYG and Netrin proteins, which are involved in synaptogenesis and axon guidance, (E) Vn and its interaction partners in the EGF and Hedgehog signaling pathways (See also Figure S4), (F) homophilic-only IgSF, including Dscams, (H) Fas2 and its interactions, and (G) others. For comparison, see Tables S3 and S4 for DroID reported interactions.
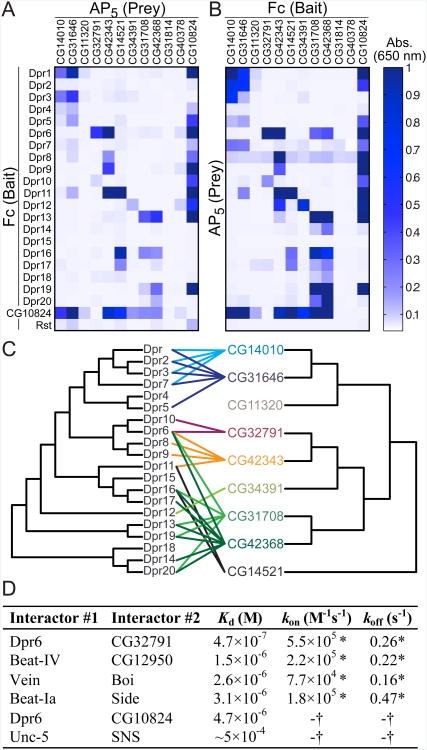
The fourth subfamily we observed is a set of closely related IgSF proteins that we found to selectively interact with members of the Dpr subfamily (Figures 3A and S3C). We name these previously uncharacterized proteins “DIPs”, for Dpr-Interacting Proteins (Figures 3A, S3D and S3E). In total, we find 17 of the 20 Dprs interacting specifically with 8 DIPs, to which we added three closely related proteins as putative DIPs to form the 11-member DIP family (See Extended Experimental Protocols: Definition of the DIP and Side families). Most members of the DIP subfamily cross-react with several members of the Dprs, and vice versa. Strikingly, we also identified CG10824, a secreted LRR-family protein, as a shared binding partner for a majority of Dprs and DIPs. We named this protein DIPc, or the “common DIP.” We performed a separate, independent mini-ECIA composed of only Dprs and DIPs, which confirmed the results from the larger ECIA (Figures 4A and 4B). This experiment also highlights the strong correlation in results when bait and prey are switched, which is apparent when Figures 4A and 4B are compared. The fact that the interacting subfamilies within these networks are comprised of proteins of related sequence, as opposed to random distributions of interacting pairs, gives us increased confidence that these are likely physiologically relevant binding partners.
Figure 4. The Dpr-ome and Biophysical Validation.
(A) Independent ECIA measurements of interactions between Dpr, DIP, DIPc and the negative control, Rst.
(B) Same as A, but with bait and prey reversed.
(C) The phylogenetic trees relating Dpr and DIP extracellular domain sequences, with interactions mapped between the Dprs and DIPs. Also see Figure S3 for the common domain topologies of the Dpr and DIP subfamilies.
(D) Binding affinities and kinetics measured with purified recombinant monomeric ECDs using SPR for a selection of the Extracellular Interactome hits. See Figure S5 for details. * indicates kinetic constants that may be inaccurate due to fast kinetics; 0.1 s−1 < koff < 0.5 s−1. † indicates that binding kinetics was too fast to be measured; koff > 0.5 s−1.
We based our definition of the four subfamilies of Drosophila IgSF on the interaction networks we experimentally identified (Figure 3), but their evolutionary relationships give insight about the origins of these interaction networks (Figure S3F). It is likely that the ancestral pairs of Dpr–DIP and Beat-Side multiplied through gene duplication events in the arthropod lineage, as we do not observe orthologous subfamilies outside arthropods. Those members of a subfamily that are most closely related to each other (e.g., Dpr1, Dpr2, Dpr3, and Dpr7) often interact with the same members of the partner subfamily that are also closest to each other (e.g., CG14010 and CG31646) (Figure 4C). Remarkably, the interaction network we identified is a powerful predictor of these evolutionary relationships, while the phylogenetic trees alone cannot predict protein interactions (Figure 4C). The evolutionary relationships between the interaction pairs we have discovered constitute an additional line of independent evidence bearing on the validity and accuracy of our interactome.
Vein, a Drosophila EGF, interacts with Hedgehog Co-receptors
The Drosophila IgSF includes Vein (Vn), a member of the Epidermal Growth Factor (EGF) family, and a ligand of the EGF receptor (EGFR) (Schnepp et al., 1996). With the ECIA, we found that Vn also binds two related Hedgehog (Hh) co-receptors, Ihog (interference Hedgehog) and Boi (Brother of Ihog). These interactions provide plausible physical connectivity between developmentally crucial EGF and Hh signaling pathways (Figure S4A), and there is evidence that the two pathways interact in Drosophila (Amin et al., 1999; Crozatier et al., 2002). For initiating EGF signaling, the EGF domain of Vn is known to engage EGFR, while the N-terminal domains of Vn, including the Ig domain, were shown to have a distinct function, and may engage an unknown ligand (Donaldson et al., 2004). To test whether this unknown ligand could be Ihog and/or Boi, we performed the ECIA using constructs composed of individual domains and several combinations of domains of Vn, Ihog and Boi (Figure S4B). We find that the Ig domain of Vein and the four Ig domains of Ihog or Boi were necessary and sufficient for this interaction. Our results suggest the possibility of a multi-component signaling complex involving EGFR, Vn, ihog, Boi and Hh (Figures S4C and S4D). Since Vn and Ihog/Boi have human orthologs (neuregulins and CDO/BOC, respectively), these interactions could potentially be relevant to the therapeutic targeting of ErbB receptors (human EGFR paralogs), neuregulins or the Hh pathway, although we have not tested the mammalian interactions. Our results will hopefully stimulate such experiments.
