Abstract
Autophagy is in principle a non-selective degradation system within cells, which is conserved in all eukaryotic cells. Autophagy is usually suppressed at low levels but can be upregulated during periods of nutrient starvation, which facilitates cell survival. In addition to this fundamental role, basal autophagy was recently revealed to be important for constitutive turnover of intracellular proteins and organelles. Autophagy has been considered to be involved also in presentation of endogenous antigens, degradation of invasive bacteria, tumor suppression, cell death and development. This review will discuss the biological significance of autophagy, particularly focusing on its implications in protein metabolism in mammals.
Keywords: protein degradation, starvation, aggregation, amino acids
Introduction
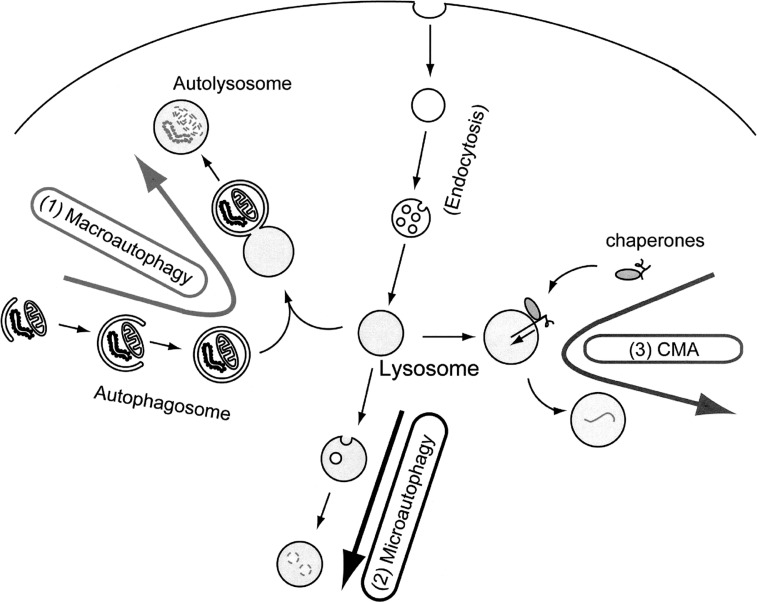
Intracellular protein degradation systems can be roughly classified into two groups: one that is selective and another that is non-selective. The selective degradation is primarily carried by the ubiquitin-proteasome system present in the cytoplasm and nucleus. On the other hand, the latter type occurs mainly in the lysosome that is an organelle specialized for degradation. The degradation and recycling of cytoplasmic content by lysosomes is generically referred to as autophagy.1)–4) Three distinct types of autophagy have been identified, macroautophagy, microautophagy and chaperone-mediated autophagy (CMA) (Fig. 1). CMA is mediated by chaperones that specifically recognize substrates. Thus, CMA is a type of selective, rather than a bulk or non-selective, degradation system.1) Microautophagy has been proposed to occur based on morphological change of lysosome, but its molecular mechanism remains unknown. Among these, the molecular mechanism and physiologic significance of macroautophagy have been best studied. Therefore, under most circumstances, autophagy refers to macroautophagy. Upon induction of autophagy, a membrane sac called the isolation membrane elongates and encloses a portion of cytoplasm. Complete sequestration, which takes about 10 minutes, results in formation of a double membrane structure called the autophagosome. The diameter is usually 0.5–1.5 μm in mammalian cells and 0.3–0.9 μm in yeast cells. Organelles such as mitochondria and ER fragments are often enclosed. When the outer membrane of autophagosome fuses with the lysosome, the inner membrane of the autophagosome and the cytoplasm-derived materials are degraded by lysosomal hydrolases.
Fig. 1.
Target to the lysosome via three types of autophagy.
(1) macroautophagy: A portion of cytoplasm is enclosed by the autophagic isolation membrane, which eventually results in the formation of a double membrane structure, called autophagosome. The outer membrane of the autophagosome then fuses with the lysosome where the cytoplasm-derived materials are degraded.
(2) microautophagy: A small portion of cytoplasm is engulfed by the lysosome membrane itself.
(3) Chaperone-mediated autophagy (CMA): Cytosolic proteins containing KFERQ-like motifs are recognized by a cytosolic chaperone Hsc70 and co-chaperones. When the resulting complexes bind to a lysosomal receptor, Lamp2a, the substrates are unfolded and transported into the lysosomal lumen for degradation.
