Abstract
The bHLH-PAS transcription factor BMAL1 (MOP3) is an essential component of the mammalian circadian pacemaker. Bmal1−/− mice lose circadian rhythmicity but also display tendon calcification and decreased activity, body weight and longevity. To investigate whether these diverse functions of BMAL1 are tissue-specific, we produced transgenic mice that constitutively express Bmal1 in brain or muscle, and examined the effects of rescued gene expression in Bmal1−/− mice. Circadian rhythms of wheel running activity were restored in brain-rescued Bmal1−/− mice in a conditional manner; however, activity levels and body weight were lower than those of wild-type mice. In contrast, muscle-rescued Bmal1−/− mice exhibited normal activity levels and body weight yet remained behaviorally arrhythmic. Thus Bmal1 has distinct tissue-specific functions that regulate integrative physiology.
Circadian rhythms control many aspects of mammalian physiology and behavior. The suprachiasmatic nuclei (SCN) act as pacemakers required for the generation of circadian behavioral rhythms as well as synchronizers of autonomous peripheral tissue clocks (1). Molecular circadian regulation engages a transcription/translation feedback loop comprised of the activating proteins CLOCK and BMAL1, which induce expression of the negative feedback elements Per and Cry (1). BMAL1 was originally characterized by its high expression in brain and muscle (2, 3) and was identified as a heterodimeric binding partner of CLOCK (4, 5). Bmal1−/− mice not only lose behavioral circadian rhythmicity, but also exhibit a variety of other phenotypes including decreased activity levels and body weight, progressive joint disease and shortened life span (6–12). Therefore, in addition to circadian rhythm regulation, BMAL1 appears to play a role in a variety of functions that are potentially dependent on the tissue type in which it is expressed. To determine whether BMAL1 has unique tissue-specific functions, we generated transgenic mice that express Bmal1 ubiquitously or in distinct tissue types. We then crossed these lines onto a Bmal1 null background and determined which phenotypes could be rescued by exogenous, tissue-specific Bmal1 expression.
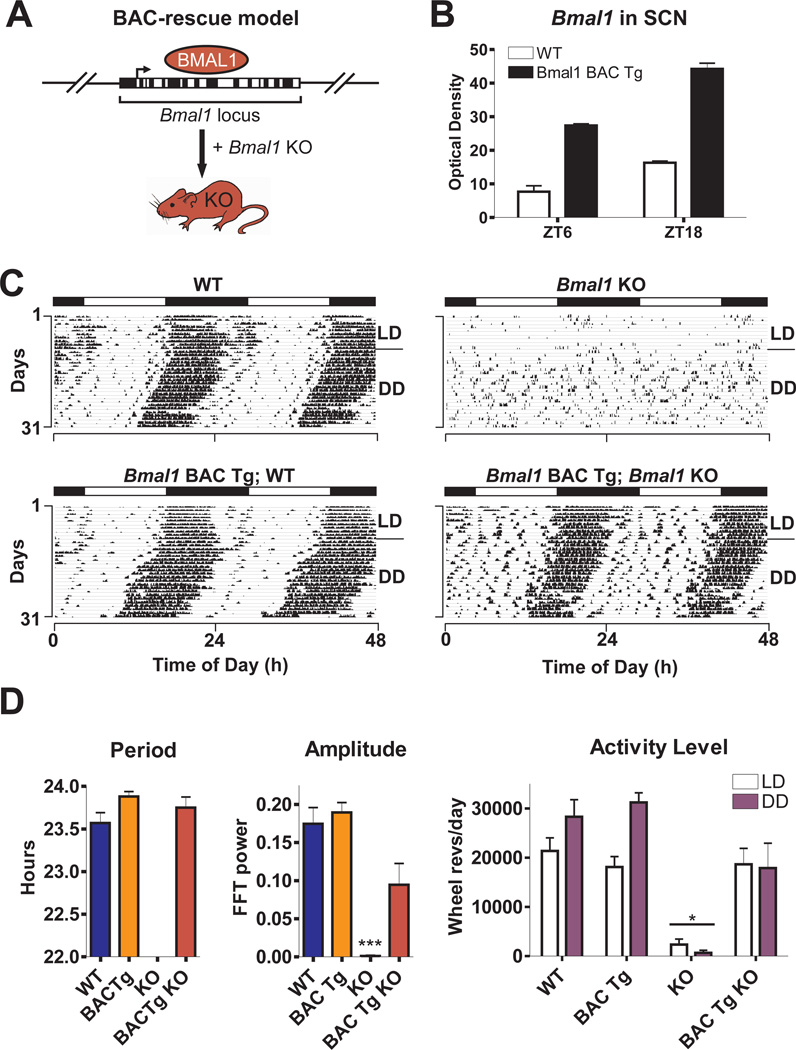