Validation of Extracellular Interactome Hits
We tested and confirmed a subset of our putative pairs using a quantitative biophysical method, Surface Plasmon Resonance (SPR), performed with purified monomeric ectodomains (Figures 4D and S5). We observe several of these interactions, including Side–Beat and Dpr–DIP interactions, to have dissociation constants (Kd) of ∼1 μM and weaker, as is typical for affinities between cell adhesion receptors (van der Merwe and Barclay, 1994). The equilibrium data for all interactions tested can be mathematically fit to a Langmuir isotherm, and therefore strongly support specificity of the interactions tested (Figure S5). These interactions exhibit fast kinetics and off-rates, which would have been difficult to capture without the avidity-enhancement or signal amplification as was used in our ECIA (Figures 4D and S5, compare with Figure S1G).
In Vivo Observation of Extracellular Interactions
We wished to determine whether these interactions could be detected in vivo. Flanagan and colleagues developed methods to visualize ligands in situ by staining vertebrate embryos with dimeric AP-tagged receptor fusion proteins (Flanagan et al., 2000). Fox and Zinn (2005) adapted these methods to live-dissected Drosophila embryos, using AP fusion proteins made with the appropriate arthropod glycosylation patterns (see Lee et al., 2009 for a video protocol). In our initial experiments with proteins produced for the ECIA, we examined if media from transiently transfected Drosophila cells containing pentamerized AP (AP5) fusion proteins could be used directly for staining embryos.
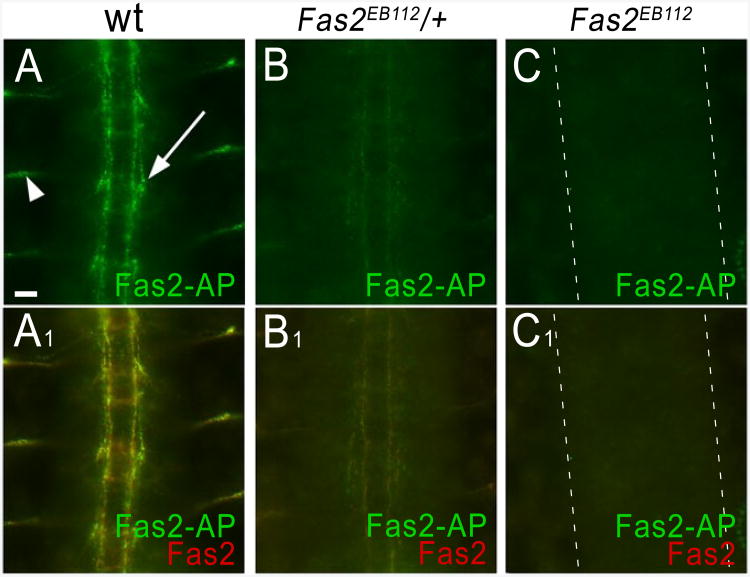
Our first experiments assessed staining for previously known interactions in order to give us confidence in the approach. Earlier work showed that muscle attachment sites in embryos mutant for the Syndecan (Sdc) failed to stain with a LAR-AP fusion protein, demonstrating that Sdc is responsible for LAR-AP binding to these sites (Fox and Zinn, 2005). In a similar manner, we used genetics to demonstrate that AP5 fusion proteins can recognize the appropriate ligands in live-dissected embryos. Fasciclin II (Fas2) is a homophilic IgSF adhesion molecule (Grenningloh et al., 1990; Schuster et al., 1996), and we detected homophilic Fas2 binding with ECIA (Figure 3H). In late embryos, Fas2 is expressed on longitudinal tracts in the CNS and on motor axons. Fas2-AP5 stains an identical pattern (Figure 5A). In embryos heterozygous for a null mutation, fas2EB112, Fas2-AP5 staining intensity is reduced (Figure 5B), and there is no staining in fas2EB112 hemizygous embryos, which completely lack Fas2 protein (Figure 5C), but have a normal pattern of CNS axons (Grenningloh et al., 1990; Figure S6). These data show that homophilic Fas2 interactions can be detected in vivo by staining with Fas2-AP5. Interestingly, we also identified two other Fas2 binding partners, CG15630 and CG33543, with ECIA (Figure 3H). However, these two genes are reportedly not expressed in late embryos (Graveley et al., 2011), and therefore fas2 null mutant embryos at stage 16 should exhibit no specific staining pattern for Fas2-AP5. We were also able to detect binding of another homophilic adhesion molecule, Fasciclin III (Fas3), at the appropriate sites (data not shown).
Figure 5. Fasciclin II-mediated homophilic adhesion can be detected by fusion protein binding to live embryos.
Live-dissected stage 16 embryos incubated with Fas2-AP5 were stained with anti-Fas2 mAb 1D4 (red) and rabbit anti-AP (green) antibodies. Three segments of the CNS and ventral periphery are shown in each image; anterior is up.
(A, A1) In wild-type (wt) embryos, the pattern of Fas2-AP5 binding is superimposable on the pattern of anti-Fas2 staining. Arrow, longitudinal tract; arrowhead, a motor axon bundle.
(B, B1) Embryos heterozygous for the null Fas2EB112 allele have only 50% of the normal amount of Fas2, and thus show less binding to Fas2-AP5 and weaker staining with anti-Fas2 mAb.
(C, C1) Homozygous Fas2EB112 embryos, which have no Fas2, fail to stain with either Fas2-AP5 or anti-Fas2 mAb. See Figure S6 for CNS patterning of these embryos. Scale bar, 10 μm.
