SUMMARY
Hepatitis B Virus (HBV) is a major cause of liver disease. Assembly of the HBV capsid is a critical step in virus production and an attractive target for new antiviral therapies. We determined the structure of HBV capsid in complex with AT-130, a member of the phenylpropenamide family of assembly effectors. AT-130 causes tertiary and quaternary structural changes, but does not disrupt capsid structure. AT-130 binds a hydrophobic pocket that also accommodates the previously characterized HAP compounds, but favors a unique quasi-equivalent location on the capsid surface. Thus, this pocket is a promiscuous drug binding site and a likely target for different assembly effectors with a broad range of mechanisms of activity. That AT-130 successfully decreases virus production by increasing capsid assembly rate without disrupting capsid structure delineates a new paradigm in antiviral design, that disrupting reaction timing is a viable strategy for assembly effectors of HBV and other viruses.
INTRODUCTION
Current antiviral therapies focus on outright inhibition of viral processes. For example, entry inhibitors have been developed to combat human immunodeficiency virus (HIV) by blocking viral entry into the target cell, by inhibiting either host-virus interactions or membrane fusion (Eggink, 2010). Protease inhibitors used against HIV block the cleavage steps necessary for viral maturation (Wensing, 2010). A wide variety of nucleoside analogues bind viral polymerases in herpes simplex virus, hepatitis C virus, HIV and hepatitis B virus (HBV) to block synthesis of the viral genomes (Menédez-Arias, 2008; Snoeck, 2000; Toniutto, 2006; Yuen, 2011). Release inhibitors have been developed against influenza that block the necessary host receptor cleavage (Jackson, 2011). Due to the mutation-prone nature of virus replication, resistance is always a serious problem for any antiviral therapy, and new drugs with new modes of action are continuously sought (Razonable, 2011).
One under-exploited target for antivirals is assembly of the virus capsid, the protein coat that encloses the viral genome and any other components necessary to virus structure or function. The capsid does play a protective role, sequestering the genomic material from outside threats, but it is not a static shell (Bothner et al., 1998; Gertsman et al., 2010; Hilmer et al., 2007; Johnson, 2003; Lewis et al., 1998; Monroe et al., 2010; Phelps and Post, 1995; Tuma et al., 2008). Rather, it is a dynamic structure comprised of flexible subunits and is capable of a wide range of movement and involvement in many cellular processes. In the case of HBV, upon viral entry the capsid must expose nuclear localization signals in order to interact with host trafficking machinery for transport to the nucleus (Yeh et al., 1990). Upon reaching the nucleus, the capsid must release its contents to initiate replication (Rabe et al., 2003). Newly-synthesized capsid subunits must assemble with high fidelity at the correct time and place and package the correct viral pregenomic RNA and associated proteins with high specificity (Porterfield, 2010). Initiation of DNA synthesis from the packaged RNA is dependent on capsid formation and core phosphorylation state (Lan et al., 1999; Lott et al., 2000). The DNA-containing virions must then be shuttled through the cell via host export pathways to the ER for secretion or to the nucleus (Lambert, 2007). Once released from the cell, the capsid must remain stable in the extracellular milieu, until it enters a new host cell, wherein it must release its genetic material in order to restart the HBV lifecycle (Ganem and Schneider, 2001).
The ability of the virion to participate in these dynamic processes is due in large part to the properties of the capsid protein. One hundred and twenty copies of the homodimeric capsid protein (Cp) form a T=4 icosahedron, the most populous species found in HBV virions, in which each monomer must adopt one of four slightly different conformations corresponding to the quasi-equivalent environments dictated by the symmetry of the icosahedral complex (Wynne et al., 1999; Zlotnick et al., 1997). Capsid assembly is initiated by the slow formation of a trimer of Cp dimers to form a nucleus, followed by the rapid stepwise addition of single dimers to form a complete capsid (Ceres P, 2002; Zlotnick et al., 1999a). Cp itself is a largely α-helical structure that consists of a central chassis domain and three distal sub-domains—the spike tip, the contact domain, and the fulcrum helix—linked to the chassis by glycine and proline hinge residues that allow each sub-domain to move independently (Packianathan et al., 2010). Assembly can be triggered by an allosteric conformational switch of Cp from an ‘open’ inactive state into the capsid-like assembly-active state (Chen et al., 2011; Packianathan, 2010). Recent studies have shown a wide range of conformations available to the capsid dimer and confirmed that the assembly-active state is indeed more compact (DiMattia MA, 2013; Tan et al., 2013). After assembly, the protein must still be flexible enough to allow exposure of peptide sequences that carry signals for intracellular trafficking (Bothner et al., 1998; Yeh et al., 1990; Zlotnick et al., 1997). Cores may then be exported from a cell, gaining an envelope, or transported to the nucleus, a process in common with a new infection. Nuclear entry requires the virus capsid to undergo further conformational change, destabilization, and/or disassembly to release its genome (Rabe et al., 2003). All of these processes are dynamic and must be tightly regulated.
