Abstract
The poly(ADP-ribose) polymerase (PARP) family of proteins use NAD+ as their substrate to modify acceptor proteins with adenosine diphosphate-ribose (ADPr) modifications. The function of most PARPs under physiological conditions is unknown. Here, to better understand this protein family, we systematically analyze the cell cycle localization of each PARP and of poly(ADP-ribose), a product of PARP activity, then identify the knock-down phenotype of each protein and perform secondary assays to elucidate function. We show that most PARPs are cytoplasmic, identify cell cycle differences in the ratio of nuclear to cytoplasmic poly(ADP-ribose), and identify four phenotypic classes of PARP function. These include the regulation of membrane structures, cell viability, cell division, and the actin cytoskeleton. Further analysis of PARP14 shows that it is a component of focal adhesion complexes required for proper cell motility and focal adhesion function. In total, we show that PARP proteins are critical regulators of eukaryotic physiology.
Introduction
Post-translational protein modifications such as phosphorylation, ubiquitination, and acetylation are critical for regulating acceptor protein function1. A less well-understood modification is ADP-ribosylation, in which units of ADP-ribose (ADPr) are added onto acceptor proteins using NAD+ as substrate2. Proteins can be modified by polymers of ADPr (poly(ADP-ribose) or PAR), that vary in length and extent of branching, or by shorter modifications such as mono(ADP-ribose) (MAR). The best known functions of ADP-ribosylation occur during regulation of cell stress responses such as DNA damage3, apoptosis4, heat shock5, cytoplasmic stress6 and the unfolded protein response7. However it has become increasingly clear that ADP-ribose modifications are critical for cell physiology under non-stress conditions, since cell division8–11, transcription, and chromatin structure regulation12,13 all require ADP-ribosylation.
Poly(ADP-ribose) polymerases (PARPs; also known as ADP-ribosyl transferases (ARTDs)) are a family of enzymes found in eukaryotes and prokaryotes that generate ADP-ribose modifications onto acceptor proteins2,12,14. Humans are thought to express 17 PARPs identified on the basis of sequence homology to the catalytic domain of PARP115,16 (for a summary of PARP/ARTD nomenclature see Table 1). The PARP family is further grouped into four subfamilies based on the presence of functionally characterized domains in regions outside the PARP domain: DNA-dependent PARPs, initially thought to require DNA binding for enzymatic activity; tankyrases, with protein-binding ankyrin repeats; CCCH zinc finger PARPs that contain CCCH zinc finger domains shown to bind viral RNA; and macro PARPs, with ADPr-binding macro domains17. The remaining PARPs are referred to as unclassified PARPs. A diagram of the PARPs, including motifs and domains, is provided in Figure 1A (a more thorough description of the PARP family is reviewed in 11, 12, 18, 19). Both the expression pattern of the various PARPs in human somatic cells and the physiological function of the majority of PARPs have not been established.
Table 1.
Summary of PARP family localization and knock-down phenotypes.
| Subfamily | PARP | Other Names | Localization | Knock-down Phenotype | ||
|---|---|---|---|---|---|---|
| Interphase | Mitosis | |||||
| Cytoplasm | Nucleus | |||||
| DNA Dependent | 1 | PARP, ARTD1 | Diffuse | Chromatin | ||
| 2 | ARTD2 | Punctate | Punctate | Diffuse, Cytoplasmic | ||
| 3 | ARTD3 | Punctate | Punctate | Punctate, Cytoplasmic | ||
| Tankyrase | 5a | TNKS1, ARTD5 | Punctate, Centrosome | Spindle Pole | Mitotic Defect, Viability Defect | |
| 5b | TNKS2, ARTD6 | Punctate | Spindle | |||
| CCCH-Zn Finger | 7 | tiPARP, ARTD14 | Punctate | Punctate | Diffuse, Cytoplasmic | Mitotic Defect |
| 12 | ARTD12 | Punctate, Golgi | Punctate, Cytoplasmic | |||
| 13 | ZAP, ARTD13 | Punctate | Punctate, Cytoplasmic | Viability Defect | ||
| Macro | 9 | BAL1, ARTD9 | Diffuse, Plasma Membrane | Diffuse | Diffuse, Cytoplasmic | Actin Cytoskeletal Def |
| 14 | BAL2, ARTD8 | Punctate, Focal Adhesions | Punctate | Punctate, Cytoplasmic | Actin Cytoskeletal Def Viability Defect | |
| 15 | BAL3, ARTD7 | Not Assayed | Not Assayed | Not Assayed | Not Assayed | |
| Unclassified | 4 | vPARP, ARTD4 | Punctate | Diffuse | Diffuse, Cytoplasmic | |
| 6 | ARTD17 | Punctate | Punctate, Cytoplasmic | |||
| 8 | ARTD16 | Punctate, Centrosome, Nuclear Envelope | Spindle Pole | Membrane Defect, Viability Defect | ||
| 10 | ARTD10 | Punctate | Punctate, Cytoplasmic | |||
| 11 | ARTD11 | Punctate | Punctate | Centriole | Not Assayed | |
| 16 | ARTD15 | Punctate, Reticular | Punctate, Cytoplasmic | Membrane Defect | ||
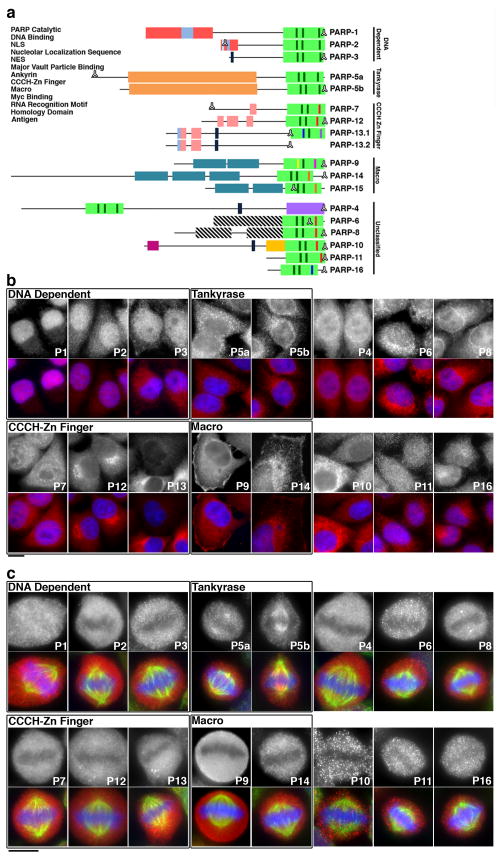
Figure 1. PARPs localize throughout the cell.
A) Domain structure of PARP proteins. Functional domains are indicated and green dashes within the catalytic domain indicate H-Y-E amino acids thought to be required for PAR synthesis activity. Dashes with different colors indicate the replacement of these amino acids with the following residues: I (red), Y (blue), V (purple), Q (yellow), T (pink), L (orange). B-C) HeLa cells were fixed then stained with affinity-purified antibodies generated against each PARP. Data is presented in PARP subfamily groupings, labeled in boxes, with each PARP labeled as P(x). A summary of localization patterns is provided in Table 1. B) Interphase localization of PARP proteins. Most PARPs are cytoplasmic (top). Merge (below) shows PARP (red) and Hoechst 33342 staining (blue). C) PARP localization in mitotic cells (top). A subset of PARPs localize to the mitotic spindle (P5a, 5b, 8,11). Merge (below) shows PARP (red), tubulin (green) and Hoechst 33342 (blue) staining. Scale bars, 10 μm. See also Supplementary Figures S1–S3 and Tables 1–2.
Based on the experimental study of a subset of PARPs combined with bioinformatic analysis, each PARP is predicted to exhibit either MAR or PAR synthesis activity, or catalytic inactivity20. Sequence analysis predicts that DNA-dependent PARPs, tankyrases, and PARP4 generate PAR; PARP9 and 13 are catalytically inactive; and all other PARPs generate MAR20. Specific amino acid residues that have been identified as targets of PARP modification include glutamic acid, aspartic acid and lysine residues21.
To better understand the PARPs and PAR, we performed a systems-level analysis of each PARP protein and the PAR polymer, examining localization and expression throughout the cell cycle. We then examined the knock-down phenotype of each PARP and performed follow up analyses to help elucidate function. This work identifies new physiological functions for the PARP family, including the regulation of cell viability, cellular membrane structures, and the actin cytoskeleton. Finally, we closely examined the function of PARP14, a member of the actin cytoskeleton-regulating MacroPARPs. We report that PARP14 is a component of focal adhesion complexes that regulates the strength and stability of cellular attachment to substrate.