We first examined the effects of rescuing Bmal1 ubiquitously by using a transgenic mouse line produced with Bmal1-containing bacterial artificial chromosome (BAC) clones (Fig. 1A, 13). Since the BAC clones contain genomic coding and promoter sequence of Bmal1, expression of the transgene should occur in all tissues that normally express Bmal1. We measured increased Bmal1 expression in the SCN of BAC transgenic mice at normal peak and trough times of Bmal1 mRNA (zeitgeber time (ZT)18 and ZT6, respectively), and also observed increased Bmal1 expression during peak times in the liver (ZT 18-2) (fig. S1, 13). BAC transgenic mice were then sequentially crossed with Bmal1+/− mice to produce BAC-rescued Bmal1−/− mice (13). Circadian rhythms of locomotor activity were then analyzed in a 12h:12h light:dark (LD) cycle followed by constant darkness (DD) conditions (Fig. 1C) (13).
Fig. 1.
Bmal1-containing BAC transgenes rescue Bmal1−/− phenotypes. (A) Bmal1 BAC clones were used to create transgenic (Tg) mice, which were consecutively crossed with Bmal1+/− mice to create BAC-rescued Bmal1−/− mice. (B) Bmal1 mRNA levels in SCN were examined by in situ hybridization in wild-type (WT) and Bmal1 BAC Tg mice, sacrificed at ZT6 and ZT18 (shown are mean ± SEM; significant effect of genotype, GLM ANOVA). (C) Representative wheel-running activity records from WT, Bmal1 BAC Tg, Bmal1−/− (Bmal1 KO) or BAC-rescued (Bmal1 BAC Tg; Bmal1 KO) mice. Mice were housed in LD then released into DD for 3 weeks. (D) Bar graphs of mean ± SEM show that BAC-rescued mice (n=6) exhibit free-running period, amplitude of circadian rhythm and activity levels that are not significantly different from WT. Amplitude is graphed as the peak amplitude of the proportion of the total variance in the time series in the circadian (~24h) range (***p<0.001, one-way ANOVA; *significant effect of genotype, GLM ANOVA).
While Bmal1−/− mice exhibited no circadian rhythm of activity in DD and showed reduced activity levels, BAC-rescued Bmal1−/− mice displayed normal circadian rhythm characteristics (free-running period and amplitude of circadian rhythm) and activity levels in LD and DD that were similar to wild-type (WT) mice (Fig. 1D). In addition, 100% of BAC-rescued Bmal1−/− mice survived until the end of experimental analysis (≥10 months old) compared to 29% of Bmal1−/− mice. Therefore, long-term survival was restored in the BAC-rescued Bmal1−/− mice, and no gross abnormalities such as low body weight or joint calcification were observed in the BAC-rescued mice. Thus, Bmal1 BAC transgenes completely rescued the phenotypes observed in Bmal1−/− mice.