Contrary to the examples of viral enzyme inhibitors described above, studies in HBV assembly have shown that the opposite effect, activation of virus assembly, is a very effective antiviral measure (Zlotnick and Stray, 2003). The heteroaryldihydropyrimidine (HAP) compounds, originally identified by Bayer, were found to be non-nucleoside assembly activators that increase the pairwise contacts between the capsid subunits, driving assembly further and faster to the point of misdirecting assembly into large, pleiomorphic non-capsid structures (Stray et al., 2005; Stray and Zlotnick, 2006; Weber et al., 2002). In cell culture and animal models, HAP treatment leads to the clearance of the capsid protein from the cell and does not produce intact virus (Deres et al., 2003; Weber et al., 2002). Determination of the 5 Å resolution crystal structure of the HBV capsid in complex with a prototypical member of the HAP family, HAP1, resulted in a cautious assignment of the HAP1 binding site to a hydrophobic pocket at the dimer-dimer interface (Bourne et al., 2006). HAP1 binding did not alter the protein shape or block its ability to self-assemble; in fact, it causes a stronger inter-dimer contact energy. However, HAP binding also changed the geometry of the contacts between capsid protein dimers at the quaternary level, resulting in an overall disruption of the spherical capsid structure and (at high stoichiometry of HAP to capsid protein) resulting in misassembly (Bourne, 2008). The low-resolution assignment of the HAP1 site has been tested with a HAP-resistant capsid protein mutant. The insertion of a tryptophan residue that partially fills the binding pocket confers HAP-like activities upon the resulting protein: assembly was faster, subunit-subunit interactions were stronger, the mutation inhibited viral DNA synthesis when expressed in culture, and mutant Cp had a dominant negative phenotype when co-expressed with wild-type HBV Cp (Tan et al., 2013).
Another class of non-nucleoside compounds, the phenylpropenamides, also had anti-HBV activity in cell culture (Delaney et al., 2002; King et al., 1998; Perni et al., 2000). Studies with these compounds suggested that the phenylpropenamides appeared to block RNA packaging, producing apparently normal capsids that lacked genetic material (Feld, 2007). Subsequent studies revealed an assembly effector mechanism underlying the apparent blocking of RNA-packaging (Katen, 2010). The effects of the phenylpropenamides are almost entirely kinetic, affecting only assembly reaction rate and timing while still producing normal capsids, with very little thermodynamic effects on capsid stability. The loss of RNA packaging is not the direct result of these compounds, but presumably a secondary consequence of initiating assembly without viral RNA (Katen, 2010).
Given that the effectiveness of the HAP compounds correlates directly with their effects on assembly rate, rather than their thermodynamic aspects (Bourne, 2008), and the fact that despite different chemistries, the HAPs and phenylpropenamides have similar size and hydrophobicity, we proposed that phenylpropenamides might act on HBV assembly in the same way and in the same site as the HAPs with the observed differences in behavior arising from the weaker effect on capsid stability of the phenylpropenamides (Katen, 2010). To explore this hypothesis, we have now solved a 4.2 Å structure of the HBV Cp assembly domain in complex with AT-130 (Protein Data Bank Code 4G93). AT-130 is the phenylpropenamide derivative with the greatest reported efficacy in cell culture, (EC50 = 0.13μM) (Perni et al., 2000). We discovered that while AT-130 does bind the same hydrophobic pocket as HAP, the pockets of the different quasi-equivalent subunits within the capsid asymmetric unit are not identical, and that AT-130 favors a different quasi-equivalent location than the one favored by HAP1. While there are some quaternary changes that were similar to those observed with HAP1, the phenylpropenamides also induced compensatory tertiary structural changes (not observed with the HAPs) that likely allow the formation of the observed normal capsids, rather than the noncapsid polymers observed with HAPs (Tan et al., 2013). This structure identifies a promiscuous binding pocket as a highly attractive target for multiple antivirals and reveals that relatively modest effects on the molecular level can still correlate with significant antiviral effects.
