Abstract
Transfer RNA genes are distributed throughout eukaryotic genomes, and are frequently found as multicopy families. In Saccharomyces cerevisiae, tRNA gene transcription by RNA polymerase III suppresses nearby transcription by RNA polymerase II, partially because the tRNA genes are clustered near the nucleolus. We have tested whether active transcription of tRNA genes might also suppress recombination, since recombination between identical copies of the repetitive tRNA genes could delete intervening genes and be detrimental to survival. The opposite proved to be the case. Recombination between active tRNA genes was elevated, but only when both genes are transcribed. We also tested the effects of tRNA genes on recombination between the direct terminal repeats of a neighboring retrotransposon, since most Ty retrotransposons reside next to tRNA genes, and the selective advantage of this arrangement is not known.
INTRODUCTION
Most tRNA genes exist in multiple copies distributed throughout the genome, and appear to have duplicated in dispersed locations through RNA-mediated transposition (Hani and Feldmann, 1998; Dujon et al., 2004). Little is known about which predicted tRNA genes in multicellular eukaryotes are transcribed, but there are indications from a limited number of studies that there might be considerable developmental regulation in the transcription of tRNA gene subclasses (Koski and Clarkson, 1982; Wilson et al., 1985). In addition to tRNA genes, vertebrates have highly repetitive DNA elements, termed SINEs (short interspersed repetitive elements) that are derived from small RNA genes with tRNA-class promoters for RNA polymerase III (pol III) (Jurka, 2004). The major SINE in humans, the Alu elements, are originally derived from the 7SL RNA found in signal recognition particles, and are found in more than 500,000 copies per haploid genome (Gilbert and Labuda, 1999). Although few of these elements appear transcriptionally active under normal conditions, extensive Alu expression can be observed during viral infection or in response to cellular stress (Fornace and Mitchell, 1986; Liu et al., 1995), and most cloned Alu repeats can be transcribed by pol III in vitro (Elder et al., 1981; Liu and Schmid, 1993).
The existence of such frequent, highly similar DNA sequences in the genome raises the question of whether there is some mechanism to protect against frequent deletion of chromosome segments between repeats by homologous recombination, since such deletions could lead to the death of single cells or developmental abnormalities in complex organisms. In adults, such deletions can lead to unregulated growth, such as human mammary tumors with deletions between Alu elements at the BRCA1 loci (Rohlfs et al., 2000; Pavlicek et al., 2004; Tournier et al., 2004).
Transcription of genes by RNA polymerase II (pol II) increases their susceptibility to recombination (Smith et al., 1996), but this has not been addressed for pol III transcription units, which are very short and almost entirely covered by the active transcription complex when present. Although transfer of genetic information between identical pol III genes is possible (e.g., tRNA) (Munz et al., 1982), recombination that deletes intervening material might be predicted to be suppressed as being counter to survival. Here we use a yeast model system to address the effects of pol III transcriptional activity on recombination between and near tRNA genes in the yeast chromosome. We examined the rate of recombination between two nearly identical tRNA genes in the yeast Saccharomyces cerevisiae that were either transcriptionally active or inactive. The results were unexpected, in that recombination increased when both tRNA genes were active.
In addition to testing the effect of pol III transcription on recombination between tRNA genes, there was reason to suspect that homologous recombination within a few hundred base pairs of active tRNA genes might also be suppressed. Most of the Ty retrotransposons in yeast (Ty1–Ty4) have evolved mechanisms for preferential insertion and retention near tRNA genes (Chalker and Sandmeyer, 1992; Hull et al., 1994; Kendall et al., 2000; Bolton and Boeke, 2003). A rare fifth class, Ty5, prefers transcriptionally silent regions such as telomeres and silent mating type loci (Zou et al., 1995). Proximity to a tRNA gene might have the selective advantage of conferring conditional pol II transcriptional silencing (Hull et al., 1994), but reduced recombination between the direct, long terminal repeats (LTRs) of the Ty retrotransposons might also have provided the selective advantage for the Ty elements to have developed this insertion preference.