In a second set of experiments with known ligand-receptor pairs, we stained embryos with AP5 fusion proteins for the three IgSF Roundabout (Robo) subfamily members, Robo, Robo2 (Lea), and Robo3. Each fusion protein stained midline glial cells and CNS axons, although there were subtle and interesting differences between the staining patterns (Figures S7A-C). These staining patterns correspond to the known distribution of the Robo family ligand Slit, which is an extracellular matrix protein that is made by midline glia and deposited on axons (Kidd et al., 1999; Long et al., 2004). We verified that the observed staining pattern is in fact due to Slit by showing that slit null mutant embryos exhibit no specific staining with any of the Robo family fusion proteins (Figures S7D-F).
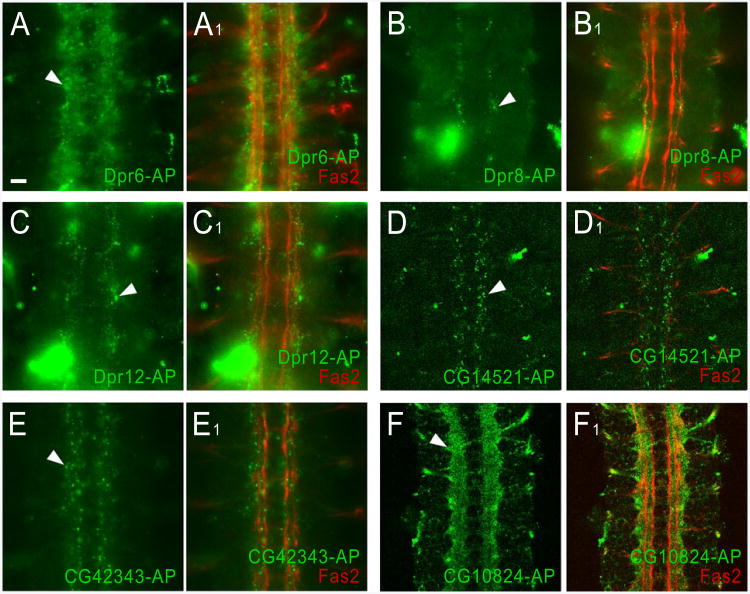
Having demonstrated that known interactions could be reliably detected by staining live-dissected embryos with AP5 fusion proteins, we used Dpr- and DIP-AP5 staining to evaluate the expression patterns of their binding partners identified with the ECIA (Figures 6A-C). Dpr6 stains a large number of neuronal cell bodies in the CNS. By contrast, Dpr8 stains a very small number of cells or cell processes located along the longitudinal tracts. Dpr12 also stains a small subset of cells, as well as the axons of the intersegmental nerve root. Dpr8 binds only to CG42343, and Dpr12 only to CG34391 (in addition to DIPc) (Figures 3A and 4), so the observed staining patterns might correspond to the expression patterns of these DIPs. Dpr6 binds to four DIPs (Figures 3A and 4), and this might account for the fact that it stains a much larger number of cells than do the other two Dprs. The DIP CG14521 stains dorsally located cell bodies along the longitudinal tracts, which could be longitudinal glia. CG42343, another DIP, stains many CNS cell bodies and the nerve roots (Figures 6D-E). Each of these binds to several Dprs (Figures 3A and 4). Finally, DIPc (CG10824) brightly stains most or all axons in the CNS (Figure 6F). Since this protein binds to most Dprs and DIPs (Figures 3A and 4), the observed staining pattern may represent the sum of many protein expression patterns.
Figure 6. Binding of Dpr-AP5 and DIP-AP5 fusion proteins to live embryos.
Staining of the longitudinal tracts and motor axons was performed as in Figure 5, except that Dpr-AP5 and DIP-AP5 fusion proteins were used for staining.
(A, A1) Dpr6-AP5 stains a fuzzy ladder-like pattern (arrowhead) in the same focal plane as the longitudinal tracts. This may represent glial or neuronal cell surfaces.
(B, B1) Dpr8-AP5 weakly stains a segmentally repeated pattern of small puncta (arrowhead) within the borders of the longitudinal tracts. These puncta, and the puncta seen with Dpr12-AP5, CG14521-AP5, and CG42343-AP5, are probably too small to represent entire cell bodies. For Dpr8-AP5, Dpr12-AP5, and CG42343-AP5, the puncta are in the focal plane of the longitudinal tracts, and may represent cross-sections through dorsoventrally projecting neuronal processes and growth cones.
(C, C1) The Dpr12-AP5 puncta are slightly lateral to the longitudinal tracts.
(D, D1) CG14521-AP5 stains puncta (arrowhead) that are dorsal to the longitudinal tracts, as shown by the fact that longitudinal tracts are not visible in (D1), but motor axons, which are more dorsal, are in the focal plane. The dorsal localization of these puncta suggests that they may represent portions of the surfaces of longitudinal glia.
(E, E1) CG42343-AP5 stains puncta (arrowhead) and a fuzzy pattern along the longitudinal tracts.
(F, F1) CG10824-AP5, which binds to all Dprs and some DIPs, brightly stains most or all longitudinal axons (arrowhead).
Scale bar, 10 μm. See Figure S6 Robo-AP5 staining of live embryos.