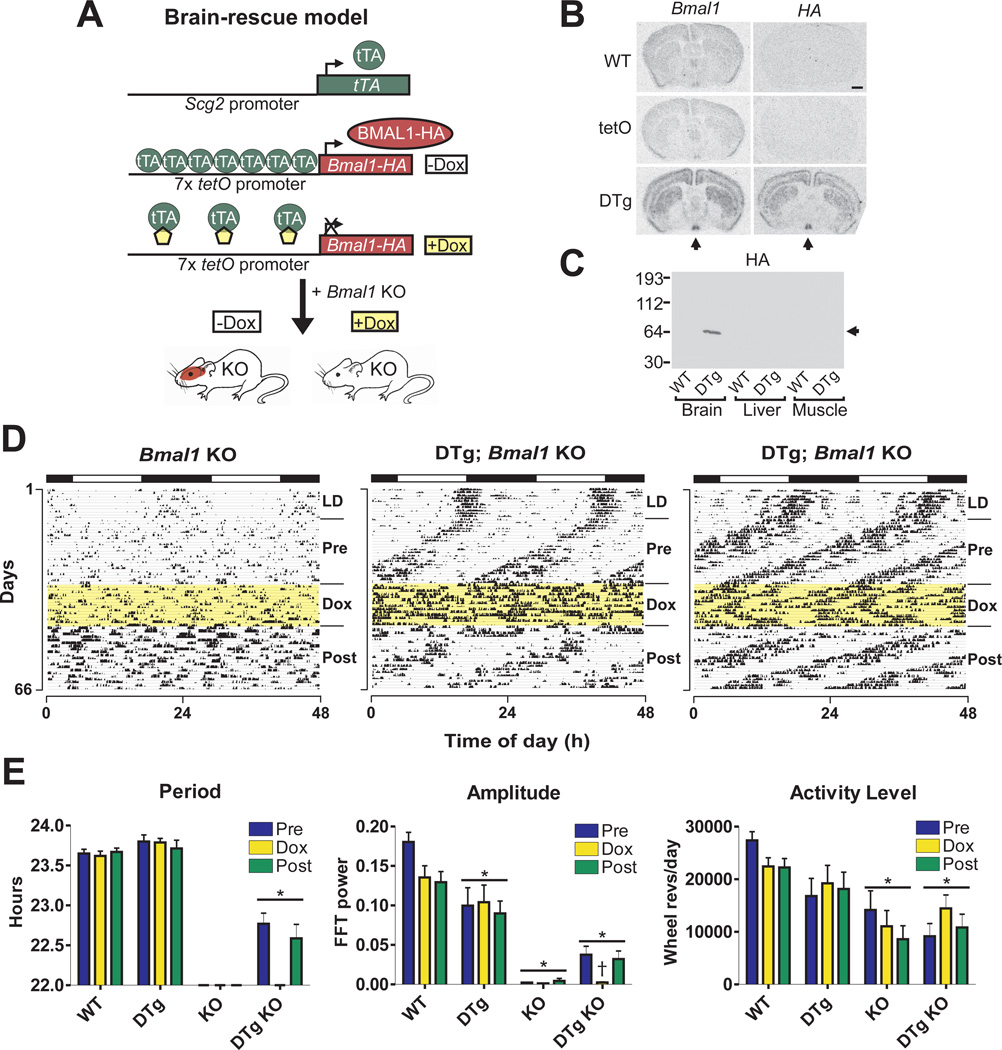
We next determined whether expression of Bmal1 in brain tissue could restore behavioral rhythms in Bmal1−/− mice as well as alleviate other phenotypes. To produce the brain-rescued line, we used the tetracycline transactivator (tTA) system, which requires two transgenes for expression of the target gene, Bmal1 (see Fig. 2A) (14, 15). We used the promoter sequence of Scg2, which is expressed in brain and enriched in the SCN (16), to drive expression of the tetracycline transactivator (tTA) (13). The tTA protein binds to the tetracycline operator (tetO) sequence and drives expression of downstream hemagglutinin (HA)-tagged Bmal1 (Bmal1-HA) cDNA. Doxycycline (Dox) inhibits tTA binding to the tetO promoter, which halts expression of Bmal1-HA. In situ hybridization showed strong, specific expression of Bmal1-HA in Scg2∷tTA x tetO∷Bmal1-HA double transgenic mice only (Fig. 2B, 13). The pattern of expression observed is consistent with high Scg2 expression in SCN. HA-tagged protein at the correct molecular weight for BMAL1 (~69kD) was produced specifically in double transgenic mouse brain extracts (Fig. 2C), and HA-tagged BMAL1 was shown to be functional by Per1∷luciferase reporter gene assays (fig. S2, 13). The double transgenic mice were crossed onto Bmal1−/− background to create brain-rescued Bmal1−/− mice, and wheel-running experiments were performed as described above (Fig. 2D, fig. S3, S4 and S5).