Results
Most human PARPs are expressed and most are cytoplasmic
To examine PARP expression levels in human somatic cells, we analyzed transcript abundance using Illumina RNA-sequencing of HeLa cells originally derived from transformed cervical cancer cells, and retinal primary epithelial (RPE1) cells immortalized via hTERT expression. Fragments Per Kilobase of transcript per Million fragments mapped (FPKM) values for each PARP, poly(ADP-ribose) glycohydrolase (PARG), a PAR hydrolyzing enzyme, and standard normalizing controls (actin and GAPDH), are shown in Table 2. With the exception of PARP15 (omitted from subsequent analyses), all PARPs were expressed in both epithelial cell lines, with relative expression levels varying by several orders of magnitude.
Table 2.
PARP Transcript Levels
| Subfamily | Gene/PARP | FPKM | |
|---|---|---|---|
| HeLa | hTERT-RPE1 | ||
| Actin | 11.61 | 12.503 | |
| GAPDH | 11.966 | 13.776 | |
| PARG | 4.298 | 4.043 | |
| DNA Dependent | 1 | 6.871 | 5.399 |
| 2 | 4.338 | 4.639 | |
| 3 | 2.848 | 3.418 | |
| Tankyrase | 5a | 2.528 | 3.553 |
| 5b | 4.271 | 4.547 | |
| CCCH-Zn Finger | 7 | 3.422 | 4.705 |
| 12 | 3.377 | 2.253 | |
| 13 | 4.983 | 3.666 | |
| Macro | 9 | 3.038 | 2.584 |
| 14 | 0.044 | 2.114 | |
| 15 | 0 | 0 | |
| Unclassified | 4 | 4.537 | 4.87 |
| 6 | 5.153 | 6.073 | |
| 8 | 0.012 | 1.914 | |
| 10 | 2.4 | 0.352 | |
| 11 | 0.055 | 0.037 | |
| 16 | 2.849 | 1.963 | |
Localization was then analyzed using a library of N-terminal GFP fusions and affinity-purified peptide antibodies generated for each PARP. Specificity of each reagent was cross-verified by staining and blotting GFP-PARP expressing HeLa cells with corresponding anti-PARP antibodies (Supplementary Figure S1). GFP-PARP expressing cells exhibited overlapping GFP and antibody staining patterns when fixed and stained with anti-PARP antibody (Supplementary Figure S1A, S1B). To further confirm that the N-terminal GFP fusions did not disrupt PARP localization, we analyzed the localization of N-terminal fusions of the much smaller streptavidin binding peptide (38 amino acid) to select PARPs. SBP-PARP and GFP-PARP localization patterns were identical (Supplementary Figure S1C). In general, expression of GFP-PARP fusions did not result in cellular phenotypes, however instances where phenotypes were detected are noted below. PARP antibodies verified in this manner were then used to stain interphase (Figure 1B) and mitotic (Figure 1C) cells. Localization was highly similar in HeLa and hTERT-RPE1 cells (Figure 1, Supplementary Figure S2).
Cell cycle localization was then analyzed in RPE1 cells arrested in Go/G1, S-phase and mitosis (Supplementary Figure S2B–D). Cell cycle state was confirmed via centrin staining, to identify single centriole pairs present in Go/G122, EdU labeling for S-phase, and tubulin staining to identify mitotic spindles present during mitosis. Most PARPs were cytoplasmic throughout the cell cycle, exhibiting additional enrichment at specific organelles. Six PARPs from three subfamilies localized to the centrosome: PARP2 and 3 in Go/G1 (Supplementary Figure S2B; centrosomal localization for PARP3 and 5a were previously identified23,24), PARP11 and PARP5b during mitosis (Supplementary Figure S3A), and PARP5a and 8 throughout the cell cycle (Supplementary Figure S3A). Multiple PARPs and the previous identification of PARG at this site25 suggest that ADPr is actively regulated there. Five PARPs from three subfamilies localized to membranous organelles: PARP8 to nuclear envelope, PARP12 to Golgi (Supplementary Figure S3B), PARP9 and 14 to plasma membrane (Supplementary Figure S3C) and PARP16 to endoplasmic reticulum (Supplementary Figure S3D). Finally, 4 PARPs from two subfamilies localized to the mitotic spindle: PARP5a and 5b and unclassified PARP8 and 11 localized to spindle poles9,10,24 (Supplementary Figure S3A). PARP localization, largely consistent with knock-down phenotypes, is summarized below and in Tables 1 and 2.
All DNA-dependent PARPs were nuclear during interphase, with PARP2 and 3 exhibiting additional cytoplasmic localization (Figure 1B). Consistent with previous studies23,26, PARP3 localization varied with the cell cycle, with centrosomal and nuclear enrichment in Go/G1 and cytoplasmic enrichment in S-phase (Supplementary Figure S2B, S2C). During mitosis, PARP1 was enriched at chromatin, and PARP2 and 3 were cytoplasmic (Figure 1C, Supplementary Figure S2D).
Both tankyrases localized to cytoplasmic puncta during interphase (Figure 1B). During mitosis, PARP5a and 5b localized to the mitotic spindle pole9,27. We identified additional localization for PARP5b throughout the spindle (Figure 1C, Supplementary Figure S2D).
The CCCH Zn finger PARPs were primarily cytoplasmic during interphase, exhibiting punctate localizations (Figure 1B). In addition, PARP7 localized to nuclear foci, PARP12 exhibited juxtanuclear localization resembling Golgi staining, and PARP13 exhibited a ~4–5 fold variability in expression between clusters of cells, suggesting clonal variability (Figure 1B). Cells expressing high concentrations of GFP-PARP12 and 13 assembled stress granules in the cytoplasm as previously described6. PARP12 localization to the Golgi was confirmed by co-localization with trans Golgi protein p230 (Supplementary Figure S3B). Overexpression of GFP-PARP12 blocked PARP12 localization to the Golgi and disrupted Golgi structure, analyzed via staining of p230, a trans Golgi marker (Supplementary Figure S4A, S4B). While p230 staining intensity was unaltered in cells overexpressing GFP-PARP12, Golgi area increased ~1.58 fold relative to control cells. No such defects were found in untransfected, GFP-overexpressing and GFP-PARP13.2 overexpressing cells suggesting that the Golgi defects were specific to PARP12 overexpression (Supplementary Figure S4B). During mitosis, all CCCH Zn finger PARPs localized to the cytoplasm (Figure 1C, Supplementary Figure S2D).
Macro PARPs 9 and 14 were primarily cytoplasmic with additional nuclear localization consistent with previous studies (Figure 1B) 28–30. Nuclear PARP9 staining was highly enriched in S-phase arrested cells (Supplementary Figure S1A, S2C) suggesting that the previously described nucleo-cytoplasmic shuttling27 of PARP9 might be cell cycle-regulated. In addition, PARP9 and 14 exhibited enriched localization at the cell periphery (Figure 1B). Both proteins were later confirmed to co-localize with actin filaments, motile elements of the actin cytoskeleton that are enriched at the cell periphery (Supplementary Figure S3C). Such enrichment was particularly prominent in cells fixed with 10% Trichloroacetic acid (TCA), conditions ideal for staining cortical proteins31 (Supplementary Figure S3C). During mitosis, all macro PARPs were cytoplasmic; PARP9 staining was diffuse while PARP14 staining was punctate (Figure 1C, Supplementary Figure S2D).
Previous reports identified cytoplasmic localization for PARP432, and cytoplasmic and nuclear localizations for PARP10, with a nuclear enrichment upon inhibition of nuclear export33. Each unclassified PARP localized to cytoplasm during interphase, with PARP4 and 11 exhibiting additional nuclear localization (Figure 1B). PARP4 staining at the nucleus is likely non-specific as knock-down of PARP4 (see below) resulted in reduction of cytoplasmic, but not nuclear signal (Figure 3). PARP8 was enriched at the nuclear envelope in fixed cells (Figure 1B) and cells expressing high concentrations of GFP-PARP8 exhibited nuclear accumulation of the protein (Supplementary Figure S1A, B). PARP10 was punctate in the cytoplasm and largely absent from the nucleus. Similar to PARP13, PARP10 expression varied ~6–7 fold among small clusters of cells (Figure 1B). Cells with increased expression of PARP10 exhibited strong cytoplasmic juxtanuclear staining. Finally, PARP16 exhibited reticular membrane localization that co-localized with DiI, consistent with our previous results identifying it as an endoplasmic reticulum protein7 (Supplementary Figure S3D). Cells expressing high concentrations of GFP-PARP16 contained DiI positive membrane structures as previously shown (Supplementary Figure S4C)7.