Molecular mechanism of autophagy
Yeast genetic studies have identified more than 20 genes (ATG genes) required for autophagy, most of which function in autophagosome formation.3), 5), 6) With these remarkable findings, studies of autophagy have been rapidly progressed for the past decade.
In yeast cells, autophagosomes seem to be generated from a structure near the vacuole, termed the pre-autophagosomal structure (PAS), where most ATG gene products (Atg proteins) are targeted.7), 8) The precise nature of PAS is still unclear and it remains also unknown whether mammalian cells contain PAS-like structures. Formation of PAS requires a PtdIns 3-kinase complex made up of Vps15, Vps30/Atg6, Atg14, and Vps34.9) Vps30/Atg6 has a mammalian counterpart called Beclin-1.10) Beclin-1 interacts with class III PtdIns 3-kinase (mammalian Vps34) and p150 (mammalian Vps15).11) Beclin-1 was originally isolated as a Bcl-2-interacting protein10) and Beclin-1 was reported to be involved in tumorigenesis12) and cell death/survival.13) Since Vps30/Atg6 is a common subunit shared with another type PtdIns 3-kinase complex made up of Vps34, Vps15, Vps30 and Vps38, which functions in the vacuolar protein sorting pathway. Thus, Beclin-1 is also considered as a multifunctional protein. Therefore, it remains to be determined which roles of Beclin-1 is indeed related to autophagy.
Many Atg proteins are likely involved in the elongation process of isolation membrane. One of them is the Atg12 conjugation system.14)–16) Atg12 and Atg5 are covalently attached to each other post-translationally and the resulting conjugated behaves as if it is a single molecule. The conjugation reaction is similar to the ubiquitin system.5) At the initial step of the conjugation reaction, the carboxy-terminal glycine residue of Atg12 is activated by Atg7 (E1-like), resulting in formation of an Atg12-Atg7 thioester intermediate. Atg12 is then transferred to Atg10 to form an Atg12-Atg10 thioester intermediate. Finally the carboxy-terminal glycine of Atg12 is covalently attached to a central lysine of Atg5 via an isopeptide bond. The Atg12 system is completely conserved in mammals.17) The Atg12-Atg5 conjugate further interacts with Atg16 (Atg16L in mammals) to form a large protein complex. 18)–20) Although most of this complex resides in the cytosol, a small fraction localizes on the isolation membrane throughout its elongation process, and then dissociates from the membrane upon completion of autophagosome formation. Analysis of Atg5−/− embryonic stem cells revealed that the Atg12-Atg5 conjugate is indeed required for the membrane elongation.16) Another system that is thought to be involved in the elongation step is Atg8 or its mammalian orthologue, LC3 (microtubule-associated protein 1 (MAP1) light chain 3).5), 21), 22) Atg8/LC3 is also a ubiquitin-like protein, which is conjugated to phosphatidylethanolamine (PE). Atg8/LC3 possesses some C-terminal extension following the conjugation-competent glycine residue. Immediately after synthesis, this C-terminal region of Atg8/LC3 (1, 22 and 5 amino acids in yeast, rat and human LC3, respectively) is cleaved by Atg4.22)–26) The processed form, having a glycine residue at the C-terminal end (called LC3-I in mammals) resides in the cytosol. After activation by Atg7, which is shared with the Atg12 system, Atg8/LC3 is transferred to a specific E2 equivalent enzyme, Atg3. Atg8/LC3 is finally attached to PE (called LC3-II in mammals). In contrast to Atg12-Atg5, Atg8/LC3 localizes on the membrane of complete spherical autophagosomes as well as on the isolation membranes. Thus LC3 is now widely used as an autophagosome marker.22), 27), 28)
Autophagy in starvation response
One of the most fundamental and evolutionarily conserved functions of autophagy is its role in the response to starvation. In budding yeast, autophagy is suppressed to undetectable levels under growing conditions. However, it is rapidly induced during nitrogen starvation.29) Similar response is also observed in whole animals. We have developed an autophagy indicator mouse model, in which autophagosomes are labeled with LC3 fused to green fluorescent protein (GFP).27), 28) Using this mouse model, we found that autophagy is upregulated in almost all organs following initiation of starvation (Fig. 2). Interestingly, autophagy is differently regulated among organs. In most tissues, the autophagic activity reaches maximal levels within 24 hours and then gradually decreases, whereas it is further accelerated even after 24 hours in some tissues such as the heart and slow twitching muscles. Some tissues show constitutively active autophagy. Thymic epithelial cells are the best example, in which autophagy actively occurs under nutrient rich conditions28) and even during embryogenesis (our unpublished observation). In this case, autophagy might be involved in presentation of cytosolic antigens onto MHC Class II proteins to establish the central tolerance.30) In contrast, autophagy is not observed in the brain even after 48-h food withdrawal. This might be because the brain is nutritionally protected under physiological conditions. For example, the brain can utilize nutrients such as glucose and ketone bodies supplied by other tissues.