MATERIALS AND METHODS
Yeast strains
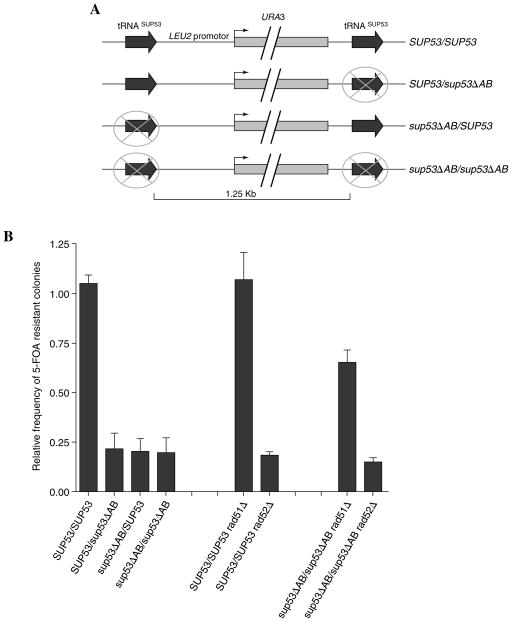
Strains were derived from Saccharomyces cerevisiae W3031A (MAT a leu2–3, 112 his3–11, 15 ade2–1 trp1–1 ura3–1 can1–100) and BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0). Growth was performed in YPD (Rose et al., 1990) except where noted. For testing recombination between identical tRNALeu genes, constructs were made in W3031A, the coding sequence of the LEU2 gene, on chromosome III, adjacent to the SUP53 tRNALeu gene was precisely replaced with the coding region of URA3. A second copy of the SUP53 gene was introduced 200 bp downstream from the 3′ end of the URA3 coding region, 1.3 kb downstream from the first tRNA gene and in the same orientation. The insertion contained SUP53 sequence from 59 bp upstream to 29 bp downstream of the tRNA coding sequence. The strains contained either a wild-type tRNASUP53 promoter at each site or a transcriptionally inactive promoter containing previously characterized point mutations in both the A box and B box (Newman et al., 1983; Hull et al., 1994). The mutated gene is designated sup53ΔAB (Fig. 1A).
FIG. 1.
(A) Recombination between tRNA genes. Four strains were modified by replacing LEU2 coding sequence with that of URA3 in strains containing either an active or transcriptionally inactive tRNA gene. An additional active or inactive tRNASUP53 gene was then placed 200 bp downstream of URA3 in each strain. Deletion events between the two tRNA genes was selected by growth on plates containing 5-FOA media and deletions were confirmed by PCR. (B) The first four bars are average values of five independent experiments. The following four values are an average from three experiments. The number of viable colonies on the 5-FOA plates was divided by the number of cells plated and normalized to recombination in the SUP53/SUP53 strain. The frequency of 5-FOA resistance in the SUP53/SUP53 strain was (9.47 ± 0.71 × 10−10). RAD51 and RAD52 were deleted in the strain constructs (Fig. 1) containing either two active tRNA genes (SUP53/ SUP53) or two inactive tRNA genes (sup53ΔAB/sup53ΔAB). Deletion of RAD51 causes a significant increase in 5-FOA resistant colonies between inactive tRNA genes, but only a modest, if any, increase between active tRNA genes. Deletion of RAD52 caused about a 10-fold decrease in the amount of 5-FOA resistant colonies in the SUP53/SUP53 strain, but only a modest decrease in the sup53ΔAB/sup53ΔAB strain.
To test recombination rates near a tRNA gene, URA3 was inserted at the Ty3–1 locus on chromosome VII in strain BY4741, (YGRWTy3–1) replacing all but 100 bp of each end of the central epsilon region. A second strain was created by then mutating the neighboring tRNACys gene promoter (triple B box mutation G52A, T53A, C55G) by homologous recombination of a PCR product with integration selected by G418 resistance and confirmed by sequencing genomic PCR fragments from the strains. The Kan gene was inserted 152 bp downstream of the tRNA gene in the opposite orientation in the strain with and without mutations to the neighboring tRNACys.
PCR fragments for deletions of RAD51 and RAD52 were produced by replacement of their coding regions with the KAN cassette from the plasmid pFA6a–KanMX6 (Longtine et al., 1998).