Fig. 2.
Reversible restoration of circadian rhythms but not activity levels in brain-rescued Bmal1−/− mice. (A) Mice were created to express Bmal1-HA conditionally in brain tissue by using the tTA system. (B) In situ hybridization was performed with HA tag or Bmal1 probes on brains from WT, tetO∷Bmal1-HA (tetO), or Scg2∷tTA x tetO∷Bmal1-HA double transgenic (DTg) mice sacrificed at ZT 6 (arrow indicates SCN, scale bar is 1mm). (C) Western blot showing HA staining in brain, liver and skeletal muscle protein extracts from WT or DTg mice sacrificed at ZT 12 (arrow indicates correct size of BMAL1). (D) Representative wheel-running activity records from one Bmal1−/− mouse and two brain-rescued Bmal1−/− (DTg; Bmal1 KO) mice. After 3 weeks in DD (Pre), mice were administered Dox for 2 weeks (Dox, highlighted yellow) and then spent an additional 3 weeks without Dox (Post). (E) Brain-rescued mice (n=10) display a free-running period of 22.8 h (Pre) and 22.6 h (Post) when Bmal1 is expressed, which are significantly different from WT and DTg groups (*significant effect of genotype, GLM ANOVA). Activity levels of KO and DTg KO mice were significantly reduced compared to WT. Amplitude of circadian rhythm was significantly different in all genotypes compared to WT, and a simultaneous loss of rhythm and decrease in amplitude were observed in DTg KO mice during Dox treatment (†significant effect of time interval). Graphs represent the mean ± SEM.
Adult (≥8 weeks old) brain-rescued Bmal1−/− mice exhibited a consistent circadian rhythm of behavior in the initial (pre-Dox) DD period, which was completely abolished after 1–2 days of Dox administration and then regained during Dox withdrawal. However, the free-running period of brain-rescued mice was approximately 1 hour shorter than that of WT (Fig. 2E). This was likely due to the constitutive bioavailability of BMAL1 protein and/or the lack of peripheral tissue feedback to the SCN (fig. S6). In support of this, Rev-Erbα−/− mice express Bmal1 in the SCN at consistently high levels and exhibit shortened period length (17). In contrast to the restoration of circadian rhythmicity in brain-rescued mice, both amplitude and activity levels were significantly lower than that seen in WT mice (Fig. 2E). Thus, brain-rescued mice exhibit restored circadian rhythms of behavior but their locomotor activity is still impaired.