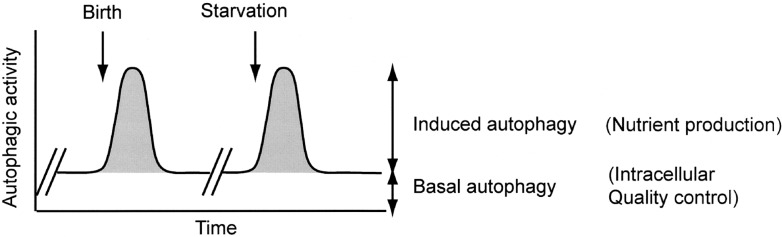
Fig. 2.
Roles of induced autophagy and baseline autophagy.
The level of autophagy is usually low but can be upregulated by starvation such as birth and fasting. The induced autophagy is important for intracellular generation of amino acids. On the other hand, the basal autophagy is crucial for intracellular quality control.
The critical role played by autophagy in maintaining viability during starvation has been shown in Sacharomyces cerevisiae,31)Dictyostelium discoideum32) and Drosophila melanogaster.33) In addition, we recently revealed that autophagy is also important to help mammals survive a unique period of starvation, the birthing process.34) It is known that neonates face severe starvation at birth due to sudden termination of the placental nutrient supply. We found that autophagy is transiently upregulated in various tissues soon after birth. We then determined the significance of this neonatal autophagy by analyzing mice deficient in Atg5, a gene essential for elongation of the isolation membrane as described above.16) Atg5−/− mice appear grossly normal at birth, but exhibit systemic amino acid insufficiency within 10 h after birth. In particular, the amino acid concentrations in plasma and tissues are significantly lower in Atg5−/− mice. Atg5−/− mice display significantly shorter survival times than wild-type mice under starvation conditions, but death could be delayed by forced milk ingestion. Therefore, these findings suggest that neonates use amino acids degraded from tissue proteins.34)
How amino acids produced by autophagy during starvation are utilized in mammals still remains to be determined. Although amino acids are not generally considered to be a good fuel source, amino acids, particularly branched-chain amino acids (BCAA), can be used as direct energy source. The activity of the branched chain α-ketoacid dehydrogenase complex, which is the most important regulatory enzyme for BCAA catabolism, is upregulated in starvation. The importance of amino acid production via autophagy was also shown in cultured cells. IL-3 dependent cells derived from mice deficient for Bax and Bak, both of which are key initiators of apoptosis, maintain cell viability even after IL-3 withdrawal.35) Due to reduced surface expression of amino acid transporters, these cells are unable to take up amino acids from the outside environment, but do still survive for a lengthy period of time. Nutrient levels in these cells are maintained by upregulation of autophagy, as RNAi-mediated inhibition of autophagy was lethal to these cells. As a final confirmation, cell death due to inhibition of autophagy could be avoided by addition of methylpyruvate, a cell permeable tricarboxylic acid (TCA) cycle substrate. These results suggest that cultured cells use amino acids as an energy source. In addition to direct energy production, amino acids produced by autophagy can be used for gluconeogenesis. During early starvation, glycogen is used for glucose homeostasis. However, in mammals, glycogen stores are consumed within one day even in adults. Subsequently, blood glucose is maintained by gluconeogenesis from pyruvate or TCA cycle intermediates such oxaloacetate in the liver. Amino acids produced by autophagy in the liver and other tissues could be utilized for this aim. Autophagy would be also important for protein synthesis required for the proper starvation response, which has been clearly demonstrated in yeast.36) Taken together, these studies emphasize that increased intracellular generation of amino acids by autophagy is a physiologically important starvation response.