Recombination assay
For each experiment three cultures were inoculated from individual colonies and were grown in SDC-Ura (Rose et al., 1990) medium to ensure retention of the URA3 genes. These cultures were used to inoculate 200 ml of YPD cultures to a starting OD600 of 0.2 and grown for 7 h to allow loss of URA3. Cells were collected and resuspended in 10 ml of 10 mM Tris-HCl pH 7.5, 1 mM EDTA. Cells are plated on SDC + 5-fluorootic acid (5-FOA) medium, to select against URA3. The number of 5-FOA-resistant colonies is expressed relative to the number of cells plated.
To test whether 5-FOA resistance arose from homologous recombination to delete URA3, 24 to 32 5-FOA-resistant colonies were tested in two or more separate experiments by PCR of genomic DNA from flanking primers.
RESULTS
Recombination between identical tRNA genes
Since many tRNA genes in yeast are duplicated up to 15 times throughout the genome, suppression of recombination between duplicates could have survival value. The question addressed here is whether transcription of tRNA genes by RNA polymerase III affects the recombination rate between identical tRNA genes. We modified the LEU2 locus on chromosome III as shown in Figure 1A. The tRNASUP53 gene is normally located upstream of the LEU2 gene, and its transcription has been extensively characterized (Newman et al., 1983; Huibregtse and Engelke, 1989). The coding sequence of LEU2 was precisely replaced with that of URA3, along with inserting a second copy of the tRNASUP53 gene (SUP53) 200 base pairs downstream from URA3. Strains were produced containing either active tRNA genes or tRNA genes inactivated by point mutations in the internal promoters (G19C, C56G; sup53ΔAB) that prevent formation of any part of the RNA polymerase III complex (Newman et al., 1983; Huibregtse et al., 1989). The four tested constructs had either both tRNA genes active (SUP53/SUP53), both inactive (sup53ΔAB/sup53ΔAB), or only one at a time active.
Loss of URA3 function was selected on media containing the 5-FOA, and the normalized rate of number of 5-FOA-resistant colonies is shown in Figure 1B. These data are averages from five different experiments, each done in duplicate.
To determine if 5-FOA resistance arose through deletion between the tRNA genes, PCR was done on genomic DNA from independent isolates using primers flanking the tRNA genes. The starting strains produce a 1.8-kb product spanning both tRNA genes and the URA3 gene, whereas precise homologous recombination between the tRNA repeats produces a 175-bp product. Sixteen 5-FOA resistant colonies from two separate experiments on each strain all showed a 175-bp product, consistent with a precise deletion between sequences in the tRNA genes.
In contrast to our original hypothesis that recombination would be repressed between transcribed tRNA genes, the highest homologous recombination rate was found to be the one where both tRNASUP53 genes were active. This strain had a recombination rate averaging five times greater than the other three strains in four separate experiments. It is particularly interesting that inactivating either tRNA gene gave the same recombination as inactivating both. The failure of only a single active tRNA transcription unit to stimulate recombination implies that the increase in recombination between active tRNA genes is the result of some direct or indirect communication between components of active pol III complexes on both genes.
Recombination path
Single-strand annealing (SSA) is one of the common mechanisms leading to deletions between direct repeats. When a break occurs between the two repeats the 5′ ends are resected. This allows the 3′ strands to anneal within the repeated elements, followed by removal of the nonhomologous 3′ tails corresponding to the sequence between the repeats, gap filling, and ligation (Liu and Schmid, 1993; Prado et al., 2003; Tournier et al., 2004;). Deletions caused by recombination between direct repeats of untranscribed regions have been shown to be reduced 10- to 100-fold in rad52 mutants consistent with the role of Rad52p in SSA (Jackson and Fink, 1981; Prado and Aguilera, 1995). In contrast, the rad52 mutation did not affect deletion rates of direct repeats in rDNA and high-copy CUP1 tandem arrays (Ozenberger and Roeder, 1991). Another participant in homologous recombination, Rad51p, acts by promoting pairing and strand exchange with an intact homologous duplex. Rad51p is required for most recombination but dispensable for SSA (Ozenberger and Roeder, 1991). To examine whether recombination between the tRNA genes was proceeding through a SSA path, RAD52 and RAD51 were individually deleted from our strains with either two active or two inactive tRNA genes flanking the URA3 gene.