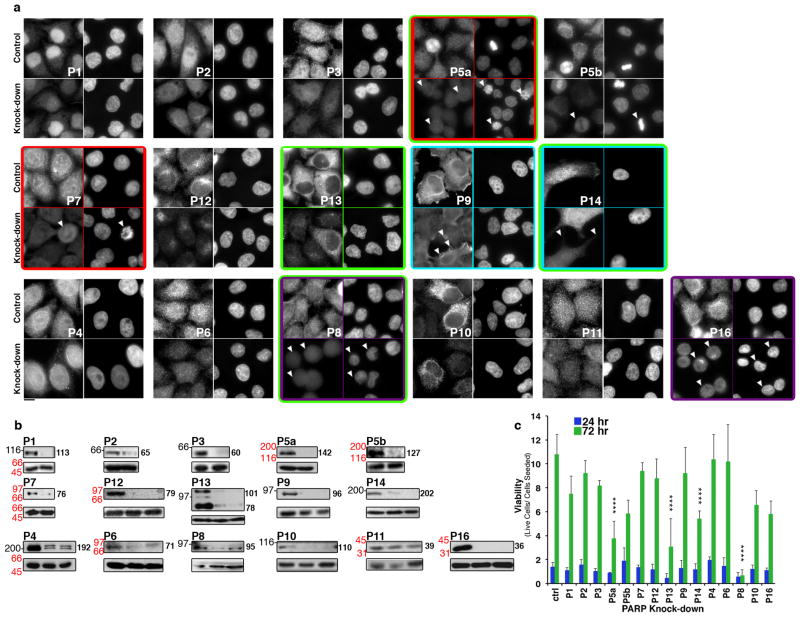
Figure 3. Knock-down phenotypes of the PARP family.
PARP knock-down results in cell viability, membrane, actin cytoskeleton, and mitosis phenotypes. PARP expression was knocked-down via siRNA transfection in HeLa cells. Cells were then stained for each PARP (A) and immunoblot analysis performed to confirm knock-down (B). A) Cells were transfected with control siRNAs (Control) or siRNAs specific for each PARP (Knock-down) then stained for PARP (left) and Hoechst 33342 (right). Knockdown identified 4 phenotypes: defects in membranes (purple boxes), actin cytoskeleton (cyan boxes), mitosis (red boxes), and cell viability (green boxes, Supplementary Figure S5A). Arrowheads indicate cells exhibiting knock-down. Scale bar, 10 μm. B) Lysates from HeLa cells transfected with control (left lane) or PARP-specific siRNA (right lane(s)) were immunoblotted with corresponding anti-PARP antibody. Positions of molecular weight markers are indicated on the right in black, molecular weights shown in italics identify the closest markers above and below the cropped region and the approximate molecular weight (kDa) of the relevant PARP in each knock-down is indicated to the left of each PARP blot. The corresponding tubulin blot is included as a loading control (lower panels). C) Cell number was analyzed 24h and 72h after knock-down of each PARP and presented as fold change relative to the initial number of cells seeded. PARP knock-downs that resulted in a decrease in viability of ≥ 2 standard deviations relative to control knock-downs where identified as defective in cell viability. Error bars represent standard deviation, n=3, ****p<0.00001, Student’s T test. See also Supplementary Figure S6 and Table 1.
During mitosis, all unclassified PARPs localized throughout the cytoplasm. With the exception of PARP4, cytoplasmic staining was punctate (Figure 1C, Supplementary Figure S2D). PARP8 and 11 exhibited strong staining at the spindle poles as shown by co-staining with centrin (Figure 1C, Supplementary Figure S3A). No centriolar staining was identified for PARP11 during interphase, suggesting the localization is specific to mitosis (Figure 1C, Supplementary Figure S3A).
Poly(ADP-ribose) is found throughout the cell
The predominant cytoplasmic localization of the PARPs and their cell cycle dependent localization patterns suggested that previously undetected cytoplasmic ADPr, generated in a cell cycle dependent manner, could exist. Since reagents to label MAR are not well characterized17,34, we focused on PAR, for which several well-characterized antibodies are available.
We examined PAR localization in asynchronous, Go/G1, S-phase, and mitosis arrested HeLa cells, and HeLa cells treated with H2O2 to induce DNA damage, resulting in upregulated PAR synthesis in the nucleus35 (Figure 2A, Supplementary Figures S5A & S5C). Cells were fixed with methanol (MeOH) or 20% TCA to minimize post-fixation PAR synthesis36 then stained with two antibodies generated against distinct PAR antigens: Tulip chicken anti-PAR IgY (Figure 2A) and BD rabbit anti-PAR (Supplementary Figure S5C). Cell cycle arrest was verified as above. Each fixation condition and antibody yielded similar results.
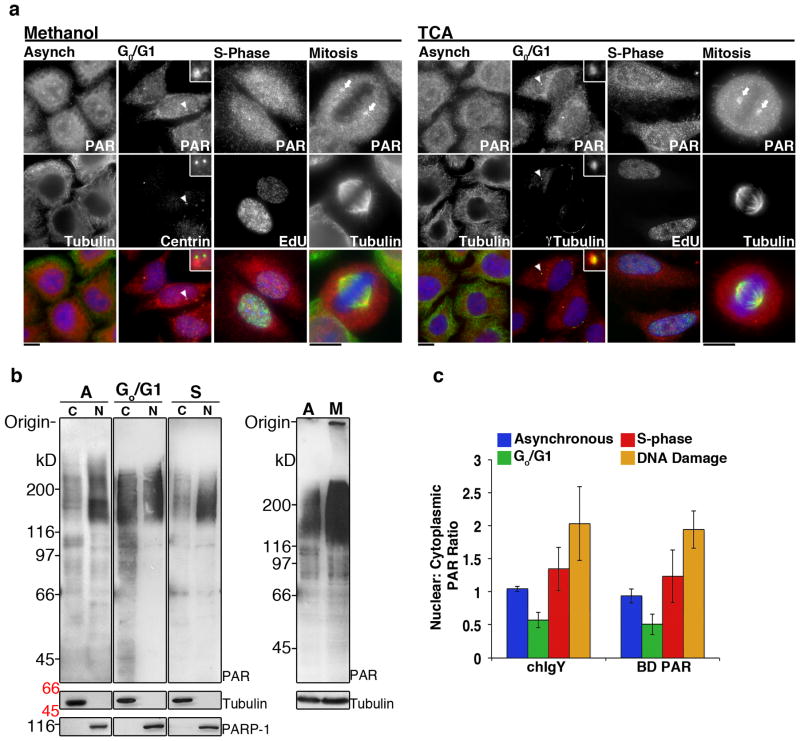
Figure 2. Poly(ADP-ribose) is localized to the cytoplasm and nucleus throughout the cell cycle.
A) Asynchronous populations of HeLa cells (Asynch) or cells arrested in G0/G1, S-phase or Mitosis were fixed with Methanol or TCA, then stained for PAR (Tulip chIgY), and Centrin or γ-Tubulin, to identify single centriole pairs found during Go/G1, EdU, incorporated during S-phase, and Tubulin, to stain mitotic spindles. In asynchronous cells and during Go/G1, S-phase, and Mitosis, PAR staining was punctate with strong staining at the centrosome (arrowhead) and poles of the mitotic spindle (arrows). S-phase cells exhibited increased punctate staining in the nucleus relative to Go/G1 cells. Merge shows PAR (red), cell cycle markers (green) and Hoechst 33342 (blue). Scale bars, 10 μm. B) Cytoplasmic (C) and nuclear (N) extracts prepared from identical cell pellets, then normalized to cell volume, were generated from asynchronous, Go/G1 and S-phase arrested cells. Extracts were immunoblotted with Tulip chIgY anti-PAR antibody. Cytoplasmic and nuclear extracts were further examined for the presence of tubulin, a cytoplasmic protein, or PARP1, a nuclear protein, to assay for contamination between the fractions. Total cell extracts were also prepared from asynchronous (A) and mitotic (M) cells and immunoblotted with Tulip chIgY anti-PAR antibody. Positions of molecular weight markers are indicated on the right in black, molecular weights shown in italics identify the closest markers above and below the cropped region. C) Quantitation of signal intensity of PAR immunoblots using chIgY and BD PAR antibodies. The ratio of nuclear to cytoplasmic PAR increases during S-phase and DNA damage. The integrated intensity over the entire lane was determined and the ratio of nuclear:cytoplasmic signal calculated, error bars represent standard deviation, n=3. See also Supplementary Figure S5.
Significant amounts of PAR were detected in both cytoplasm and nucleus of asynchronous cells, H2O2 treated cells, and during all cell cycle stages (Figure 2A, Supplementary Figure S5A & S5C). Staining in the interphase cytoplasm and nucleus appeared both diffuse and punctate, with increased nuclear PAR staining during S-phase. Such staining was punctate and distinct from nuclear staining during other cell cycle stages suggesting S-phase specific functions for these puncta. In addition, PAR was also significantly enriched at the interphase centrosome and mitotic spindle poles, consistent with PARP localization described above, and previous results8–10,37 (Figure 2A, Supplementary Figure S5C).