RESULTS
Co-crystallization with AT-130 altered crystal packing and capsid morphology
The crystallization properties, crystal packing and unit cell dimensions of HBV capsids crystallized in the presence of AT-130 (hereafter the +AT-130 structure) were markedly different from those of the analogous apo (lacking drug) structure (Protein Data Bank Code 2G33). The +AT-130 unit cell dimensions were similar to those of the structure of the HBV capsid co-crystallized with HAP1 (Protein Data Bank Code 2G34) (Table 1, Table S1, Figure S1) (Bourne et al., 2006).
Table 1.
Data Collection and Refinement Statistics
| HBV 3CA Cp150 + AT-130 | |
|---|---|
| Parameter | Valuea |
| X-ray source | APS 14-BMC |
| Space group | C2 |
| Unit cell constants | |
| a = 527.4 Å | |
| b = 362.8 Å | |
| c = 538.2 Å | |
| α = γ = 90°, β = 105.1° | |
| Resolution (Å) | 40.0-4.20 (4.27-4.20)b |
| R sym | 0.123 (0.882) |
| Average I /σ | 10.2 (1.1) |
| Completeness | 99.1 (97.5) |
| Redundancy | 3.0 (2.9) |
| No. of Unique Reflections | 700,096 (34,333) |
| 60-fold NCS averaging | |
| R factor | 0.24 |
| Correlation coefficient | 0.941 |
| Torsion dynamics and positional refinement | |
| Number of atomsc | 4,542 |
| RMSD bond length (Å) | 0.005 |
| RMSD bond angle (°) | 1.00 |
| Ramachandran preferred, disallowed (%) | 64.32, 12.43 |
| R factord | 0.37 |
| Mean atomic temperature factor, B (Å2) | |
| Protein | 59.8 |
| AT-130 | 57.1 |
See also Table S1.
Highest resolution shell statistics in parentheses
There are visible reflections to 3.9 Å, but the large number of weak reflections results in a high R factor and a low I/σ. Imposing a 4.2 Å cutoff results in an I/σ of 1.1 in the highest resolution shell, and 60-fold NCS averaging allows these data to make an observable contribution to density.
Number of atoms in the icosahedral asymmetric unit
Due to the 60-fold NCS averaging, the R free value is correlated with and identical to the R factor, so is not reported here
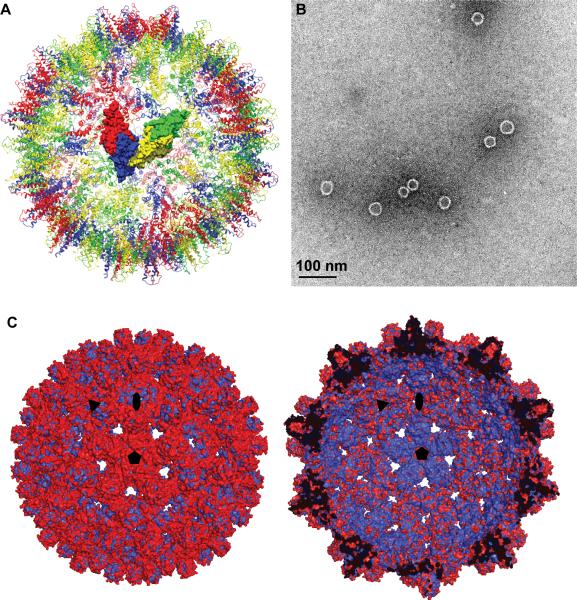
Phenylpropenamide-bound HBV capsids retained their T=4 morphology with two quasi-equivalent dimers in the asymmetric unit (Figure 1A); previous studies showed that co-assembly with AT-130 still yielded what appeared to be normal capsids (Figure 1B), so we had expected that they would be compatible with T=4 symmetry (Feld, 2007; Katen, 2010). However, global differences between the apo and drug-bound structure were readily apparent. Superposition of the density of the two structures (Figure 1C and S2) showed that the drug-bound capsid was swollen compared to the native form. The inner surface of the superimposed capsids is dominated by the native structure, while the outer surface is dominated by the greater diameter of the drug-bound structure. The diameter difference was most prominent around the five-fold axes. From the density superposition of capsid viewed at the A/B dimer, which forms the five-fold vertex, it was evident that the increase in capsid diameter resulted from an elevation of the entire dimer with respect to that of the apo capsid (Figure S2A). From spike tip to spike tip around the five-fold axis, there is an approximately 8 Å increase in capsid diameter due to the five-fold bulging. In contrast, the density superposition of the C/D dimer showed no consistent elevation (Figure S2B).
Figure 1. Capsid Structure in the Presence of AT-130.