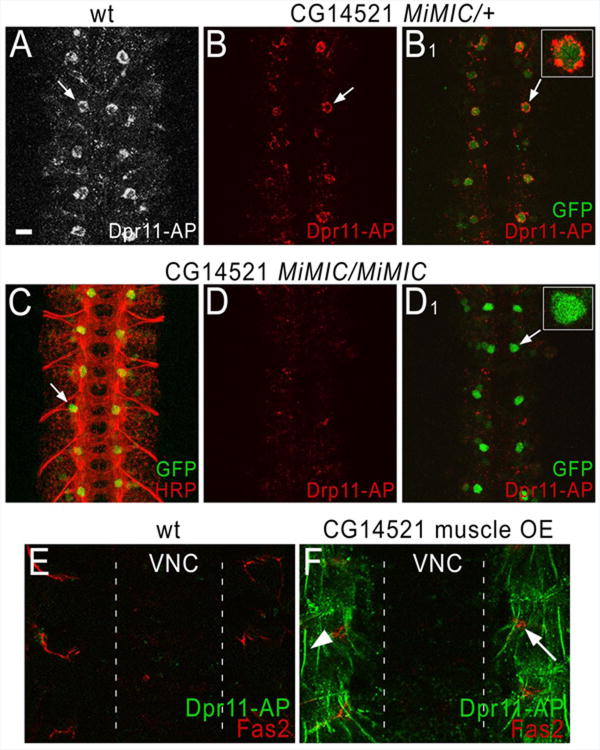
For Dpr11, we were able to demonstrate that CG14521, a DIP we identified as a partner in vitro, also binds to it in live embryos, using both LOF and GOF genetics. Dpr11 stains a pair of large dorsal cell bodies of unknown identity in each CNS segment (Figure 7A). To evaluate whether this staining pattern is due to binding to CG14521, we examined embryos that were heterozygous or homozygous for a lethal insertion mutation in the 5′ UTR of the CG14521 gene. This insertion is of a MiMIC element, which encodes eGFP with its own ATG. When MiMICs are inserted in 5′ UTRs, intact (unfused) GFP is expressed by all cells that transcribe the mRNA (Venken et al., 2011). The presence of the upstream MiMIC ATG usually prevents use of the normal ATG for the gene, so most 5′ UTR MiMICs are LOF mutations.
Figure 7. Genetic evidence for a specific interaction between Dpr11 and CG14521.
Staining was performed as in Figures 5 and 6, using Dpr11-AP5 fusion proteins, anti-Fas2 mAb 1D4, anti-GFP, and anti-HRP (a marker that labels all Drosophila neurons).
(A) In wild-type embryos, Dpr11-AP5 (white) stains a segmentally repeated pattern of large dorsal cell bodies within the CNS (one cell per hemisegment; arrow).
(B) In heterozygotes for the CG14521 MiMIC element, Dpr11-AP5 (red) stains the same cell body pattern (arrow). Staining is weaker than in (A).
(B1) The same image as in B, but showing both the Dpr11-AP5 (red) and anti-GFP (green) signals. Note that the green cells are ringed by red staining (arrow); one is shown at higher magnification in the inset.
(C) Double-staining of a CG14521 MiMIC homozygote with anti-HRP (red) and anti-GFP (green) shows a regular pattern of axons, which is indistinguishable from wild-type (see Figure S6A1 for anti-HRP staining of a wild-type embryo).
(D) A CG14521 MiMIC homozygote stained with Dpr11-AP5 (red). No staining of the cell bodies is observed.
(D1) The same image as in (D), but showing both the Dpr11-AP5 (red) and anti-GFP (green) signals. Note that the large dorsal cell bodies are present (arrow), and are brighter than in the heterozygote (B1), but there is no red staining associated with them. One cell body is shown at higher magnification in the inset. Note also that in the homozygote the brighter GFP expression reveals other cell bodies that stain more weakly with anti-GFP, implying that they express CG14521 at lower levels. Cell bodies at these positions are not detectably stained by Dpr11-AP5 in (A) or (B).
(E) A wild-type embryo double-stained with Dpr11-AP5 and anti-Fas2 mAb, showing the focal plane of the muscles, which are dorsal to the CNS focal plane in a live-dissected embryo. No CNS staining is visible in this focal plane, and the muscles do not stain with Dpr11-AP5. Motor axons are visible, however (red). The borders of the CNS are indicated by dotted lines.
(F) An embryo in which CG14521 was overexpressed in muscles. Note that these embryos display bright staining of the outlines of the ventrolateral and ventral muscle fibers (arrowhead indicates the muscle 6/7 border). Arrow, a motor axon bundle. Scale bar, 10 μm.
Staining of embryos heterozygous for this MiMIC element reveals that the large dorsal cell bodies that bind to Dpr11-AP5 express the CG14521 gene (Figure 7B). The entire cell body is filled with GFP, while Dpr11 stains the rim of the cell (inset). Embryos homozygous for the CG14521 MiMIC insertion still have these GFP-labeled dorsal cell bodies, but they are not stained by Dpr11 (Figure 7D). Dpr11 also faintly stains neuronal processes in the CNS, and this staining is unaffected by loss of CG14521. Although the CG14521 mutation is homozygous lethal, it has no visible effects on CNS axon ladder morphology as visualized with anti-HRP (Figure 7C) or anti-Fas2 (data not shown).
We used a UAS-containing insertion upstream of CG14521 to overexpress the gene in muscles, which normally do not stain with Dpr11-AP5 (Figure 7E), and do not express the CG14521 gene, since they are not green in CG14521 MiMIC embryos. Figure 7F shows that muscles ectopically expressing CG14521 stain brightly with Dpr11. Note that Dpr11 binds to both CG14521 and CG42343 in vitro (Figures 3A and 4), but there is no decrease in staining with Dpr11 in embryos homozygous for a deficiency removing CG42343, and we do not have an insertion that allows us to overexpress CG42343.
Overall Network Properties
A common feature of protein interaction networks is the presence of interaction hubs, molecules that bind to many other partners (Barabási and Oltvai, 2004). Despite the limited size of our protein network, of which 85 had any interactions, we observed one hub molecule: DIPc (CG10824). DIPc connects to 19 molecules, where the average number of edges per node in the network is 0.55. The evolution of DIPc as a hub molecule is likely due to the expansion of Dpr and DIP subfamilies through gene duplication, which is the current paradigm for the emergence of hub molecules in protein interaction networks (Pastor-Satorras et al., 2003). DIPc may have originally arisen to interact with the ancestral progenitor to Dprs and DIPs. Yet, while both subfamilies have retained their affinity for DIPc by virtue of gene duplication events, they diverged so as to acquire distinct Dpr/DIP binding specificities. It is unclear why the DIPc gene did not correspondingly expand to generate paralogs with private Dpr and DIP binding specificities. Insight into this question will likely be gained as the functional consequences of the DIP/Dpr interactions and the structural basis of DIPc engagement are revealed.