Cell size is determined by the relative rate of cell growth and cell division,37) and both growth factor signaling (e.g. insulin) and nutrient signaling pathways control cell growth. The primary cause of cell atrophy following suppression of growth factor and nutrient signaling is thought to be a reduction in macromolecular synthesis. However, both nutrient limitation and growth factor withdrawal are well known inducers of autophagy.38) Additionally, inhibition of TOR signaling by rapamycin induces autophagy. 39) Since protein synthesis and autophagy are tightly coupled, the specific inhibition of autophagy is required to determine whether autophagy contributes to cell size reduction. For this purpose, we established mouse fibroblast lines coupling the Tet-off system with an Atg5−/− mouse embryonic fibroblast line to artificially regulate autophagic ability. 40) In the presence of doxycycline, Atg5 expression was completely suppressed and these cells were autophagy-defective. After removal of doxycycline, autophagic ability was restored. Using this novel tool, we found that cell size reduction in response to starvation was significantly inhibited in cells unable to undergo autophagy.40)
Autophagy in intracellular clearance
Autophagy occurs constitutively at low levels even under normal growth conditions (Fig. 2). It was recently revealed that this basal autophagy is critical for intracellular protein quality control particularly in neurons and hepatocytes.41)–43)
The role of basal autophagy was first demonstrated in liver-specific Atg7−/− mice.43) In these mice, the Atg7 gene can be deleted in adult mice by intraperitoneal pIpC injection. After twenty days, they show hepatomegaly and some abnormal organelles such as deformed mitochondria and endoplasmic reticulum accumulated in hepatocytes. However, the most unexpected results with these mice are that many ubiquitin-positive aggregates are generated in hepatocytes. Finally, liver-specific Atg7−/− mice at 90 days after pIpC injection show severe hepatomegaly with disorganized hepatic lobules, cell swelling and cell death. Serum alanine aminotransferase and aspartate aminotransferase are significantly elevated at this stage.
We also observed that protein aggregates already accumulated in the liver of Atg5−/− neonates.41) Systematic analysis of Atg5−/− neonates revealed that the amount of aggregate accumulation differs among cell and tissue types. Ubiquitin-positive aggregates accumulate vigorously only in hepatocytes, a subset of neurons, the anterior lobe of the pituitary gland, and the adrenal gland.41) In contrast, very few aggregates are seen in skeletal muscle, heart, and kidney. Atg5−/− mice show a suckling defect at birth.34) The reason for this has not yet been determined, but is probably attributable to central nervous system (CNS) dysfunction due to abnormal protein accumulation. These findings suggest that baseline autophagy is critical for intracellular clearance (Fig. 2).
To further understand the role of basal autophagy in neurons, neural cell-specific Atg5 and Atg7 knockout mice were generated and analyzed. 41), 42) These mice were born normally but exhibited growth retardation. After three weeks, they developed progressive motor and behavioral deficits, including ataxic gait, impaired motor coordination, abnormal limb clasping reflexes, and systemic tremor. Sporadic death of some animals was observed after three-weeks of age. Histological examination showed partial loss of Purkinje and cerebral pyramidal cells and axonal swelling. Ubiquitin-positive protein aggregates (inclusion bodies) also accumulated in neurons. Therefore, autophagy is required to prevent neurodegeneration, even if animals do not express disease-associated mutant (aggregate prone) proteins.
In contrast, autophagy-defective yeast cells,31) embryonic stem (ES) cells16) and embryonic fibroblasts34) are quite healthy and show no apparent abnormalities under growing conditions. In rapidly dividing cells, abnormal proteins may be quickly diluted even if they are not degraded. Therefore, intracellular clearance by autophagy should be much more important in post-mitotic cells.
The mechanism underlying the accumulation of ubiquitin-positive aggregates is unknown. Loss of autophagy first leads to accumulation of diffuse ubiquitinated proteins in the cytoplasm followed by the generation of protein aggregates.41) Therefore, the aggregate formation is likely a secondary result of a general protein turnover defect. If turnover of most cytosolic proteins is impaired, proteins would have more opportunities to be damaged or misfolded. In that situation, they have a greater chance to be both ubiquitinated and aggregated. This is consistent with the general agreement that autophagic sequestration is random, and non-selective. It has been suggested that large aggregates themselves may not be pathogenic, whereas mutant proteins diffusely present in the cytosol could be the primary source of toxicity.44) Consistent with this, Purkinje cells, which are partially lost in neural cell-specific Atg5−/− and Atg7−/− mice, do not exhibit ubiquitin-positive inclusions. Thus, one beneficial effect of basal autophagy is the elimination of diffused abnormal proteins, not protein aggregates themselves.