(A) A Hepatitis B capsid structure showing quasi-equivalence and asymmetric unit. The A/B dimer (red and blue respective monomers) forms icosahedral fivefold, while the C/D dimer (yellow and green respective monomers) forms icosahedral threefold. Color scheme is maintained for remaining figures.
(B) Electron micrograph of Cp149 assembly with AT-130 showing morphologically normal 35nm capsids.
(C) +AT-130 crystal structure (red) overlaid with apo structure (blue), showing drug-induced capsid expansion. Left: Exterior of overlaid structures. Right: Cross-section of overlaid structures. Icosahedral symmetry axes are indicated. See also Figure S2.
AT-130 binding causes differential quaternary and tertiary changes in the quasi-equivalent subunits
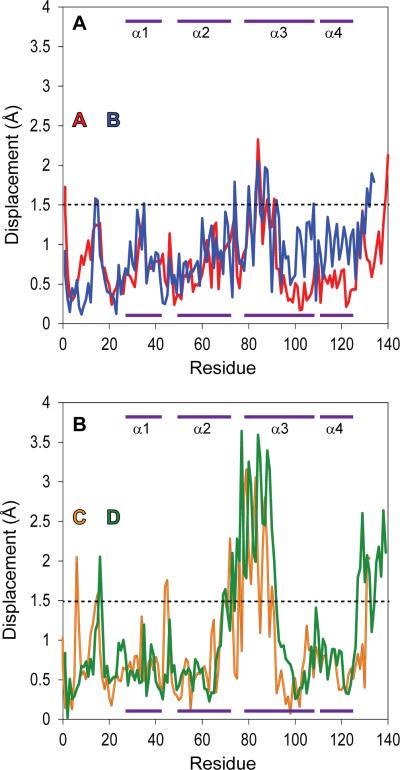
Detailed comparison of dimers extracted from the highest available resolution (3.3 Å) native (apo) capsid (Protein Data Bank Code 1QGT) (Wynne et al., 1999), with the refined +AT-130 structure was performed to identify structural changes due to AT-130 binding. The global comparisons obscured tertiary structural differences. The RMS deviation for the C-alphas of the +AT-130 noncrystallographic asymmetric unit (the A/B and C/D dimer complex, which is repeated 60 times to form a T=4 icosahedron) compared to the 1QGT non-crystallographic asymmetric unit was 0.9 Å, indicating that the overall structures of the subunits were maintained. When the two individual dimers within the non-crystallographic asymmetric unit were overlaid independently, the RMSD for the A/B dimer was only 0.8 Å. From a plot of the displacements of the C-alphas of the +AT-130 A/B subunits compared to those of the apo structure, it was apparent that there was no region of distinct difference (Figure 2A). In contrast, while the overall RMSD for the C/D dimers was slightly elevated at 1.0 Å, a minimal overall difference, a plot of C-alpha displacements (Figure 2B) showed a significant number of individual atoms had shifted by more than 2.0 Å indicating regions where there were systematic changes in the capsid protein structure.
Figure 2. C-alpha Displacement of Overlaid Dimeric Subunits.
(A) Displacement of the C-alphas in the A/B dimer from residues 1-139, showing regions of tertiary structural change. Dimers from the +AT-130 structure were overlaid on the equivalent dimers from 1QGT (Wynne et al., 1999) to minimize the RMSD of the dimer. Purple bars at the top and bottom of the panel indicate helical regions (as labeled in Figure S3).
(B) Displacement of the C-alphas in the C/D dimer, overlaid and displayed as described above. See also Figure S3.
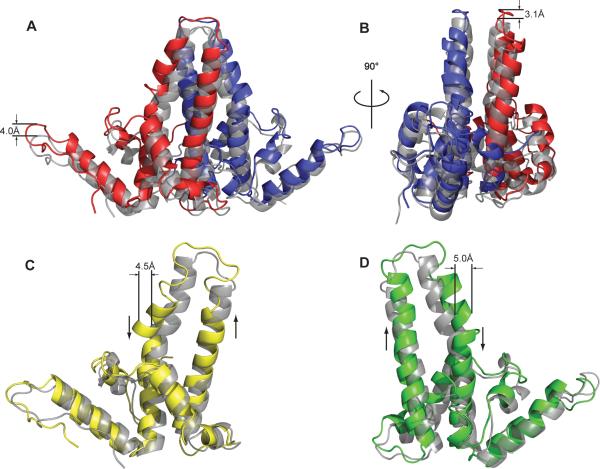
In the context of an HBV capsid, there are multiple levels of quaternary structure: an A/B dimer, a C/D dimer, the two-dimer icosahedral asymmetric unit, and the complete capsid. The change in A/B structure is almost entirely due to changes in capsid quaternary structure. Examining the overlay of the A/B dimers from the apo and +AT-130 structures in the context of the capsid, it is clear that though the tertiary structures of the individual monomers were essentially unchanged (Figure 2A), it is readily apparent that they are spatially out of register (Figure 3B). AT-130 binding induces an upward motion of the A/B dimer as a rigid body (Figures 3A and S2A). The entire dimer pivots as a rigid body, with residue Y132 of the B-chain, which is deeply buried in the inter-dimer interface, as the apparent fulcrum (Figure S3B). The upward displacement is minimal at the extreme of the B-chain, because this was near the center of the pivot, but the entire subunit is elevated from this point, resulting in a full 4.0 Å upward displacement at the opposite end of the dimer. This is also readily apparent in the upward shift of the A-monomer spike tip and assembly domain (Movie S1). This pivoting motion of the A/B dimer is the basis of the bulging at the five-fold axes seen in the completed capsid.