Estimates of Accuracy and Weaknesses
We have demonstrated that the ECIA can be a powerful tool for discovering extracellular protein-protein interactions accurately and in a high-throughput manner. We further examined whether ECIA could identify known interactions. Since no high quality databases exist for the Drosophila proteome, we manually curated a positive reference set from published literature for interactions within the set of proteins studied in this work (Table S2; also in Table S1 column T). Out of 44 interactions in the positive reference set, we detected 23 of them, for a true positive rate of 52%, where the combined, existing large datasets had 0%. For the subset of ECIA-derived interactions we tested biochemically, we found that none of them were false positives (Figure 4D). Overall, our estimates of accuracy point to excellent data quality, and to a reliable collection of new receptor-ligand interactions.
We identified and addressed some possible problems that could impact the false negative and false positive rates of the ECIA. One issue that might lead to false positives is the presence of “sticky” proteins that appear to bind to an excessive number of unrelated proteins. Such proteins also give rise to elevated background averages, causing false negatives. We can account for these undesirable effects during data analysis through simple normalization of the data matrix within every bait and prey protein (See Extended Experimental Protocols: Interactome Data Analysis and Other Properties of the ECIA data).
The major contributing factor to our false negative rate is that certain proteins only express at very low levels. We see very weak to undetectable expression for 27 out of 202 protein constructs (13%) (Table S1). Out of our 21 false negatives, 8 involved proteins that expressed very weakly or not at all (Table S2, column H).
The axon guidance complex of Robo–Slit is one of our false negatives due to lack of Slit expression. However, we can demonstrate this interaction by labeling Slit in embryos with our Robo-AP5 (prey) reagent (Figure S7). Slit is known to bind Robo only with an N-terminal cleavage product (Wang et al., 1999). To test whether cleavage was the culprit, we cloned the N- and C-terminal Slit fragments, and tested them against our complete prey collection. We saw that all fragments of Slit expressed, and all tested splice variants of Slit-N were able to bind the three Robo paralogs. Addition of these positives to our list of detected interactions increases our true positive rate to 59%.
The ECIA is sensitive enough that low expression levels are not uniformly detrimental to detecting interactions. An illustrative example is the Vein–Ihog/Boi interactions, despite nearly undetectable Vein expression. Overall, our results suggest that time-consuming standardization of protein concentrations for bait and prey, and the removal of weak or non-expressers are not necessary, and may not even be desirable. Since expression levels range over three orders of magnitude, normalization of protein quantities would require either heavy dilutions of strong expressers, resulting in loss of sensitivity, or removal altogether of weak expressers, resulting in false negatives. The oligomerization of the prey, as well as AP, results in signal amplification such that interactions of weak expressers can still be detected.
Discussion
Cell surface receptors and secreted ligands mediate cellular adhesions of all types, serve as immune and neural receptors, connect the extracellular matrix to the cytoskeleton, determine cell shape, polarity and motility, relay extracellular signals to the cytoplasm, and regulate processes that can lead to disease. For our basic understanding of the development and functioning of complex organisms, determining protein-protein interactions within the extracellular milieu is crucial. Our study reports the complete interactome of extracellular IgSF, FnIII and LRR protein families of D. melanogaster, classes of proteins known to mediate important homeostatic functions and neural wiring, and shows that a majority of interactions (78%) were previously unidentified. D. melanogaster offers the advantage of rapid and accessible experimental tools to now assess the functions of these interactions by the scientific community.
High-throughput approaches to revealing extracellular interactomes can succeed only if the special requirements for the production and treatment of these proteins and the general properties of these interactions are factored into the strategy (X). Established high-throughput methods such as Y2H and AP/MS, which have been applied to complete proteomes with overall success, largely failed to identify interactions of the extracellular proteome, with true positive rates approaching zero. In contrast, the ECIA successfully identified previously unknown, as well as known interactions. Interestingly, the new interactions we identified in D. melanogaster do not bear obvious relationships to IgSF and LRR partners from zebrafish (Bushell et al., 2008; Söllner and Wright, 2009). An important feature of our study was the near completeness of the set of proteins screened due to the technically manageable number of IgSF, FnIII and LRR proteins in D. melanogaster, and this in large part accounted for our high yield of new interactions. We feel that completeness of the test set is important to minimize the chances of missing interactions (i.e. “holes in the repertoire”) that likely would be a consequence of screening incomplete collections or selected members of a family.
A large portion of our Extracellular Interactome Network is made up of protein subfamilies that likely function in nervous system development. These include the Beats, Sides, Dprs and DIPs. Beat-Ia and Side have central roles in the control of motor axon guidance (Fambrough and Goodman, 1996; Siebert et al., 2009; Sink et al., 2001), and mutants for some of the other Beats have subtle motor axon guidance defects. Each of the Beats is expressed in a different subset of CNS cells (Pipes et al., 2001). These results, combined with our findings that Dprs and DIPs bind to subsets of cells in the CNS, suggest that these subfamilies of interacting proteins may have evolved to facilitate the formation of neuronal circuits. However, such roles may be difficult to uncover through standard LOF genetics, because none of the Dprs, DIPs, Beats, or Sides (a total of 54 genes) were identified in any published LOF screen for lethality or for axon guidance/synaptogenesis defects. The only LOF phenotype associated with any of the Dprs or DIPs is a defective salt aversion response in dpr1 mutants. Dpr1 is expressed in a subset of gustatory neurons (Nakamura et al., 2002). It is likely that LOF mutations in these genes will have subtle phenotypes, and analysis of such defects will require examination of the specific neuronal subsets that express these proteins. Another approach is to use GOF genetics, obtaining insights into the roles of the genes by expressing them at high levels in neurons or neuronal targets. This often produces stronger phenotypes than LOF mutations. Indeed, Dpr6, Dpr8, Dpr12, Dpr20, and the DIPs CG31708 and CG32791 were all found to produce defects in formation of motor neuron synapses when overexpressed in larval muscle fibers (Kurusu et al., 2008).