The rad52Δ strains consistently showed reduced 5-FOA resistant colonies. In the SUP53/SUP53 strain there was a 20-fold decrease, and in the sup53ΔAB/sup53ΔAB strain there was a 33-fold decrease in homologous recombination (Table 1). This sensitivity to rad52 deletion distinguishes recombination between these direct repeats transcribed by pol III from the rDNA and CUP1 repeats transcribed by RNA polymerases I and II. In contrast, rad51Δ caused little change in the number of 5-FOA resistant colonies in the SUP53/SUP53 strain, and caused a slight increase in 5-FOA resistance in sup53ΔAB/sup53ΔAB strains (Fig. 1B). This is consistent with previous demonstrations that recombination between direct repeats can occur in the absence of strand exchange (McDonald and Rothstein, 1994; Rattray and Symington, 1995; Ivanov et al., 1996). These results suggest that recombination between tRNA genes is occurring in the absence of strand exchange regardless of their transcriptional activity, with Rad52p facilitating SSA.
Table 1.
Rates of Homologous Recombination
| Strain | Rate of 5-FOA Resistance (× 10−10) | % Undergone homologous recombination |
|---|---|---|
| SUP53/SUP53 | 9.47 ± 0.71 | 100 |
| sup53ΔAB/sup53ΔAB | 1.76 ± 0.75 | 100 |
| SUP53/SUP53 rad52Δ | 1.23 ± 0.28 | 39 |
| sup53ΔAB/sup53ΔAB rad52Δ | 1.04 ± 0.19 | 5 |
| SUP53/SUP53 rad51Δ | 10.17 ± 2.61 | NDa |
| sup53ΔAB/sup53ΔAB rad51Δ | 5.68 ± 1.23 | ND |
ND = not determined.
To test whether the residual 5-FOA resistant colonies arose through recombination between the tRNA genes or some other defect, we again tested genomic numerous 5-FOA isolates by PCR. This analysis showed that only 39% of the SUP53/SUP53 and 5% of the sup53ΔAB/sup53ΔAB 5FOA-resistant colonies underwent deletion by homologous recombination in the Δrad52 strain, compared to 100% in RAD52 strains. In the SUP53/SUP53–Δrad52 strain 22% of the 5FOA resistant colonies came through a probable URA3 mutation, since deletions were not detected, and 39% appear to have a large chromosomal deletion in the region. In the sup53ΔAB/sup53ΔAB-Δrad52 strain most (95%) of the 5-FOA-resistant colonies represented a probable small mutation in URA3 (Table 1).
Recombination between retrotransposon LTR elements
We also tested the effect of a nearby tRNA gene on the recombination between LTR elements of a well-characterized yeast retrotransposon locus Ty3–1, in its native chromosomal position on chromosome VII (Bilanchone et al., 1993). We replaced the interior epsilon region of the retrotransposon Ty3–1 with URA3. We then either left the neighboring tRNACys gene intact or mutated three base pairs in the B box internal promoter (Fig. 2), which dramatically decreases pol III transcription (Allison et al., 1983; Kinsey and Sandmeyer, 1991; Aguilera, 2002).
FIG. 2.
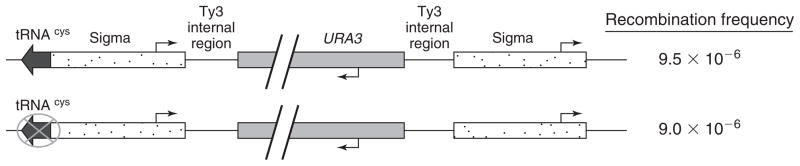
Recombination between Ty3 sigma direct repeats. The URA3 gene was inserted at the Ty3–1 locus, replacing all but 100 bp of either end of the epsilon region. Multiple B box promoter mutations were then created in the neighboring tRNACys gene in a second strain, inactivating the transcription promoter. Recombination between the two sigma elements was identified by section against the URA3 gene through growth on 5-FOA media and screening for precise recombination between sigmas by PCR from genomic DNA. Recombination frequency is shown as number of recombinants divided by the total number of cells plated.
The strains with active versus inactive tRNA genes were tested for their relative frequency of URA3 loss. The averages from four separate experiments demonstrated that the frequencies for the active tRNA gene and inactive gene were essentially identical (9.5 × 10−6 and 9.0 × 10−6, respectively) within error for these experiments. Recombination between the LTR direct repeats therefore appears to be unaffected by the presence of a nearby active tRNA gene.