To biochemically validate cell cycle dependent differences in PAR concentrations between the cellular compartments, the nuclear to cytoplasmic ratio of PAR was determined by generating nuclear and cytoplasmic extracts from H2O2 treated, asynchronous, and Go/G1 and S phase-arrested cells. Extracts were immunoblotted with chIgY and BD anti-PAR (Figure 2B, Supplementary Figure S5B & S5D), and the nuclear to cytoplasmic PAR ratios determined by integrating signal intensities of each lane of the blots (Figure 2C, n=3). During Go/G1, the ratio was 0.57 +/− 0.11 and 0.51 +/− 0.15 for the chIgY and BD PAR antibodies respectively, and during S-phase this ratio increases to 1.3 +/− 0.33 and 1.2 +/− 0.39 respectively for each antibody. During DNA damage the ratio of nuclear to cytoplasmic PAR is 2.03 +/−0.55 and 1.94+/−0.29 for chIgY and BD PAR antibodies respectively. Additionally, immunoblotting for PAR in asynchronous and mitotically arrested cells demonstrated increased PAR concentrations during mitosis, consistent with previous reports10 (Figure 2B, Supplementary Figure S5D). Thus significant amounts of PAR exist in the cytoplasm, and the relative amounts of PAR in the cytoplasm and nucleus change with the cell cycle.
PARP knock-down phenotypes
We next examined the phenotypes of PARP knock-down via RNAi, focusing on changes in cell viability and morphology. For PARPs with unpublished phenotypes, at least eight siRNAs were generated. To verify phenotypes, at least two siRNAs demonstrating > 80% protein knock-down via immunoblot and cell staining were required to result in similar phenotypes (Figure 3A, 3B, Supplementary Table S1). For PARPs 1, 2, 3, 4, 5a, 5b and 7, published siRNA sequences were utilized9,38. Published shRNA sequences for PARP10 knock-down were ineffective in our siRNA assays, therefore new siRNAs were screened39. In all cases non-targeting siRNAs were used as control.
Cell viability was examined via seed assay in which PARPs were knocked-down, total cell number quantitated, and cells replated at the same density. Cell number was determined 24 or 72h post seeding and the ratio of cell number at each time point to initial seed number used to assess defects in cell viability. PARP knock-downs that resulted in a fold change of at least two standard deviations were considered to cause viability defects (Figure 3C).
Cell morphology was assayed by comparing PARP and DNA staining in control and knock-down cells, then verified by follow-up assays. In total, both assays identify four phenotypes upon PARP knockdown: decreased cell viability, defects in the actin cytoskeleton, defects in internal membrane structure, and defects in mitosis.
Knock-down of individual DNA-dependent PARPs did not result in observable changes in cell viability or morphological defects suggesting that each is non-essential (Figure 3A, 3C).
PARP5a knock-down resulted in mitotic arrest, increased mitotic index confirmed by phosphoH3 staining (~13% mitotic index in knock-down cells vs. ~3% in control) and decreased cell viability (Figure 3A & 3C), consistent with our previous results and work by others9–11,40. PARP5b knock-down did not result in observable phenotypes or decreased viability (Figure 3A, 3C).
Consistent with previous reports, PARP7 knock-down resulted in increased mitotic index confirmed by phosphoH3 staining (~6% in knock-down cells vs. ~3% in control)37 (Figure 3A, Supplementary Figure S6). A significant decrease in cell viability was not observed in PARP7 knock-downs, suggesting that knock-down cells are able to undergo mitosis (Figure 3C). Neither PARP12 nor 13 knock-down resulted in morphological phenotypes, however PARP13 knock-down resulted in significantly decreased cell viability (Figure 3A,C).
Knock-down of each macro PARP resulted in cell morphology defects (Figure 3A). Roughly 25% of PARP9 knock-down cells exhibited pronounced membrane blebbing phenotypes. This phenotype was not likely due to apoptosis, as a substantial increase in apoptotic nuclei, identified by the presence of highly condensed DNA via Hoescht staining, was not observed (~3.5% in PARP9 knock-downs vs. 1% in control) (Figure 3A). PARP14 knock-down resulted in decreased cell viability (Figure 3C) and dramatic alterations in cell morphology, as ~60% of cells had elongated processes extending far from the cell body (Figure 3A, see below).
Knock-down of PARP4, 6 and 10 did not result in obvious morphological defects or decreased cell viability (Figure 3A, 3C). In contrast, PARP8 and 16 knock-down resulted in cell morphology defects, with ~50% of PARP8 knockdowns and ~30% of PARP16 knock-down cells appearing completely round (Figure 3A). PARP16 knock-down cells exhibiting the round cell phenotype were always found in pairs while round PARP8 knock-downs were individual cells. PARP8 knock-down resulted in the most pronounced decrease in cell viability of all PARPs (Figure 3C). PARP11 siRNAs were ineffective for knock-down and therefore we cannot report on phenotype.
PARP knock-down phenotypes identify new cellular functions
Cell morphology phenotypes identified in initial RNAi screens led us to design secondary assays to better understand function. Macro PARP knockdown phenotypes were reminiscent of actin cytoskeletal misregulation41,42. We therefore stained control, PARP9 and 14 knock-down cells with phalloidin, a filamentous actin (F-actin) label (PARP9 knockdowns), or an actin antibody (PARP14 knock-downs) (Figure 4). In control cells, PARP9 strongly co-localized with F-actin at the cell cortex, while PARP14 localized to the ends of actin fibers specifically at cell protrusions in a staining pattern consistent with focal adhesion structures (Figure 4, arrowheads). Both PARP9 and 14 knock-downs exhibited highly abnormal F-actin staining suggesting functions in actin cytoskeletal regulation; PARP9 knockdowns contained multiple actin-rich membrane blebs and PARP14 knock-down cells exhibited highly elongated actin-rich processes (Figure 4, arrows; Supplementary Figure S3C). These results are consistent with previous reports implicating this subfamily in the regulation of cell migration in B-cell lymphomas43.
Figure 4. Analysis of knock-down phenotypes identifies new PARP functions.
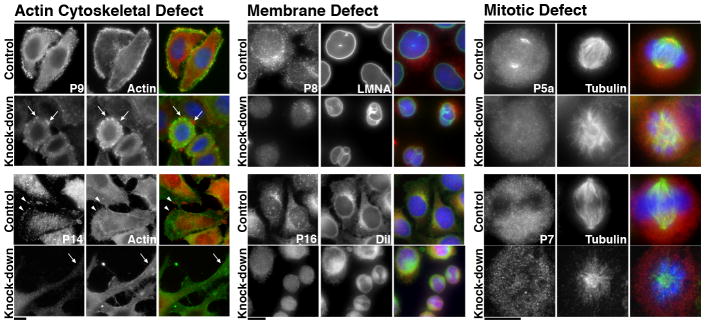
Morphological phenotypes identified upon PARP knock-down were grouped into actin cytoskeletal, membrane or mitotic defects then analyzed to determine biological function. Control and Knock-down cells stained with Hoechst 33342 (blue) and antibodies against the knocked-down PARP (red). Cells exhibiting Actin Cytoskeletal Defects were co-stained with phalloidin (P9) or an actin antibody (P14) to stain filamentous Actin (green), Membrane Defects for Lamin A/C (LMNA) or DiI (green) and mitotic defects for Tubulin (green). PARP9 (P9) co-localized with actin in control cells. Knock-down resulted in actin rich blebs shown by Arrows. PARP14 (P14) localized to cell protrusions in control cells (Arrowheads), identified in Figure 5 as focal adhesions. P14 knockdowns exhibited severe morphological defects with assembly of extended cellular protrusions (Arrowheads). PARP8 (P8) co-localized with LMNA in control cells, but not knockdown cells. PARP8 knockdown resulted in abnormal, bilobed nuclei. PARP16 (P16) co-localized with the membrane dye DiI in control cells. Knock-down resulted in pairs of round cells. PARP5a (P5a), but not PARP7 (P7) localized to the mitotic spindle. Knockdown of P5a resulted in multipolar spindles, while P7 knockdown resulted in an increase in pre-metaphase spindles. Actin cytoskeletal defects are further examined in Figure 5–6, and mitotic defects in Supplementary Figure S6. Scale bars, 10 μm.