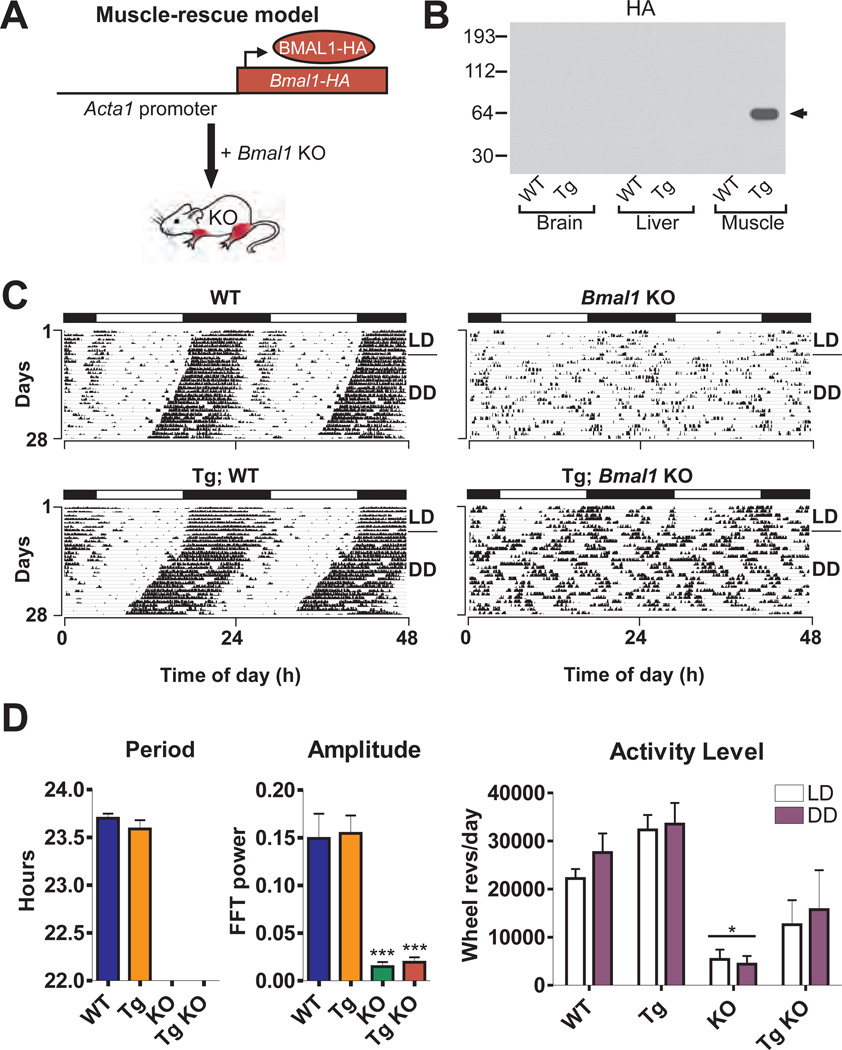
Because Bmal1 is highly expressed in muscle, we investigated whether muscle-specific rescue might restore activity levels in Bmal1−/− mice. We produced muscle-specific Bmal1 transgenic mice using a DNA construct consisting of human alpha actin-1 (Acta1) promoter sequence positioned upstream of Bmal1-HA (Fig. 3A, 13). HA-tagged protein was specifically detected in transgenic muscle extracts (Fig. 3B, 13). Adult muscle-rescued Bmal1−/− mice did not express circadian rhythmicity of activity (Fig. 3C, D); however, their level of locomotor activity was not significantly different from WT mice (Fig. 3D).
Fig. 3.
Muscle-rescued mice exhibit restored activity level but not circadian rhythms. (A) Muscle-specific Bmal1 Tg mice were created by fusing the Acta1 promoter sequence to Bmal1-HA. (B) Western blot shows HA staining in brain, liver and skeletal muscle protein extracts from WT or Tg mice sacrificed at ZT 12 (arrow indicates correct size of BMAL1). (C) Representative wheel-running activity records are shown from WT, Tg, Bmal1 KO, and muscle-rescued (Tg; Bmal1 KO) mice. (D) Muscle-rescued mice (n=6) are arrhythmic in DD with significantly reduced amplitude of rhythm (**p<0.01, one-way ANOVA), but display activity levels that are not significantly different from WT mice. Graphs show mean ± SEM (*significant effect of genotype, GLM ANOVA).