Our interactome results also highlighted that 53 of the 129 extracellular IgSF proteins have likely evolved from just two pairs of interacting proteins. We expect that large extracellular protein subfamilies with “interaction codes” between them fulfill a need for specificity in cell-to-cell adhesion and communication events during development. In other words, expression of a certain set of these receptors may differentiate specific sets of cells, axonal projections, synapses, and other cellular junctions, by virtue of their specific interactions to receptors expressed on neighboring cells or subcellular structures. Yet, such evolutionary relationships are not limited to these large subfamilies producing dense interaction codes; the four-member family of proteins related to C. elegans SYG-1 and SYG-2 (Rst, Kirre, SNS and Hbs), and the two novel binding partners of Fas2 (CG33543 and CG15630) are likewise products of more recent gene duplications (Figure S4), resulting in multiplication of their interactions, and highlighting the importance of gene duplication in the evolution of multicellular organisms.
Expansion of extracellular interactomes to complete receptomes and secretomes is the next logical step in extanding our understanding of protein function in the extracellular space. Interactomes of special cellular structures (such as the neurological or the immune synapse), and of cell types (such as neurons or leukocytes) will have direct positive effects to those studying extracellular processes. Furthermore, we expect that animal genomes contain many unrecognized and orphan protein families (like DIPs), whose evolutionary origins can only be revealed by virtue of their associations with ligands, which must be experimentally determined. Interactome studies in other species and especially humans, which have a very large IgSF with ∼800 members, should reveal such interacting protein subfamilies.
Experimental Procedures
Molecular Cloning of Drosophila cDNA and Protein Expression
A majority of ECDs (140 out of 202) were cloned from cDNAs available from the BDGP (Stapleton et al., 2002a, 2002b) and fly genetics community, while the remaining ECDs (62) were cloned from Drosophila adult and embryonic mRNAs using RT-PCR. Cloning into Drosophila expression vectors for bait and prey was achieved using TOPO TA Cloning followed by Gateway Recombination (Invitrogen). All bait and prey constructs were expressed using transient transfection of S2 cells. See Extended Experimental Procedures for details.
Interaction Assay and Data Analysis
Protein A-coated plates (96-well format, Thermo Fisher Scientific, 15130) were used to capture bait proteins from S2 transfection media. Prey binding was detected by alkaline phosphatase activity using the colorigenic BluePhos Phosphatase Substrate (KPL, 50-88-02), read at 650 nm using a VersaMax microplate reader (Molecular Dimensions). See Extended Experimental Procedures for details.
Biophysical Studies of Interacting Protein Pairs
Extracellular domains (ECDs) of interest were cloned into pAcGP67A (BD Biosciences) with C-terminal hexahistidine tags for monomeric expression in Trichoplusia ni High Five cells (Invitrogen) using baculoviruses. Proteins were purified with immobilized metal affinity chromatography, followed by size exclusion chromatography. Surface Plasmon Resonance was performed with SA (Streptavidin) chips on a Biacore T100 (GE Healthcare); ligands for the stationary phase were tagged with Biotin C-terminal to the ECDs in order to be captured by on-chip SA. See Extended Experimental Procedures for details.
In Vivo Staining of Drosophila Embryos
We have used protocols established as in (Lee et al., 2009); also see Extended Experimental Procedures for details.
Supplementary Material
Highlights.
Avidity-based method detects interactions among Drosophila extracellular proteins.
202 immunoglobulin and leucine-rich repeat proteins show 106 interactions.
Four families, comprising 53 proteins, create two dense interaction networks.
Biophysical methods and in vivo staining experiments validate interactions.
Acknowledgments
We would like to thank Natalia Goriatcheva for technical help, Kevin J Mitchell and Karsten Hokamp for sharing their superb and detailed list of Drosophila LRR proteins, Claudia Y. Janda for technical discussions on interaction assays, Liqun Luo, Xiaomeng M. Yu, Weizhe Hong and Marena Tynan La Fontaine for discussions on Drosophila genes and genetics, Nick V. Grishin for discussions on bioinformatics, and Demet Araç and Michael E. Birnbaum for critical reading of the manuscript. Work at Caltech and at LBNL were supported by NIH RO1 grants NS62821 and NS28182 to K.Z., and by NHGRI grant P41HG3487 to S.E.C. through the Department of Energy under contract no DE-AC02-05CH11231, respectively. K.C.G. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberle H. Searching for guidance cues: follow the Sidestep trail. Fly (Austin) 2009;3:270–273. doi: 10.4161/fly.9790. [DOI] [PubMed] [Google Scholar]
- Almén MS, Nordström KJV, Fredriksson R, Schiöth HB. Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009;7:50. doi: 10.1186/1741-7007-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin A, Li Y, Finkelstein R. Hedgehog activates the EGF receptor pathway during Drosophila head development. Development. 1999;126:2623–2630. doi: 10.1242/dev.126.12.2623. [DOI] [PubMed] [Google Scholar]