The round cell phenotypes suggested that PARP8 and 16 knockdowns exhibited defects in membrane structure. Due to its nuclear envelope localization, we stained control and PARP8 knock-down cells with an antibody against the nuclear envelope protein Lamin A/C. PARP8 staining co-localized with Lamin A/C in control cells, and was largely undetectable at the nuclear envelope after PARP8 knockdown. The average size of nuclei in PARP8 knock-down cells was slightly smaller than control cells (~180 +/− 50 μm2 in knock-downs, n=360 cells vs. 205 +/− 54 μm2 in controls, n=282 cells). Lamin A/C staining identified multilobed nuclei in the knockdowns (~14% in knock-downs vs. 0.7% in controls, n>400 cells) suggesting structural defects in the nuclear envelope, or a misregulation of nuclear envelope biogenesis (Figure 4). Together PARP8 localization to the nuclear envelope and the defects in nuclear envelope structure upon knockdown suggest that it is a critical nuclear envelope protein. PARP16 knockdown resulted in the accumulation of pairs of round cells, reminiscent of defective cytokinesis. PARP16 localization to membrane prompted us to examine membrane structure in the PARP16 knock-downs by staining with DiI (Figure 4). Total intensity of DiI staining was unchanged, however PARP16 knock-downs exhibited a disruption of organized reticular membrane staining (Figure 4), perhaps a result of the dramatic change in cell morphology. It is unclear if this phenotype relates to PARP16 function in the unfolded protein response7, however activation of the unfolded protein response is known to result in cytokinesis defects in S. cervisiae44.
PARP5a and 7 knock-downs were stained for tubulin to examine spindle structure. Consistent with previous results, control cells exhibited PARP5a staining at spindle poles, and knock-down cells lacking PARP5a staining at spindle poles contained multiple microtubule foci and improperly aligned chromosomes (~58% of mitotic cells with improperly aligned chromosomes for knock-downs vs. 3% in controls, n>30 mitotic cells) 9,10,40 (Figure 4, Supplementary Figure S6). In contrast, PARP7 did not localize to the mitotic spindle in control cells. Tubulin staining of PARP7 knock-downs revealed a disproportionate increase in pre-metaphase cells among mitotic cells (~57% in PARP7 knock-down vs. ~19% in controls, n>50 mitotic cells), a mitotic defect distinct from that of PARP5a knock-down (Figure 4, Supplementary Figure S6). Since the increase in mitotic index was mild (~6% vs. ~3% for control cells) and PARP7 knock-down did not significantly decrease cell viability, our results suggest that PARP7 is not required for spindle function, but that its knock-down increases the length of pre-metaphase mitosis.
PARP14 is a focal adhesion protein
The PARP14 knock-down phenotype was particularly striking prompting us to closely examine its function in actin cytoskeletal regulation. Both PARP14 siRNAs resulted in cells that were elongated and contained dendritic-like membrane protrusions emanating from the cell body (Figure 5A, B). These extended cellular protrusions were substantial- PARP14 knock-downs measured ~3 times longer than control cells (n=20 cells) (Figure 5B). Greater than 60% of PARP14 knock-down cells exhibited these extended protrusions in contrast to control knock-down cells that contained similar protrusions in ~5% of the population (n=3, 200 cells counted for each condition) (Figure 5B).
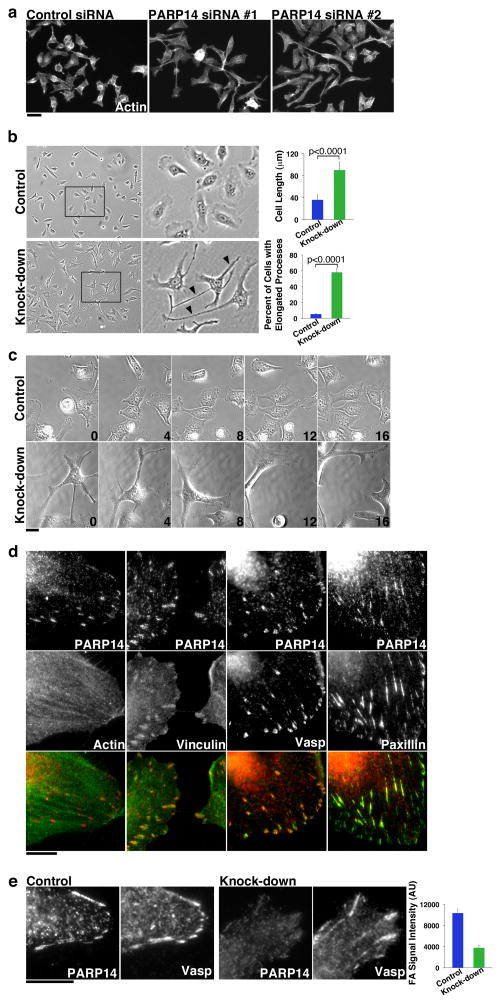
Figure 5. PARP14 is a focal adhesion protein whose knock-down results in abnormal cell morphology and cell migration defects.
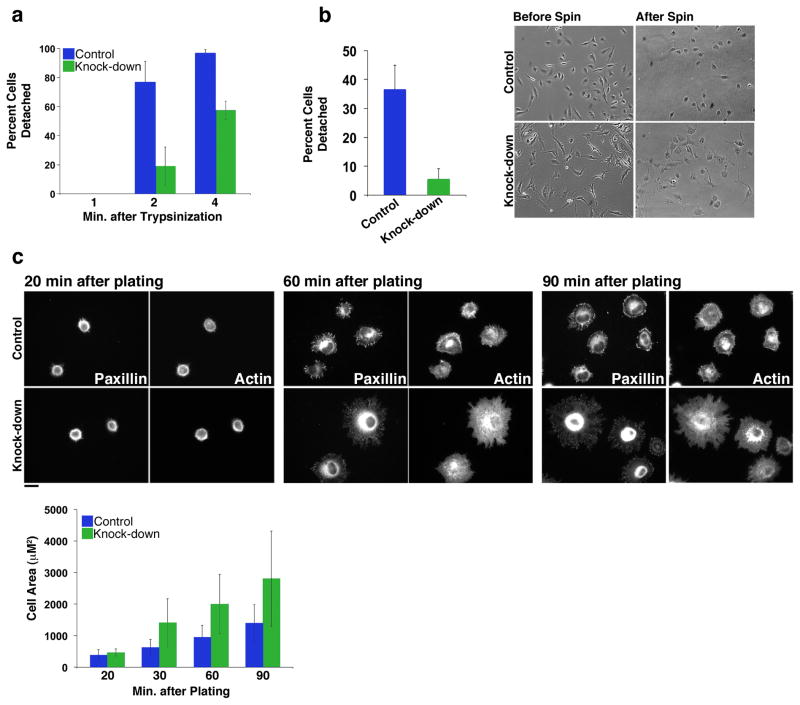
A) HeLa cells were transfected with control siRNA or siRNA 1 or 2 directed against distinct PARP14 sequences, then stained with Phalloidin. Both PARP14 siRNAs result in similar cell phenotypes containing extended cellular protrusions. Scale Bar, 50 μm. B) Bright field images of representative fields of control and PARP14 siRNA treated cells. Extended cellular protrusions in PARP14 siRNA treated cells are marked with Arrowheads. Quantitation of maximum cell length (n=20 cells) and percent cells displaying extended cellular protrusions (n=3, 200 cells counted per condition) shown at left. Error bars represent standard deviation. Significance determined by student’s t-test. C) Still images of the indicated time points of movies taken of control and PARP14 siRNA treated cells undergoing random migration on the substrate fibronectin. See also Supplementary Movie 1 (Control cells) and Supplementary Movie 2 (PARP14 depleted cells). Scale bar, 25 μm D) HeLa cells plated on fibronectin fixed with TCA and stained for PARP14 (red), and the indicated focal adhesion proteins (green). Scale bar, 10 μm. E) Control and PARP14 knock-down cells fixed with TCA and co-stained for PARP14 and Vasp. PARP14 knock-down results in loss of signal at focal adhesions demonstrating the specificity of focal adhesion staining (arrows). The intensity of PARP14 signal at focal adhesions was quantified in both control and PARP14 knock-down cells. Scale bar, 10 μm.
To better understand how these extended protrusions were generated, we analyzed control and PARP14 knock-downs using real-time imaging. In control cells, small membrane protrusions were generated and quickly retracted as cells moved in different directions (Figure 5C, Supplementary Movie 1). In PARP14 knock-downs, cells appeared unable to retract protrusions efficiently, resulting in highly elongated extensions (Figure 5C, Supplementary Movie 2). Thus, abnormal membrane protrusions appeared to result from defective retraction of protrusions normally generated in migrating cells. Retraction of protrusions was so defective in PARP14 knock-downs that cells stretched into severely elongated shapes as they explored the substrate (Figure 5C, Supplementary Movie 2).
A major control point in cell migration is the assembly, maturation and disassembly of focal adhesions (FA)s, multi-protein complexes that serve as connections between the substrate and the actin cytoskeleton45. FA regulation requires the ordered recruitment of FA proteins and the strength and stability of FAs depends on the protein composition of the FA46. Cells must balance FA strength so that adhesions are strong enough to pull the cell forward, but sufficiently labile so that they can be rapidly disassembled as the cell moves. The PARP14 knock-down phenotype suggested defects in FA function.