However, there is yet another possibility that ubiquitinated proteins are recognized via the inner membrane of autophagosomes and thus preferentially degraded. Recently, it was proposed that p62/SQSTM1 mediates the specific recognition of ubiquitinated aggregates by autophagosomes.45) p62/SQSTM1 can bind both ubiquitin and LC3, allowing it to function as an adaptor protein linking ubiquitinated proteins and autophagosomes. Indeed, p62/SQSTM1 is selectively degraded by the autophagy pathway: p62/SQSTM1 is extensively enriched in various autophagy-deficient cells and organs46) (M. Komatsu and our unpublished observation). However, it remains to be determined how much this pathway contributes to the total degradation of ubiquitinated proteins under physiological conditions.
In contrast to the ubiquitin-proteasome system, autophagy can degrade not only proteins but also intracellular organelles. The importance of peroxisome degradation by autophagy was recently shown using liver-specific Atg7−/− mice.47) In addition to the normal turnover of intracellular organelles, dramatic degradation of organelles is observed in the processes of lens and erythroid development. The lens contains two types of cells, the epithelial cells covering the anterior surface of the lens and the fiber cells that differentiate from the epithelial cells. During late embryogenesis, membrane-bound organelles within the epithelial cells are rapidly lost, which allows fiber cells to be transparent.48), 49) Similarly, intracellular organelles are eliminated during erythroid cell maturation. However, organelle degradation in lens and erythroid cells occurred normally in autophagy-deficient Atg5−/− mice.50) Therefore, a degradation system(s) other than autophagy appear to play major roles in organelle degradation during these processes.
Concluding remarks
I have reviewed the physiological role of autophagy in protein metabolism. Autophagy has at least two different modes: induced autophagy and basal autophagy. The former is important for nutrient regulation and the latter for intracellular protein and organelle quality control. Therefore, understanding how autophagy switches these two functions would be very important. Although the involvements of amino acids and insulin in the autophagy regulation have been considered, physiological significance of these factors is still unclear. Besides the role in protein metabolism, autophagy is thought to be involved in many processes such as killing of intracellular bacteria, survival of some viruses, antigen presentation, tumor suppression, caspase-independent cell death, development and anti-aging. Because autophagy is one of the major degradation systems, these functions are eventually linked to the degradation of intracellular components. In addition, selectivity of autophagy will be another important topic of this field. Like p62, there may be various substrates that are preferentially incorporated into autophagosomes. Further studies will provide important insights into this conserved degradation system.
Acknowledgments
This work was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Kato Memorial Bioscience Foundation and Toray Science Foundation.
Profile
Noboru Mizushima was born in 1966. He graduated from Faculty of Medicine at Tokyo Medical and Dental University in 1991, and finished the resident program in 1993 in the department of internal medicine at Tokyo Medical and Dental University. Then he started his research career with studies on molecular immunology and received Ph.D. in 1996. He then moved to National Institute for Basic Biology and started works on autophagy in yeast and mammals in Dr. Yoshinori Ohsumi’s laboratory. His discovery of the Atg12 conjugation system was the first demonstration of a set of Atg proteins being involved in yeast autophagy. He moved to Tokyo Metropolitan Institute of Medical Science in 2004 as a laboratory head and made extensive studies on the physiological role of autophagy using mouse genetics. He was promoted to Professor of Physiology and Cell Biology, in Graduate School and Faculty of Medicine at Tokyo Medical and Dental University in 2006. He was awarded the Young investigator award of the Japanese Biochemical Society in 2001, Mitsubishi Chemical Award of the Molecular Biology Society of Japan in 2005, and The Commendation for Science and Technology by the Minister of Education, Culture, Sports, Science and Technology, The Young Scientists’ Prize in 2006.