DISCUSSION
Contrary to our original hypothesis, the work presented here showed that the occurrences of recombination between two identical tRNA genes is higher when both tRNA genes are being actively transcribed. In this respect, pol III transcription appears to behave similarly to pol II transcription in that transcription stimulates recombination (Aguilera, 2002). However, there are also distinct differences. It is unclear why both tRNA genes needed to be actively transcribed in order for the increase in homologous recombination to occur. One possibility is having two active tRNA genes causes an increase in breakage, consistent with tRNA genes causing replication fork pause (Deshpande and Newlon, 1996), which have been shown to cause increased homologous recombination (Bilanchone et al., 1993; Aguilera, 2002). If this is the case, it is not clear why a single active tRNA gene might not give an intermediate level of recombination increase. A second possibility is that having two active tRNA genes changes the type of repair that is used. If this is the case, perhaps the spatial organization of these tRNA genes is playing a part in their repair. However, the rad52Δ data suggest that the path of this increased recombination is still through SSA.
Recent findings showing spatial clustering of the linearly dispersed tRNA genes suggest a possible explanation for our results. Yeast tRNA genes are largely localized to the nucleolus when actively transcribed, but not when inactivated by promoter point mutations (Thompson et al., 2003). The mechanism of this colocalization is not currently known, but it is conceivable that tRNA transcription complexes associate with some sort of framework that brings them into proximity, increasing the likelihood of physical interactions. This would be consistent with evidence that the frequency of disease-specific chromosomal translocations are nonrandom, and have been correlated to the spatial proximity of the sites involved (Lukasova et al., 1997; Neves et al., 1999; Roix et al., 2003).
The increased recombination between identical tRNA genes when they are transcriptionally active could help explain why families of tRNA genes in yeast are not found tandemly repeated, or even in close proximity. The closest pair of identical tRNA genes occurs on chromosome IX, where two tRNA genes Asp are about 12 kb apart from each other. Studies using plasmid constructs and non-tRNA repeats have shown that increasing the distance between repeats decreases the efficiency of SSA in competition with gene conversion (Fishman-Lobell et al., 1992; Haber and Leung, 1996; Puget et al., 1999).
The results presented here might be pertinent in considering the occurrence of deletions, inversions, and duplications at Alu elements throughout the human genome. Alu elements contain tRNA-class internal promoters from the 7SL RNA genes (Gilbert and Labuda, 1999). While this pol III promoter in cloned Alu repeats is generally found to be transcriptionally competent in vitro, most of the hundreds of thousands of Alu repeats in human cells are inactive in cells, and little is known as to which of the elements are activated in response to cellular insults (Fornace and Mitchell, 1986; Liu et al., 1995; Gilbert and Labuda, 1999). An example of recombination between Alu repeats has been studied is the human tumor suppressor gene, BRCA1. BRCA1 genomic sequence is composed of 41.5% Alu sequence, corresponding to an Alu element every 650-bp average (Smith et al., 1996). Alu recombination seems to be the main source of genomic rearrangements in patients with a hereditary predisposition to breast and ovarian cancers (Puget et al., 1999; Rohlfs et al., 2000; Pavlicek et al., 2004). The data in this report suggests that the transcriptional activation of these Alu repeats, possibly through stress response or viral infection, might increase their ability to recombine.
Acknowledgments
This work was supported by the National Institutes of Health (GM063142 to D.R.E. and CA102563 to T.E.W.). M.J.P.-H. is a predoctoral fellow supported by the Cellular and Molecular Biology Training Grant at the University of Michigan (T32 GM007315).