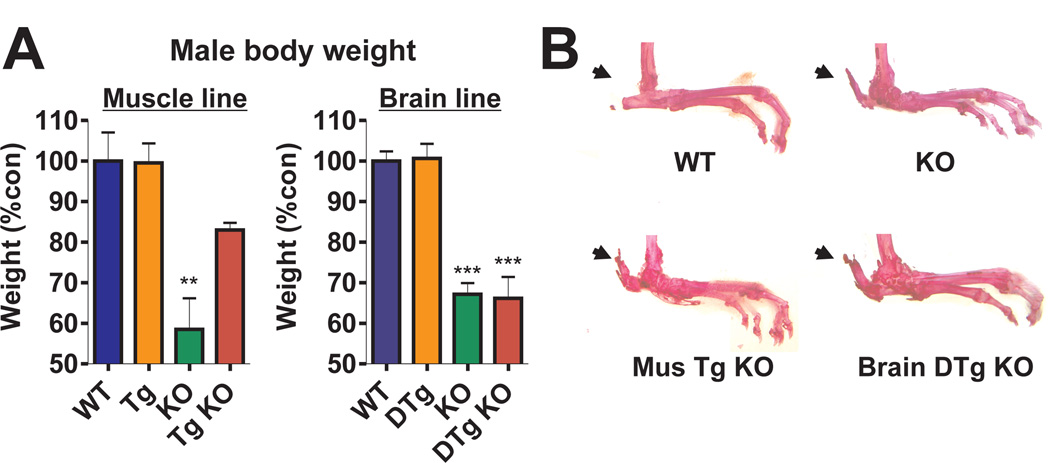
We also found that at 4–6 months of age, the Bmal1−/− and brain-rescued mice weighed significantly less than WT mice. In contrast, the body weight of muscle-rescued mice was restored to a level not significantly different from that of WT mice (Fig. 4A, 13). Only 75% of brain-rescued mice survived to the end of the experiment, whereas 100% of muscle-rescued mice survived. These results suggest that BMAL1 function in muscle is important for activity as well as body weight maintenance and longevity. In addition, bone phenotypes were examined by Alizarin Red stain: both brain and muscle-rescued mice showed significant tendon calcification similar to that seen in Bmal1−/− mice (Fig. 4B). This suggests that Bmal1-HA was not expressed in bone in either line and that the calcification observed in Bmal1−/− mice was not improved by restoring BMAL1 expression in muscle or brain. Thus, three distinct patterns of rescue could be observed in these mice distinguishing among: 1) circadian activity rhythms; 2) activity level and body weight; and 3) tendon calcification.
Fig. 4.
Effects of tissue-specific Bmal1 expression on body weight and tendon calcification. (A) Brain-rescued mice and KO mice in both lines have significantly reduced body weight, while muscle-rescued mice exhibit similar body weight to WT mice (graphs represent mean + SEM; **p<0.01 ***p<0.001, one-way ANOVA). (B) Photographs of Alizarin Red-stained hind limbs from WT, KO, muscle and brain-rescued KO mice are shown. Arrows indicate calcaneal tendon calcification in all but WT mice.
Unlike the BAC transgenic line, the brain and muscle transgenic lines were designed to constitutively express BMAL1-HA. To verify this, we measured similar levels of Bmal1 mRNA and BMAL1-HA protein at normal peak and trough times in the brain and muscle transgenic lines (fig. S6, S7, 13). We then examined mRNA levels of key BMAL1 target genes Per1 and Per2 (6). Compared to WT and brain double transgenic mice, Bmal1−/− mice exhibited consistently low expression levels of Per1 in the SCN (fig. S7). In contrast, the brain-rescued mice had increased amplitude of Per1 expression, although peak levels remained significantly lower than WT (fig. S7). Per2 expression was measured in both muscle and liver from the brain and muscle transgenic lines at normal peak time for Per2 (ZT12) (fig. S8, 13). Per2 in muscle of Bmal1−/− mice was significantly reduced to below 50% WT levels at ZT12. This decreased expression was restored to WT levels in the muscle-rescued mice but not in the brain-rescued mice. These data suggest that the presence of BMAL1 is important for proper expression of Per1 genes in brain and muscle tissue (but not liver, see fig. S8). Both Rev-Erbα and Dbp exhibited dramatic downregulation in liver and muscle of Bmal1−/− mice at ZT12 and showed increased expression only in muscle of muscle-rescued mice (fig. S8).