A previous proteomic study identified PARP14 protein in purified FAs isolated from HFF-1 cells46, consistent with the PARP14 staining at cell protrusions we observed (Figure 4, Supplementary Figure S3C). We therefore examined PARP14 localization at FAs in HeLa cells grown on fibronectin coated coverslips (to mimic extra-cellular substrate). Cells were fixed and stained with antibodies against PARP14 and known FA components. PARP14 localized to the ends of actin stress fibers, similar to other FA proteins, and co-localized with Vinculin, VASP and Paxillin (Figure 5D). The specificity of PARP14 localization at FAs was confirmed by examining PARP14 antibody staining at FAs after targeted knock-down (Figure 5E). PARP14 knock-down resulted in near complete depletion of PARP14 staining at FAs, while VASP staining was maintained. In addition to validating the specificity of PARP14 localization to the FA, this result also indicates that PARP14 is not required for general FA assembly (Figure 5E). Importantly, such siRNA treatments did not deplete cytoplasmic or nuclear PARP14 staining, suggesting that staining at these locations is either non-specific or represents siRNA-resistant populations of PARP14.
PARP14 regulates focal adhesion function
For cells to properly retract membrane protrusions, they must be able to efficiently breakdown FAs in a process called FA turnover47. The inability of PARP14 knock-down cells to properly retract membrane protrusions, combined with its localization to FAs, suggested that PARP14 functions in FA turnover. Therefore we examined FA adhesive force in control and PARP14 knock-downs using three independent assays. First we utilized a trypsin-based detachment assay measuring time required to detach cells from substrate48. PARP14 knockdown cells were more resistant to trypsinization than controls, with ~ 2 times more cells remaining adhered after similar periods of trypsinization (Figure 6A). Second we employed a centrifugation assay comparing the number of cells remaining adhered to substrate after constant application of centrifugal force. Approximately 5 times more PARP14 knock-down cells remained adhered to substrate after centrifugation compared to controls (Figure 6B). Third, we used a cell-spreading assay measuring the area of cell spreading per unit time (Figure 6C). At early time points when cells adhered to substrate but had not yet formed defined FAs, control and PARP14 knock-downs were of similar size suggesting that the morphological defects observed in PARP14 knock-downs were dependent on FA formation and not simply a property of the cells themselves. As cells assembled FAs and spread onto substrate, PARP14 knock-down cells covered ~twice the surface area of control cells at each time point (Figure 6C). These three independent assays suggest that the adhesive strength between FAs and substrate is stronger in PARP14 knock-downs relative to control. It is likely that this increased adhesive strength leads to slowed FA disassembly and prevents cellular protrusions from properly retracting.
Figure 6. PARP14 depletion from focal adhesions results in increased adhesive strength.
A) Control or PARP14 siRNA treated cells plated on fibronectin were treated with trypsin for the indicated times and percent cells detached quantified at each time point. Error bars represent standard deviation. B) Control or PARP14 siRNA treated cells plated on fibronectin were subjected to a constant centrifugal force (2,000 g) for 30 min and the number of cells detached during centrifugation quantified. Representative images of control and PARP14 knock-down cells before and after centrifugation shown at right. Error bars represent standard deviation. C) Control or PARP14 siRNA treated cells were allowed to adhere to a fibronectin coated plate for the indicated times, fixed and stained with anti-paxillin and phalloidin, and the area of individual cells quantified at each timepoint. Error bars represent standard deviation. Representative images of control or PARP14 knock-down cells at each time point are shown. Scale bar, 25 μm.
Discussion
The initial bioinformatic identification of 17 human PARP genes suggested that PARPs function in more biological pathways than previously imagined14,49. Our analysis confirms the diversity of PARP function and leads to several important conclusions regarding the protein family. First, nearly all (16 of 17) of the PARPs predicted to exist in humans are expressed in epithelial cells collected from the retina and cervical tumors- tissues of diverse origin. This suggests ubiquitous expression of most PARPs among human tissues. Second, PARPs are predominantly cytoplasmic proteins, and a significant amount of one product of PARP activity, PAR, is present in the cytoplasm. Third, PAR synthesis/hydrolysis appears to be cell cycle regulated as the ratio of PAR synthesis/hydrolysis in the nucleus and cytoplasm changes during specific cell cycle stages. This suggests cell cycle specific functions for PAR. Finally, PARP proteins are important regulators of somatic cell physiology and function in multiple diverse pathways.
The importance of nuclear PARP function in the physiology of the cell is well established, PARP1/PARP2 regulate critical physiological functions in the nucleus, and while neither PARP1 nor PARP2 are essential for life50, PARP1/2 double knockout mice are non-viable suggesting that the activity of a least one of these PARPs is essential51. Our data suggests that PARP function in the cytoplasm is equally important. All multicellular eukaryotes encode both nuclear and cytoplasmic PARPs with vertebrates containing the most PARPs52. The additional PARPs found in vertebrates are cytoplasmic, and each PARP identified as essential for cell viability is found in the cytoplasm of all vertebrates suggesting that vertebrate evolution led to an increased requirement for cytoplasmic PARP function52.
Each of the PARP knock-down phenotypes points to a remarkable diversity of function among this family of proteins. Such diversity is likely conferred by the diverse functional domains present outside of the conserved PARP domain, such as the DNA binding domains of DNA dependent PARPs, and the ADP-ribose binding Macro domains of the Macro PARPs17. The similarity of localization and function of subfamily members demonstrates the importance of these functional domains and suggest that in addition to previously described roles in regulation of PARP enzymatic activity53,54 they can target PARPs to specific cellular locations (Figure 1A and Table 1).
The defects in cell viability in PARP5a, 8, and 14 knockdowns are not surprising given their phenotypes. In contrast, PARP13 knock-downs did not result in obvious morphological defects, thus its requirement for cell viability is unclear. PARP13 has been shown to be an important regulator of mRNA, targeting viral RNA for degradation, and regulating cellular mRNA post-transcriptionally by regulating miRNA activity6. Since cell cycle progression is regulated by miRNA activity55 it is possible that the decreased cell viability resulting from PARP13 knock-down results from misregulation of miRNA function or that the cell viability defect is due to general misregulation of mRNA metabolism.
Our PARP8, 12 and 16 data suggest potential functions for PARPs in the assembly or maintenance of membranous organelles. Each of the structures to which PARP8, 12 and 16 localize are derived from endoplasmic reticulum, and require trafficking from the endoplasmic reticulum for function, processes regulated by glycosylation56,57. Perhaps ADP-ribose could serve a similar function as glycosylation in vesicle trafficking either by functioning as a trafficking signal, or by regulating glycosylation of target proteins.
Finally, Macro PARPs had been postulated to regulate B cell motility via unknown mechanisms, with overexpression of PARP9 resulting in increased migration of B-cell lymphomas43. Our results provide a potential explanation for these observations by demonstrating that macro PARPs localize to the motile elements of the actin cytoskeleton and exhibit cytoskeletal defects upon knockdown. Interestingly, PARP14 expression is upregulated in multiple myelomas and has been shown to regulate the expression of IL-4 and B-cell survival factors58,59. As such, PARP14 has been mentioned as a possible target for therapeutic inhibition of multiple myelomas. It is unclear if there is a connection between this specific PARP14 function in these specialized cell types and PARP14 function at FAs. However, it is possible that inhibiting the enzymatic activity of PARP14 could also affect cellular adhesiveness, making PARP14 an attractive target for the therapeutic inhibition of metastatic cancers in addition to multiple myelomas.
Materials and Methods
Cell Culture and Reagents
Cells were grown at 37°C and 5% CO2. HeLa cells (ATCC) were passaged in DMEM + 10% fetal bovine serum (FBS) (Tissue Culture Biologicals) + penicillin/streptomycin (Invitrogen); hTERT-RPE1 cells (ATCC) in Ham’s F12/DMEM (1:1) (Mediatech) + 10% FBS, penicillin/streptomycin, and 0.01mg/mL hygromycin B (Invitrogen). Anti-PARP peptide antibodies were generated and affinity purified using a standard protocol60. Antibodies (stored at 1mg/ml) were used at 1:100 for IF and 1:1000 for IB. Additional antibodies used were anti-pADPr (chIgY, Tulip Biolabs, 1:100 IF, 1:500 IB; 51-811KD, BD Pharmingen, 1:100 IF, 1:500 IB), anti-PARP1 (PA3-951, Affinity Bioreagents, 1:1000 IB), anti-PARP5a (H350, Santa Cruz, 1:100 IF, 1:1000 IB), anti-PARP14 (HPA012063, 1:100 IB; HPA008846, 1:100 IF, Sigma Aldrich), anti-GFP (ab1218, Abcam 1:100 IF; JL8, Clontech 1:2500 IB), anti-Tubulin (YL1/2, Abcam, 1:1000 IF, 1:5000 IB), anti-lamin A/C (ab8984, Abcam, 1:100 IF), anti-actin (A3853, Sigma Aldrich, 1:100 IF), anti-vasp (a kind gift from F. Gertler, 1:500 IF), anti-zyxin (ab71842, Abcam, 1:200 IF), anti-paxillin (610051, BD Biosciences, 1:1000 IF), anti-vinculin (Sigma Aldrich, 1:200 IF), anti-p230 (611280, BD Transduction Laboratories, 1:200), anti-gamma tubulin (GTU88, Sigma-Aldrich, 1:10000 IF), anti pH2AX (05-636, Millipore, 1:100 IF), anti-centrin (a kind gift from J. Salisbury, 1:1000 IF), anti-SBP (ab119491, Abcam, 1:500 IF). Secondary antibodies, phalloidin, DiI, EdU labeling kit and Hoechst 33342 were from Invitrogen. siRNA oligonucleotides were purchased from Invitrogen (Stealth or Silencer Select) or Dharmacon (siGenome) (Table S1).