References
- AGUILERA A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALLISON DS, GOH SH, HALL BD. The promoter sequence of a yeast tRNA tyr gene. Cell. 1983;34:655–664. doi: 10.1016/0092-8674(83)90398-7. [DOI] [PubMed] [Google Scholar]
- BILANCHONE VW, CLAYPOOL JA, KINSEY PT, SANDMEYER SB. Positive and negative regulatory elements control expression of the yeast retrotransposon Ty3. Genetics. 1993;136:685–700. doi: 10.1093/genetics/134.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOLTON EC, BOEKE JD. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: A genomic point of view. Genome Res. 2003;13:254–263. doi: 10.1101/gr.612203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHALKER DL, SANDMEYER SB. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992;6:117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- DESHPANDE AM, NEWLON CS. DNA replication fork pause sites dependent on transcription. Science. 1996;272:1030–1033. doi: 10.1126/science.272.5264.1030. [DOI] [PubMed] [Google Scholar]
- DUJON B, SHERMAN D, FISCHER G, DURRENS P, CASAREGOLA S, LAFONTAINE I, DE MONTIGNY J, MARCK C, NEUVEGLISE C, TALLA E, et al. Genome evolution in yeast. Nature. 2004;430:35–44. [Google Scholar]
- ELDER JT, PAN J, DUNCAN CH, WEISSMAN SM. Transcriptional analysis of interspersed repetitive polymerase III transcription units in human DNA. Nucleic Acids Res. 1981;9:1171–1189. doi: 10.1093/nar/9.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FISHMAN-LOBELL J, RUDIN N, HABER JE. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992;12:1291–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FORNACE AJ, JR, MITCHELL JB. Induction of B2 RNA polymerase III transcription by heat shock: Enrichment for heat shock induced sequences in rodent cells by hybridization subtraction. Nucleic Acids Res. 1986;14:5793–5811. doi: 10.1093/nar/14.14.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GILBERT N, LABUDA D. Evolutionary inventions and continuity of CORE-SINEs in mammals. J Mol Biol. 2000;5:365–377. doi: 10.1006/jmbi.2000.3695. [DOI] [PubMed] [Google Scholar]
- HABER JE, LEUNG WY. Lack of chromosome territoriality in yeast: Promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci USA. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANI J, FELDMANN H. tRNA genes and retroelements in the yeast genome. Nucleic Acids Res. 1998;26:689–696. doi: 10.1093/nar/26.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUIBREGTSE JM, ENGELKE DR. Genomic footprinting of a yeast tRNA gene reveals stable complexes over the 5′-flanking region. Mol Cell Biol. 1989;9:3244–3252. doi: 10.1128/mcb.9.8.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HULL M, ERICKSON J, JOHNSTON M, ENGELKE DR. tRNA genes as transcriptional repressor elements. Mol Cell Biol. 1994;14:1266–1277. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IVANOV EL, SUGAWARA N, FISHMAN-LOBELL J, HABER JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACKSON JA, FINK GR. Gene conversion between duplicated genetic elements in yeast. Nature. 1981;292:306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- JURKA J. Evolutionary impact of human Alu repetitive elements. Curr Opin Genet Dev. 2004;14:603–608. doi: 10.1016/j.gde.2004.08.008. [DOI] [PubMed] [Google Scholar]
- KENDALL A, HULL MW, BERTRAND E, SINGER PD, ENGELKE DR. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc Natl Acad Sci USA. 2000;97:13108–13113. doi: 10.1073/pnas.240454997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KINSEY PT, SANDMEYER SB. Adjacent pol II and pol III promoters: Transcription of the yeast retrotransposon Ty3 and a target tRNA gene. Nucleic Acids Res. 1991;19:1317–1324. doi: 10.1093/nar/19.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOSKI RA, CLARKSON SG. Synthesis and maturation of Xenopus laevis methionine tRNA transcripts in homologous cell-free extracts. J Biol Chem. 1982;257:4514–4521. [PubMed] [Google Scholar]