References
- 1).Cuervo, A.M. (2004) Autophagy: in sickness and in health. Trends Cell Biol. 14, 70–77 [DOI] [PubMed] [Google Scholar]
- 2).Levine, B. and Klionsky, D.J. (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 3).Klionsky, D.J. (2005) The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118, 7–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Mizushima, N. (2005) The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell Death Differ. 12 (Suppl. 2), 1535–1541 [DOI] [PubMed] [Google Scholar]
- 5).Ohsumi, Y. (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2, 211–216 [DOI] [PubMed] [Google Scholar]
- 6).Klionsky, D.J., Cregg, J.M., Dunn, W.A.Jr., Emr, S.D., Sakai, Y., Sandoval, I.V., Sibirny, A., Subramani, S., Thumm, M., Veenhuis, M.et al. (2003) A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545 [DOI] [PubMed] [Google Scholar]
- 7).Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T. and Ohsumi, Y. (2001) The preautophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Noda, T., Suzuki, K. and Ohsumi, Y. (2002) Yeast autophagosomes: de novo formation of a membrane structure. Trends Cell Biol. 12, 231–235 [DOI] [PubMed] [Google Scholar]
- 9).Kihara, A., Noda, T., Ishihara, N. and Ohsumi, Y. (2001) Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Liang, X.H., Kleeman, L.K., Jiang, H.H., Gordon, G., Goldman, J.E., Berry, G., Herman, B. and Levine, B. (1998) Protection against fatal Sindbis virus encephalitis by Beclin, a novel Bcl-2-interacting protein. J. Virol. 72, 8586–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Kihara, A., Kabeya, Y., Ohsumi, Y. and Yoshimori, T. (2001) Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Reports 2, 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Liang, X.H., Jackson, S., Seaman, M., Brown, K., Kempkes, B., Hibshoosh, H. and Levine, B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 13).Pattingre, S., Tassa, A., Qu, X., Garuti, R., Liang, X.H., Mizushima, N., Packer, M., Schneider, M.D. and Levine, B. (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939 [DOI] [PubMed] [Google Scholar]
- 14).Mizushima, N., Noda, T., Yoshimori, T., Tanaka, Y., Ishii, T., George, M.D., Klionsky, D.J., Ohsumi, M. and Ohsumi, Y. (1998) A protein conjugation system essential for autophagy. Nature 395, 395–398 [DOI] [PubMed] [Google Scholar]
- 15).Mizushima, N., Sugita, H., Yoshimori, T. and Ohsumi, Y. (1998) A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 273, 33889–33892 [DOI] [PubMed] [Google Scholar]
- 16).Mizushima, N., Yamamoto, A., Hatano, M., Kobayashi, Y., Kabeya, Y., Suzuki, K., Tokuhisa, T., Ohsumi, Y. and Yoshimori, T. (2001) Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Mizushima, N., Yoshimori, T. and Ohsumi, Y. (2003) Role of the Apg12 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 35, 553–561 [DOI] [PubMed] [Google Scholar]
- 18).Mizushima, N., Noda, T. and Ohsumi, Y. (1999) Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 18, 3888–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Kuma, A., Mizushima, N., Ishihara, N. and Ohsumi, Y. (2002) Formation of the ∼350-kDa Apg12-Apg5 · Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 277, 18619–18625 [DOI] [PubMed] [Google Scholar]
- 20).Mizushima, N., Kuma, A., Kobayashi, Y., Yamamoto, A., Matsubae, M., Takao, T., Natsume, T., Ohsumi, Y. and Yoshimori, T. (2003) Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 116, 1679–1688 [DOI] [PubMed] [Google Scholar]
- 21).Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., Mizushima, N., Tanida, I., Kominami, E., Ohsumi, M.et al. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 [DOI] [PubMed] [Google Scholar]
- 22).Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., Kominami, E., Ohsumi, Y. and Yoshimori, T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., Ohsumi, M., Takao, T., Noda, T. and Ohsumi, Y. (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Mariño, G., Uria, J.A., Puente, X.S., Quesada, V., Bordallo, J. and López-Otin, C. (2003) Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J. Biol. Chem. 278, 3671–3678 [DOI] [PubMed] [Google Scholar]
- 25).Kabeya, Y., Mizushima, N., Yamamoto, A., Oshitani-Okamoto, S., Ohsumi, Y. and Yoshimori, T. (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell. Sci. 117, 2805–2812 [DOI] [PubMed] [Google Scholar]