Transfections
HeLa cells were transfected using Lipofectamine 2000 (Invitrogen) according to manufacturer protocols. For RNAi of PARPs-1–4, HeLa cells were transfected with 20 nM siRNA for at least 24 h prior to splitting onto coverslips. Knock-down analysis was performed 48 h post transfection. For PARPs 5a-16, HeLa cells were transfected with 20 nM siRNA for at least 24 h. Cells were split and re-transfected the following day with identical concentrations of siRNA for 24 h prior to splitting. Knock-down analysis was performed 96 h after initial transfection. AllStars negative control siRNA (Qiagen) was used as control. For further validation of PARP14 knock-down phenotype, HeLa cells were transfected with 25nM siRNA and allowed to incubate in transfection mixture for at least 48 hr prior to splitting.
Immunofluorescence
For all transfections, cells were plated on coverslips 6 h post transfections. For G0/G1 arrests, cells were plated at 50% confluence and serum starved for 72 h. For S phase arrest, cells were treated with 5 μg/mL aphidicolin for 16 h. For mitotic arrest, cells were treated with either 200 or 400 nM nocodazole for 16 h. Coverslips were fixed in either −20° MeOH for 5 min, 10 or 20% TCA (Fluka) for 15 min at 4°, or 4% formaldehyde (Polysciences) in PBS (Invitrogen) for 15 min followed by extraction with 0.5% Triton-X100 (Sigma) in PBS for 10 min. For validation of PARP14 localization, cells were grown on fibronectin coated coverslips and fixed in 10% TCA. Coverslips were blocked in abdil (PBS containing 4% BSA, 0.1% Triton X-100, and 0.02% sodium azide) then incubated with primary and secondary antibodies for 45 min. All antibody dilutions were generated in abdil. For quantitation of golgi are in PARP12 overexpression and nuclear area in PARP8 knock-downs, NIS-Elements software was used to generate a region of interest around the golgi/nucleus and the area of the ROI was determined.
Viability Assay
Following RNAi knock-down, cells were counted using a Cellometer automated cell counter then normalized and split in duplicate. Living cells were then counted after 24 and 72 h using trypan blue exclusion and a Cellometer automated cell counter. Viability was assessed by calculating the fold change in cell number relative to cell number initially seeded. PARPs whose knock-down resulted in a decrease in fold change compared to control knock-down of at least two standard deviations were considered to have an effect on viability. Three independent experiments were performed for viability analysis.
Real Time Imaging
Control and PARP14 knock-down HeLa cells were split into 6 well glass bottom plates (Greiner Bio-One) coated with fibronectin and grown overnight. Cells were rinsed with PBS and changed into CO2-independent medium containing 10% FBS (Invitrogen) for imaging. Brightfield images were collected every 5 min for 16 hrs. All live imaging was preformed using a Nikon TiEclipse microscope equipped with an environmental chamber with CO2 and temperature control and Hamamatsu ORCA R2 digital camera.
Cell adhesion assays
For trypsin-based assays, cells were sparsely seeded onto fibronectin coated plates and then incubated overnight. The next morning, the number of cells in each well was calculated. Cells were washed 2x with PBS and then treated with 0.025% trypsin in DMEM. At the indicated time points, the number of cells released into the media and the number of cells remaining on the plate was counted.
For centrifugal-based assays, cells were cells were sparsely seeded onto fibronectin coated plates and then incubated overnight. The next morning, the number of cells in each well was calculated. Cells were washed 3x with PBS and then centrifuged inverted for 30 min at 2,000 g. The number of cells remaining in each well after centrifugation was then counted.
For cell spreading assays, plated cells were gently trypsinized, resuspended in DMEM, and then allowed to recover for 10 min at 37°C. Cells were then very sparsely seeded onto fibronectin coated-coverslips. At the indicated time points after plating, coverslips were gently washed with PBS, then fixed and stained.
RNA sequencing
Total RNA was purified from HeLa and hTERT-RPE1 cells using a Qiagen RNeasy kit. The samples were poly-A purified and cDNA libraries were synthesized using the Illumina Tru-Seq protocol. The libraries were sequenced on Illumina HiSeq2000 obtaining 40nt single end reads. The reads were mapped to the Human genome (hg19) using bowtie2 version 2.0.0-beta6, samtools version 0.1.18 and tophat version 2.0.4.
Supplementary Material
Acknowledgments
The authors wish to dedicate this paper to the memory of Officer Sean Collier, for his caring service to the MIT community and for his sacrifice. We apologize to those that we were unable to cite due to space limitations. We thank A. West, and T. Sangpo for technical support; C. Whittaker of the Bioinformatics and Computing Facility at the Swanson Biotechnology Center, David H. Koch Institute for Integrative Cancer Research at MIT, for computational analysis; J. Rood, P. Ohi, J. Saeij, M. Vander Heiden and F. Solomon for comments. Funding for this project was provided while P.C. was a Rita Allen Foundation Scholar, a Kimmel Foundation for Cancer Research Scholar, and a Howard S. and Linda B. Stern Career Development Assistant Professor. This work was partially supported by Cancer Center Support (core; grant P30-CA14051), RO1GM087465 from the National Institutes of Health to PC, and funds from the Jeptha H and Emily V. Wade Fund, and Curt and Kathy Marble to P.C. M.C-C. was partially supported by NIH Grant 1F32GM103089-01.