- LIU WM, SCHMID CW. Proposed roles for DNA methylation in Alu transcriptional repression and mutational inactivation. Nucleic Acids Res. 1993;21:1351–1359. doi: 10.1093/nar/21.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU WM, CHU WM, CHOUDARY PV, SCHMID CW. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LONGTINE MS, MCKENZIE A, III, DEMARINI DJ, SHAH NG, WACH A, BRACHAT P, PHILIPPSEN A, PRINGLE JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- LUKASOVA E, KOZUBEK S, KOZUBEK M, KJERONSAK J, RYZNAR L, HORAKOVA J, KRAHULCOVA E, HORNECK G. Localization and distance between ABL and BCR genes in interphase nuclei of bone marrow cells of control donors and patients with chronic myeloid leukemia. Hum Genet. 1997;100:525–535. doi: 10.1007/s004390050547. [DOI] [PubMed] [Google Scholar]
- MCDONALD JP, ROTHSTEIN R. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1, RAD52-independent recombination. Genetics. 1994;137:393–405. doi: 10.1093/genetics/137.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUNZ P, AMSTUTZ H, KOHLI J, LEUPOLD U. Recombination between dispersed serine tRNA genes in Schizosac-charomyces pombe. Nature. 1982;300:225–231. doi: 10.1038/300225a0. [DOI] [PubMed] [Google Scholar]
- NEVES H, RAMOS C, DA SIVA MG, PARREIRA A, PARREIRA L. The nuclear topography of AMB, BCR, PML, and RAR genes: Evidence for gene proximity in specific phases of the cell cycle and stages of hematopoietic differentiation. Blood. 1999;93:1197–1207. [PubMed] [Google Scholar]
- NEWMAN AJ, OGDEN RC, ABELSON J. tRNA gene transcription in yeast: effects of specified base substitutions in the intragenic promoter. Cell. 1983;35:117–125. doi: 10.1016/0092-8674(83)90214-3. [DOI] [PubMed] [Google Scholar]
- OZENBERGER BA, ROEDER GS. A unique pathway of double-strand break repair operates in tandemly repeated genes, Mol. Cell Biol. 1991;11:1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAVLICEK A, NOSKOV VN, KOUPRINA N, BARRETT JC, JURKA J, LARIONOV V. Evolution of the tumor suppressor BRCA1 locus in primates: implication for cancer predisposition. Hum Mol Genet. 2004;13:2727–2751. doi: 10.1093/hmg/ddh301. [DOI] [PubMed] [Google Scholar]
- PRADO F, AGUILERA A. Role of reciprocal exchange, one ended invasion crossover and single-strand annealing on inverted and direct repeat recombination in yeast: Different requirements for the RAD1, RAD10. Genetics. 1995;139:109–123. doi: 10.1093/genetics/139.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRADO F, CORTES-LEDESMA F, HUERTAS P, AGUILERA A. Mitotic recombination in Saccharomyces cerevisiae. Curr Genet. 2003;42:185–198. doi: 10.1007/s00294-002-0346-3. [DOI] [PubMed] [Google Scholar]
- PUGET N, STOPPA-LYONNET D, SINILNKOVA OM, PAGE S, LYNCH HT, LENOIR S, MAZOYER GM. Screening for germ-line rearrangements and regulatory mutations in BRCA1 led to the identification of four new deletions. Cancer Res. 1999;59:455–461. [PubMed] [Google Scholar]
- RATTRAY AJ, SYMINGTON LS. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROHLFS EM, PUGET N, GRAHAM ML, WEBER BL, GARBER JE, SKRZYNIA C, HALPERIN JL, LENOIR GM, SILVERMAN LM, MAZOYER S. An Alu-mediated 7.1 kb deletion of BRCA 1 exon 8 and 9 in breast and overian cancer families that results in alternative splicing of exon 10. Genes Chromosomes Cancer. 2000;28:300–307. doi: 10.1002/1098-2264(200007)28:3<300::aid-gcc8>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- ROIX JJ, MCQUEEN PG, MUNSON PJ, PARADA LA, MISTELI T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- ROSE MD, WINSTON F, HIETER P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1990. [Google Scholar]
- SMITH TM, LEE MK, SZABO CI, JEROME N, McEUEN M, TAYLOR M, HOOD L, KING MC. Complete genomic sequence and analysis of 117 kb of human DNA containing the gene BRCA1. Genome Res. 1996;6:1029–1049. doi: 10.1101/gr.6.11.1029. [DOI] [PubMed] [Google Scholar]
- THOMPSON M, HAEUSLER RA, GOOD PD, ENGELKE DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOURNIER I, PAILLERETS BB, SOBOL H, STOPPA-LYON-NET D, LIDEREAU R, BARROIS S, MAZOYER M, COULET F, HARDOUIN A, CHOMPRET A, et al. Significant contribution of germline BRCA2 rearrangements in male breast cancer families. Cancer Res. 2004;64:8143–8147. doi: 10.1158/0008-5472.CAN-04-2467. [DOI] [PubMed] [Google Scholar]
- WILSON ET, LARSON D, YOUNG LS, SPRAGUE KU. A large region controls tRNA transcription. J Mol Biol. 1985;183:153–163. doi: 10.1016/0022-2836(85)90209-8. [DOI] [PubMed] [Google Scholar]
- ZOU S, WRIGHT DA, VOYTAS DF. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 1995;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]