- Barabási AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Braun P, Tasan M, Dreze M, Barrios-Rodiles M, Lemmens I, Yu H, Sahalie JM, Murray RR, Roncari L, de Smet AS, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell KM, Söllner C, Schuster-Boeckler B, Bateman A, Wright GJ. Large-scale screening for novel low-affinity extracellular protein interactions. Genome Res. 2008;18:622–630. doi: 10.1101/gr.7187808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozatier M, Glise B, Vincent A. Connecting Hh, Dpp and EGF signalling in patterning of the Drosophila wing; the pivotal role of collier/knot in the AP organiser. Development. 2002;129:4261–4269. doi: 10.1242/dev.129.18.4261. [DOI] [PubMed] [Google Scholar]
- Da Cunha JPC, Galante PAF, de Souza JE, de Souza RF, Carvalho PM, Ohara DT, Moura RP, Oba-Shinja SM, Marie SKN, Silva WA, Jr, et al. Bioinformatics construction of the human cell surfaceome. Proc Natl Acad Sci U S A. 2009;106:16752–16757. doi: 10.1073/pnas.0907939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Bhattacharya R, Botstein D, Brown PO. Genome-scale identification of membrane-associated human mRNAs. PLoS Genet. 2006;2:e11. doi: 10.1371/journal.pgen.0020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan J, Walshe K, Alsbury S, Hokamp K, O'Keeffe S, Okafuji T, Miller SFC, Tear G, Mitchell KJ. The extracellular leucine-rich repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genomics. 2007;8:320. doi: 10.1186/1471-2164-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson T, Wang SH, Jacobsen TL, Schnepp B, Price J, Simcox A. Regulation of the Drosophila epidermal growth factor-ligand vein is mediated by multiple domains. Genetics. 2004;167:687–698. doi: 10.1534/genetics.103.019588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrough D, Goodman CS. The Drosophila beaten path gene encodes a novel secreted protein that regulates defasciculation at motor axon choice points. Cell. 1996;87:1049–1058. doi: 10.1016/s0092-8674(00)81799-7. [DOI] [PubMed] [Google Scholar]
- Flanagan JG, Cheng HJ, Feldheim DA, Hattori M, Lu Q, Vanderhaeghen P. Alkaline phosphatase fusions of ligands or receptors as in situ probes for staining of cells, tissues and embryos. Meth Enzymol. 2000;327:19–35. doi: 10.1016/s0076-6879(00)27264-9. [DOI] [PubMed] [Google Scholar]
- Fox AN, Zinn K. The heparan sulfate proteoglycan syndecan is an in vivo ligand for the Drosophila LAR receptor tyrosine phosphatase. Curr Biol. 2005;15:1701–1711. doi: 10.1016/j.cub.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenningloh G, Bieber AJ, Rehm EJ, Snow PM, Traquina ZR, Hortsch M, Patel NH, Goodman CS. Molecular genetics of neuronal recognition in Drosophila: evolution and function of immunoglobulin superfamily cell adhesion molecules. Cold Spring Harb Symp Quant Biol. 1990;55:327–340. doi: 10.1101/sqb.1990.055.01.034. [DOI] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. A Protein Complex Network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler N, Kataoka T, Bodmer JL, Romero P, Romero J, Deperthes D, Engel J, Tschopp J, Schneider P. Development of improved soluble inhibitors of FasL and CD40L based on oligomerized receptors. J Immunol Methods. 2000;237:159–173. doi: 10.1016/s0022-1759(99)00239-2. [DOI] [PubMed] [Google Scholar]
- Hong W, Zhu H, Potter CJ, Barsh G, Kurusu M, Zinn K, Luo L. Leucine-rich repeat transmembrane proteins instruct discrete dendrite targeting in an olfactory map. Nat Neurosci. 2009;12:1542–1550. doi: 10.1038/nn.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- Kurusu M, Cording A, Taniguchi M, Menon K, Suzuki E, Zinn K. A screen of cell-surface molecules identifies leucine-rich repeat proteins as key mediators of synaptic target selection. Neuron. 2008;59:972–985. doi: 10.1016/j.neuron.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HKP, Wright AP, Zinn K. Live dissection of Drosophila embryos: streamlined methods for screening mutant collections by antibody staining. J Vis Exp. 2009 doi: 10.3791/1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H, Sabatier C, Ma L, Plump A, Yuan W, Ornitz DM, Tamada A, Murakami F, Goodman CS, Tessier-Lavigne M. Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron. 2004;42:213–223. doi: 10.1016/s0896-6273(04)00179-5. [DOI] [PubMed] [Google Scholar]
- Van der Merwe PA, Barclay AN. Transient intercellular adhesion: the importance of weak protein-protein interactions. Trends Biochem Sci. 1994;19:354–358. doi: 10.1016/0968-0004(94)90109-0. [DOI] [PubMed] [Google Scholar]
- Miller JP, Lo RS, Ben-Hur A, Desmarais C, Stagljar I, Noble WS, Fields S. Large-scale identification of yeast integral membrane protein interactions. Proc Natl Acad Sci U S A. 2005;102:12123–12128. doi: 10.1073/pnas.0505482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali T, Pacifico S, Yu J, Guest S, Roberts GG, 3rd, Finley RL., Jr DroID 2011: a comprehensive, integrated resource for protein, transcription factor RNA and gene interactions for Drosophila. Nucleic Acids Res. 2011;39:D736–743. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Baldwin D, Hannaford S, Palka J, Montell C. Defective proboscis extension response (DPR), a member of the Ig superfamily required for the gustatory response to salt. J Neurosci. 2002;22:3463–3472. doi: 10.1523/JNEUROSCI.22-09-03463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özkan E, Chia PH, Wang RR, Goriatcheva N, Borek D, Otwinowski Z, Walz T, Shen K, Garcia KC. Instructive roles for SYG-1/SYG-2 complex geometry and affinity in synaptogenesis. 2013 in review. [Google Scholar]
- Pålsson-McDermott EM, O'Neill LAJ. Building an immune system from nine domains. Biochem Soc Trans. 2007;35:1437–1444. doi: 10.1042/BST0351437. [DOI] [PubMed] [Google Scholar]