Footnotes
Author Contributions
S.V. designed and performed PARP localization and knock-down experiments and PAR cell cycle analysis. S.V. and M.C-C designed and performed experiments for PARP14 specific experiments. T.T. determined the effects of PARP12 overexpression on Golgi structure. Y-H.H. generated the full length GFP-PARP14 construct. P.C., S.V. and M.C-C conceived and designed experiments. S.V., M.C-C, and P.C. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 2.Bürkle A. Poly(ADP-ribose): The most elaborate metabolite of NAD+ FEBS J. 2005;272:4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 3.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 4.Koh DW, Dawson TM, Dawson VL. Mediation of cell death by poly(ADP-ribose) polymerase-1. Pharmacol Res. 2005;52:5–14. doi: 10.1016/j.phrs.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;11:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-Ribose) Regulates Stress Responses and MicroRNA Activity in the Cytoplasm. Molecular Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jwa M, Chang P. PARP16 is a tail-anchored endoplasmic reticulum protein required for the PERK- and IRE1α-mediated unfolded protein response. Nat Cell Biol. 2012;14:1223–30. doi: 10.1038/ncb2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 9.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 10.Chang P, Coughlin M, Mitchison TJ. Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol Biol Cell. 2009;20:4575–4585. doi: 10.1091/mbc.E09-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ha GH, Kim HS, Go H, Lee H, Seimiya H, Chung DH, Lee CW. Tankyrase-1 function at telomeres and during mitosis is regulated by Polo-like kinase-1-mediated phosphorylation. Cell Death Differ. 2012;19:321–332. doi: 10.1038/cdd.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 14.Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, Dixon N, Ahel M, Leys D, Ahel I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–20. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amé JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–89. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 16.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends in Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. The EMBO Jour. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer-Ficca ML, Meyer RG, Jacobson EL, Jacobson MK. Poly(ADP-ribose) polymerases: managing genome stability. Int J Biochem Cell Biol. 2005;37:920–6. doi: 10.1016/j.biocel.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Berkauskaite E, Jankevicius G, Ladurner A, Ahel I, Timinszky G. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J. 2013 doi: 10.1111/febs.12358. [DOI] [PubMed] [Google Scholar]
- 22.Vorobjev IA, Chentsov Yu S. Centrioles in the cell cycle. I Epithelial cells. J Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Augustin A, et al. PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression. J Cell Sci. 2003;116:1551–1562. doi: 10.1242/jcs.00341. [DOI] [PubMed] [Google Scholar]
- 24.Smith S, de Lange T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci. 1999;112:3649–3656. doi: 10.1242/jcs.112.21.3649. [DOI] [PubMed] [Google Scholar]
- 25.Ohashi S, Kanai M, Hanai S, Uchiumi F, Maruta H, Tanuma S, Miwa M. Subcellular localization of poly(ADP-ribose) glycohydrolase in mammalian cells. Biochem Biophys Res Commun. 2003;307:915–921. doi: 10.1016/s0006-291x(03)01272-5. [DOI] [PubMed] [Google Scholar]
- 26.Rouleau M, McDonald D, Gagne P, Ouellet ME, Droit A, Hunter JM, Dutertre S, Prigent C, Hendzel MJ, Poirier GG. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J Cell Biochem. 2007;100:385–401. doi: 10.1002/jcb.21051. [DOI] [PubMed] [Google Scholar]
- 27.Kaminker PG, Kim S-H, Taylor RD, Zebarjadian Y, Funk WD, Morin GB, Yaswen P, Campisi J. TANK2, a new TRF1-associated PARP, causes rapid induction of cell death upon overexpression. J Biol Chem. 2001;276:35891–35899. doi: 10.1074/jbc.M105968200. [DOI] [PubMed] [Google Scholar]
- 28.Juszczynski P, Kutok JL, Li C, Mitra J, Aguiar RC, Shipp MA. BAL1 and BBAP are regulated by a gamma interferon-responsive bidirectional promoter and are overexpressed in diffuse large B-cell lymphomas with a prominent inflammatory infiltrate. Mol Cell Biol. 2006;26:5348–5359. doi: 10.1128/MCB.02351-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goenka S, Boothby M. Selective potentiation of Stat-dependent gene expression by collaborator of Stat6(CoaSt6), a transcriptional cofactor. Proc Natl Acad Sci USA. 2006;103:4210–5. doi: 10.1073/pnas.0506981103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanagawa T, Funasaka T, Tsutsumi S, Hu H, Watanabe H, Raz A. Regulation of phosphoglucose isomerase/autocrine motility factor activities by the poly(ADP-ribose) polymerase family-14. Cancer Res. 2007;67:8682–9. doi: 10.1158/0008-5472.CAN-07-1586. [DOI] [PubMed] [Google Scholar]
- 31.Yonemura S, Hirao-Minakuchi K, Nishimura Y. Rho localization in cells and tissues. Exp Cell Res. 2004;295:300–14. doi: 10.1016/j.yexcr.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Kickhoefer VA, Siva AC, Kedersha NL, Inman EM, Ruland C, Streuli M, Rome LH. The 193-kD vault protein, vPARP, is a novel poly(ADP-ribose) polymerase. J Cell Biol. 1999;146:917–928. doi: 10.1083/jcb.146.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu M, Schreek S, Cerni C, Schamberger C, Lesniewicz K, Poreba E, Vervoorts J, Walsemann G, Grotzinger J, Kremmer E, et al. PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005;24:1982–1993. doi: 10.1038/sj.onc.1208410. [DOI] [PubMed] [Google Scholar]
- 34.Meyer T, Hilz H. Production of anti-(ADP-ribose) antibodies with the aid of a dinucleotide-pyrophosphate resistant hapten and their application for the detection of mono(ADP-ribosyl)ated polypeptides. Eur J Biochem. 1986;155:157–65. doi: 10.1111/j.1432-1033.1986.tb09471.x. [DOI] [PubMed] [Google Scholar]
- 35.Junod AF, Jornot L, Petersen H. Differential Effects of Hyperoxia and Hydrogen Peroxide on DNA Damage, Polyadenosine Diphosphate-Ribose Polymerase Activity, and Nicotinamide Adenine Dinucleotide and Adenosine Triphosphate Contents in Cultured Endothelial Cells and Fibroblasts. J Cell Physiol. 1989;140:177–85. doi: 10.1002/jcp.1041400121. [DOI] [PubMed] [Google Scholar]
- 36.Kirsten E, Kun E, Mendeleyev J, Ordahl CP. Activity Assays for Poly-ADP Ribose Polymerase. In: Tollefsbol TO, editor. Methods in Mol Biol vol 287: Epigenetics Protocols. Totowa, NJ: Humana Press; 2004. pp. 137–150. [DOI] [PubMed] [Google Scholar]
- 37.Kanai M, Tong WM, Sugihara E, Wang ZQ, Fukasawa K, Miwa M. Involvement of poly(ADP-Ribose) polymerase 1 and poly(ADP-ribosyl)ation in regulation of centrosome function. Mol Cell Biol. 2003;23:2451–2462. doi: 10.1128/MCB.23.7.2451-2462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou HY, Chou HT, Lee SC. CDK-dependent activation of poly(ADP-ribose) polymerase member 10 (PARP10) J Biol Chem. 2006;281:15201–7. doi: 10.1074/jbc.M506745200. [DOI] [PubMed] [Google Scholar]
- 40.Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304:97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 41.Fackler OT, Grosse R. Cell motility through plasma membrane blebbing. J Cell Biol. 2008;181:879–884. doi: 10.1083/jcb.200802081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aguiar RC, Yakushijin Y, Kharbanda S, Salgia R, Fletcher JA, Shipp MA. BAL is a novel risk-related gene in diffuse large B-cell lymphomas that enhances cellular migration. Blood. 2000;96:4328–4334. [PubMed] [Google Scholar]
- 44.Bicknell AA, Babour A, Federovitch CM, Niwa M. A novel role in cytokinesis reveals a housekeeping function for the unfolded protein response. J Cell Biol. 2007;177:1017–1027. doi: 10.1083/jcb.200702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burridge K, Fath K, Kelly T, Nuckolls G, Turner C. Focal adhesions: transmembrane junctions between the extracellular matrix and cytoskeleton. Annu Rev Cell Biol. 1988;4:487–525. doi: 10.1146/annurev.cb.04.110188.002415. [DOI] [PubMed] [Google Scholar]
- 46.Kuo JC, Han X, Hsiao CT, Yates JR. 3rd, Waterman, C.M. Analysis of myosin-II-responsive focal adhesion proteome reveals a rolse for β-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signaling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 48.Bhattacharya R, Gonzalez AM, Debiase PJ, Trejo HE, Goldman RD, Flitney FW, Jones JC. Recruitment of vimentin to the cell surface by beta3 integrin and plectin mediates adhesion strength. J Cell Sci. 2009;122:1390–1400. doi: 10.1242/jcs.043042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genomic. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menissier de Murcia J, Niedergang C, Ricoul M, Dutrillaux B, Mark M, Oliver J, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, Sabatier L, Chambon P, de Murcia G. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22:2255. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Citarelli M, Teotia S, Lamb RS. Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol Biol. 2010;10:308. doi: 10.1186/1471-2148-10-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 54.Loseva O, Jemth AS, Bryant HE, Schuler H, Lehtio L, Karlberg T, Helleday T. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J Biol Chem. 2010;285:8054–60. doi: 10.1074/jbc.M109.077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramirez CM, Chamorro-Jorganes A, Wanschel AC, Lasuncion MA, Morales-Ruiz M, Suarez Y, Balden A, Espluques E, Fernandez-Hernando C. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle. 2012;11:1–12. doi: 10.4161/cc.11.5.19421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatsuzawa K, Hirose H, Tani K, Yamamoto A, Scheller RH, Tagaya M. Syntaxin 18, a SNAP receptor that functions in the endoplasmic reticulum, intermediate compartment, and cis-Golgi vesicle trafficking. J Biol Chem. 2000;275:13713–20. doi: 10.1074/jbc.275.18.13713. [DOI] [PubMed] [Google Scholar]
- 57.Sciaky N, Presley J, Smith C, Zaal KJ, Cole N, Moreira JE, Terasaki M, Siggia E, Lippincott-Schwartz J. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J Cell Biol. 1997;139:1137–55. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barbarulo A, Iansante V, Chaidos A, Naresh K, Rahemtulia A, Franzoso G, Karadimitris A, Haskard DO, Papa S, Bubici C. Poly(ADP-ribose) polymerase family member 14 (PARP14) is a novel effector of the JNK2-dependent pro-survival signal in multiple myeloma. Oncogene. 2012 doi: 10.1038/onc.2012.448. [DOI] [PubMed] [Google Scholar]
- 59.Cho SH, Ahn AK, Bhargava P, Lee CH, Eischen CM, McGuinness O, Boothby M. Glycolytic rate and lymphomagenesis depend on PARP14, an ADP ribosyltransferase of the B aggressive lymphoma (BAL) family. Proc Natl Acad Sci USA. 2011;108:15972. doi: 10.1073/pnas.1017082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.