- 26).Tanida, I., Sou, Y.S., Ezaki, J., Minematsu-Ikeguchi, N., Ueno, T. and Kominami, E. (2004) HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 279, 36268–36276(Epub 2004 Jun 8) [DOI] [PubMed] [Google Scholar]
- 27).Mizushima, N. (2004) Methods for monitoring autophagy. Int. J. Biochem. Cell Biol. 36, 2491–2502 [DOI] [PubMed] [Google Scholar]
- 28).Mizushima, N., Yamamoto, A., Matsui, M., Yoshimori, T. and Ohsumi, Y. (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Takeshige, K., Baba, M., Tsuboi, S., Noda, T. and Ohsumi, Y. (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Munz, C. (2006) Autophagy and antigen presentation. Cell. Microbiol. 8, 891–898 [DOI] [PubMed] [Google Scholar]
- 31).Tsukada, M. and Ohsumi, Y. (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174 [DOI] [PubMed] [Google Scholar]
- 32).Otto, G.P., Wu, M.Y., Kazgan, N., Anderson, O.R. and Kessin, R.H. (2003) Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J. Biol. Chem. 278, 17636–17645 [DOI] [PubMed] [Google Scholar]
- 33).Scott, R.C., Schuldiner, O. and Neufeld, T.P. (2004) Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7, 167–178 [DOI] [PubMed] [Google Scholar]
- 34).Kuma, A., Hatano, M., Matsui, M., Yamamoto, A., Nakaya, H., Yoshimori, T., Ohsumi, Y., Tokuhisa, T. and Mizushima, N. (2004) The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 35).Lum, J.J., Bauer, D.E., Kong, M., Harris, M.H., Li, C., Lindsten, T. and Thompson, C.B. (2005) Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120, 237–248 [DOI] [PubMed] [Google Scholar]
- 36).Onodera, J. and Ohsumi, Y. (2005) Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 280, 31582–31586 [DOI] [PubMed] [Google Scholar]
- 37).Fingar, D.C. and Blenis, J. (2004) Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23, 3151–3171 [DOI] [PubMed] [Google Scholar]
- 38).Codogno, P. and Meijer, A.J. (2005) Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 12 (Suppl. 2), 1509–1518 [DOI] [PubMed] [Google Scholar]
- 39).Noda, T. and Ohsumi, Y. (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966 [DOI] [PubMed] [Google Scholar]
- 40).Hosokawa, N., Hara, Y. and Mizushima, N. (2006) Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 580, 2623–2629 [DOI] [PubMed] [Google Scholar]
- 41).Hara, T., Nakamura, K., Matsui, M., Yamamoto, A., Nakahara, Y., Suzuki-Migishima, R., Yokoyama, M., Mishima, K., Saito, I., Okano, H.et al. (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- 42).Komatsu, M., Waguri, S., Chiba, T., Murata, S., Iwata, J.I., Tanida, I., Ueno, T., Koike, M., Uchiyama, Y., Kominami, E.et al. (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 [DOI] [PubMed] [Google Scholar]
- 43).Komatsu, M., Waguri, S., Ueno, T., Iwata, J., Murata, S., Tanida, I., Ezaki, J., Mizushima, N., Ohsumi, Y., Uchiyama, Y.et al. (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Arrasate, M., Mitra, S., Schweitzer, E.S., Segal, M.R. and Finkbeiner, S. (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 45).Bjorkoy, G., Lamark, T., Brech, A., Outzen, H., Perander, M., Overvatn, A., Stenmark, H. and Johansen, T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46).Wang, Q.J., Ding, Y., Kohtz, S., Mizushima, N., Cristea, I.M., Rout, M.P., Chait, B.T., Zhong, Y., Heintz, N. and Yue, Z. (2006) Induction of autophagy in axonal dystrophy and degeneration. J. Neurosci. 26, 8057–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47).Iwata, J., Ezaki, J., Komatsu, M., Yokota, S., Ueno, T., Tanida, I., Chiba, T., Tanaka, K. and Kominami, E. (2006) Excess peroxisomes are degraded by autophagic machinery in mammals. J. Biol. Chem. 281, 4035–4041 [DOI] [PubMed] [Google Scholar]
- 48).David, L.L. and Shearer, T.R. (1989) Role of proteolysis in lenses: a review. Lens Eye Toxic. Res. 6, 725–747 [PubMed] [Google Scholar]
- 49).Bassnett, S. (2002) Lens organelle degradation. Exp. Eye Res. 74, 1–6 [DOI] [PubMed] [Google Scholar]
- 50).Matsui, M., Yamamoto, A., Kuma, A., Ohsumi, Y. and Mizushima, N. (2006) Organelle degradation during the lens and erythroid differentiation is independent of autophagy. Biochem. Biophys. Res. Commun. 339, 485–489 [DOI] [PubMed] [Google Scholar]