- Pastor-Satorras R, Smith E, Solé RV. Evolving protein interaction networks through gene duplication. J Theor Biol. 2003;222:199–210. doi: 10.1016/s0022-5193(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Pipes GC, Lin Q, Riley SE, Goodman CS. The Beat generation: a multigene family encoding IgSF proteins related to the Beat axon guidance molecule in Drosophila. Development. 2001;128:4545–4552. doi: 10.1242/dev.128.22.4545. [DOI] [PubMed] [Google Scholar]
- Ramani SR, Tom I, Lewin-Koh N, Wranik B, Depalatis L, Zhang J, Eaton D, Gonzalez LC. A secreted protein microarray platform for extracellular protein interaction discovery. Anal Biochem. 2012;420:127–138. doi: 10.1016/j.ab.2011.09.017. [DOI] [PubMed] [Google Scholar]
- Schnepp B, Grumbling G, Donaldson T, Simcox A. Vein is a novel component in the Drosophila epidermal growth factor receptor pathway with similarity to the neuregulins. Genes Dev. 1996;10:2302–2313. doi: 10.1101/gad.10.18.2302. [DOI] [PubMed] [Google Scholar]
- Schuster CM, Davis GW, Fetter RD, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. I. Fasciclin II controls synaptic stabilization and growth. Neuron. 1996;17:641–654. doi: 10.1016/s0896-6273(00)80197-x. [DOI] [PubMed] [Google Scholar]
- Ben-Shlomo I, Yu Hsu S, Rauch R, Kowalski HW, Hsueh AJW. Signaling receptome: a genomic and evolutionary perspective of plasma membrane receptors involved in signal transduction. Sci STKE. 2003;2003:RE9. doi: 10.1126/stke.2003.187.re9. [DOI] [PubMed] [Google Scholar]
- Siebert M, Banovic D, Goellner B, Aberle H. Drosophila motor axons recognize and follow a Sidestep-labeled substrate pathway to reach their target fields. Genes Dev. 2009;23:1052–1062. doi: 10.1101/gad.520509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sink H, Rehm EJ, Richstone L, Bulls YM, Goodman CS. sidestep encodes a target-derived attractant essential for motor axon guidance in Drosophila. Cell. 2001;105:57–67. doi: 10.1016/s0092-8674(01)00296-3. [DOI] [PubMed] [Google Scholar]
- Söllner C, Wright GJ. A cell surface interaction network of neural leucine-rich repeat receptors. Genome Biol. 2009;10:R99. doi: 10.1186/gb-2009-10-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M, Carlson J, Brokstein P, Yu C, Champe M, George R, Guarin H, Kronmiller B, Pacleb J, Park S, et al. A Drosophila full-length cDNA resource. Genome Biol. 2002a;3:RESEARCH0080. doi: 10.1186/gb-2002-3-12-research0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M, Liao G, Brokstein P, Hong L, Carninci P, Shiraki T, Hayashizaki Y, Champe M, Pacleb J, Wan K, et al. The Drosophila gene collection: identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 2002b;12:1294–1300. doi: 10.1101/gr.269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valanne S, Wang JH, Rämet M. The Drosophila Toll signaling pathway. J Immunol. 2011;186:649–656. doi: 10.4049/jimmunol.1002302. [DOI] [PubMed] [Google Scholar]
- Venken KJT, Schulze KL, Haelterman NA, Pan H, He Y, Evans-Holm M, Carlson JW, Levis RW, Spradling AC, Hoskins RA, et al. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat Methods. 2011;8:737–743. doi: 10.1038/nmeth.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Chothia C. Protein family expansions and biological complexity. PLoS Comput Biol. 2006;2:e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Teichmann SA, Chothia C. The immunoglobulin superfamily in Drosophila melanogaster and Caenorhabditis elegans and the evolution of complexity. Development. 2003;130:6317–6328. doi: 10.1242/dev.00848. [DOI] [PubMed] [Google Scholar]
- Voulgaraki D, Mitnacht-Kraus R, Letarte M, Foster-Cuevas M, Brown MH, Barclay AN. Multivalent recombinant proteins for probing functions of leucocyte surface proteins such as the CD200 receptor. Immunology. 2005;115:337–346. doi: 10.1111/j.1365-2567.2005.02161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Brose K, Arnott D, Kidd T, Goodman CS, Henzel W, Tessier-Lavigne M. Biochemical purification of a mammalian slit protein as a positive regulator of sensory axon elongation and branching. Cell. 1999;96:771–784. doi: 10.1016/s0092-8674(00)80588-7. [DOI] [PubMed] [Google Scholar]
- Williams AF, Barclay AN. The immunoglobulin superfamily--domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- De Wit J, Hong W, Luo L, Ghosh A. Role of leucine-rich repeat proteins in the development and function of neural circuits. Annu Rev Cell Dev Biol. 2011;27:697–729. doi: 10.1146/annurev-cellbio-092910-154111. [DOI] [PubMed] [Google Scholar]
- Wojtowicz WM, Wu W, Andre I, Qian B, Baker D, Zipursky SL. A vast repertoire of Dscam binding specificities arises from modular interactions of variable Ig domains. Cell. 2007;130:1134–1145. doi: 10.1016/j.cell.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GJ, Martin S, Bushell KM, Söllner C. High-throughput identification of transient extracellular protein interactions. Biochem Soc Trans. 2010;38:919–922. doi: 10.1042/BST0380919. [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR, Weiner JA. Synaptic adhesion molecules. Curr Opin Cell Biol. 2003;15:621–632. doi: 10.1016/s0955-0674(03)00107-8. [DOI] [PubMed] [Google Scholar]
- Yu J, Pacifico S, Liu G, Finley RL., Jr DroID: the Drosophila Interactions Database, a comprehensive resource for annotated gene and protein interactions. BMC Genomics. 2008;9:461. doi: 10.1186/1471-2164-9-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn K. Choosing the road less traveled by: a ligand-receptor system that controls target recognition by Drosophila motor axons. Genes Dev. 2009;23:1042–1045. doi: 10.1101/gad.1803009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.