In conclusion, we have shown that all phenotypes of Bmal1−/− mice are alleviated only when Bmal1 is rescued ubiquitously, whereas different parameters of behavioral activity (circadian rhythm and activity level), body weight and gene expression can be rescued separately by distinct spatial expression patterns of Bmal1. Genome-wide profiling experiments suggest that ~10% of the transcriptome is under circadian regulation; however, the majority of these cycling transcripts are tissue-specific (18–22). Our results are consistent with this tissue-specific diversity of circadian expression and further suggest that core circadian clock components may play distinct roles in different tissues, perhaps in addition to their function in regulating circadian rhythms. The restoration of circadian activity rhythms in brain-rescued Bmal1−/− mice is consistent with previous SCN transplant studies in rodents (23, 24). However, the transgenic approach used here has the advantages of preserving the anatomical integrity of the brain as well as allowing the conditional manipulation of the rescue via Dox treatment. The use of tissue-specific and conditional regulation of circadian clock gene expression should be a valuable method for understanding the molecular-, cellular- and systems-level regulation of circadian rhythms in mammals.
Supplementary Material
Acknowledgments
We thank N. Lampl, A. Jyawook and A. Falk for technical assistance; members of the Takahashi laboratory and F. Davis for expert advice and K. Esser, E. Hardeman and M. Mayford for the kind gifts of plasmids. Supported by NIH grants R01 ES005703 to C.A.B. and P50 MH074924 to J.S.T.
References
- 1.Lowrey PL, Takahashi JS. Annu Rev Genomics Hum Genet. 2004;5:407. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikeda M, Nomura M. Biochem Biophys Res Commun. 1997;233:258. doi: 10.1006/bbrc.1997.6371. [DOI] [PubMed] [Google Scholar]
- 3.Hogenesch JB, et al. J Biol Chem. 1997;272:8581. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 4.Gekakis N, et al. Science. 1998;280:1564. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 5.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. Proc Natl Acad Sci U S A. 1998;95:5474. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunger MK, et al. Cell. 2000;103:1009. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudic RD, et al. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunger MK, et al. Genesis. 2005;41:122. doi: 10.1002/gene.20102. [DOI] [PubMed] [Google Scholar]
- 9.Laposky A, et al. Sleep. 2005;28:395. doi: 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- 10.Shimba S, et al. Proc Natl Acad Sci U S A. 2005;102:12071. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Genes Dev. 2006;20:1868. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, et al. J Biomed Sci. 2006 [Google Scholar]
- 13.Please see Supplementary Online Material
- 14.Gossen M, Bujard H. Proc Natl Acad Sci U S A. 1992;89:5547. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furth PA, et al. Proc Natl Acad Sci U S A. 1994;91:9302. doi: 10.1073/pnas.91.20.9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong H, et al. Society for Research on Biological Rhythms Abstracts. 2006;10:30. [Google Scholar]
- 17.Preitner N, et al. Cell. 2002;110:251. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 18.Kornmann B, Preitner N, Rifat D, Fleury-Olela F, Schibler U. Nucleic Acids Res. 2001;29:E51-1. doi: 10.1093/nar/29.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akhtar RA, et al. Curr Bio. 2002;12:540. [Google Scholar]
- 20.Panda S, et al. Cell. 2002;109:307. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 21.Ueda HR, et al. Nature. 2002;418:534. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 22.Storch KF, et al. Nature. 2002;417:78. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 23.Ralph MR, Foster RG, Davis FC, Menaker M. Science. 1990;247:975. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 24.Sujino M, et al. Curr Bio. 2003;13:664. doi: 10.1016/s0960-9822(03)00222-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.