Figure 3. Tertiary and Quaternary Structural Changes in the HBV Capsid upon AT-130 Binding.
(A) An A/B dimer superposition showing 4.0 Å upward movement as a rigid body. All structures in the figure are overlaid in the context of the complete +AT-130 and 1QGT capsids; the apo structure is shown in gray, +AT-130 in color.
(B) A 90° rotation of the A/B dimer more clearly shows the out-of-register alignment of the spike helices.
(C) C-monomer superposition showing unraveling and displacement of the spike. Ascending and descending helices marked with arrows.
(D) D-monomer superposition showing skewing of the ascending helix and kink in the descending helix. Ascending and descending helices marked as in (C). See also Movies S1 and S2
In contrast to the radial shifting observed with HAP1 (Bourne et al., 2006), the +AT-130 structure shows no apparent pivoting or sideways motion of the C/D dimer. The overlaid densities have no vertical mismatch, and the helices within the structure are largely in-register (Figure 3C-D). However, the density overlay reveals divergence between the two structures in the upper region of the spike (Figure S2B). The plot of displacement of subunit C-alphas compared to the native structure shows a sharp increase in displacement of the atoms of the spike tip (Figure 2C and 3C, D), particularly in the upper region of the descending spike helix. The upper portion of the spike is delineated by two conserved glycines located in the middle of the ascending and descending helices at positions 63 and 94, respectively (Figure S3B). These glycines, atypical residues for an α-helix, have been proposed to serve as a hinge between the upper portion of the spike and the central chassis region of the protein, facilitating movement of the spike as an independent sub-domain within the capsid protein (Packianathan, 2010).
Examination of the +AT-130 structure reveals a significant tertiary change in the upper spike regions of the C/D dimer as delineated by these two glycines (Movie S2). In the C-monomer, the lower portion of ascending helix is very closely aligned with the native structure (Figure 3C). However, the region of the ascending helices above glycine 63 “drifts” from the native structure, tilting towards the D subunit. As the chain progresses upward, it loses its α-helical character (i.e. hydrogen bonding and ϕ–ψ angles) a full turn before that of the native structure, and does not return to an α-helix for a full two turns below the equivalent helix in the apo structure. It appears that the entire spike tip has unraveled and splayed outward. Approaching the second glycine hinge in the descending helix, there is a visible kink in the helix that displaces the C-alpha backbone by 4.5 Å. Beyond glycine 94, the structures are again in close alignment. While the overall RMSD for the C-monomer is only 0.9 Å, the RMSD for the upper portion of the spike (residues 69 to 93) is 1.7 Å.
In the D-monomer, the entire ascending helix appears to be skewed. The base of the helix is slightly tilted to the right (orientations with respect to Figure 3) of the native structure, yet remains closely aligned with the native structure up until the hinge glycine 63. Above that it slants to the left of the native helix so that the upper portion of the spike is similar to the C-monomer. The tilting makes the helices appear out of register with the apo structure (Figure 3D and S2B). The ascending helix of the D-monomer completes the same number of turns as the native structure, but after the loop at the tip of the spike, the descending helix begins a complete turn later than in the apo structure; the spike tip of the D-monomer is unraveled to a lesser extent than in the C-monomer. However, with a substantial kink at glycine at position 94, the upper portion of the D descending helix is even more severely displaced than in the C-monomer, displacing the C-alpha backbone by a full 5.0 Å (Figures 2B and 3D). The RMSD for the D-monomer as a whole is 1.1 Å, but for the upper spike region (residues 69 to 93, Figure S3B), the RMSD is 2.3 Å.
Definition of the AT-130 binding site
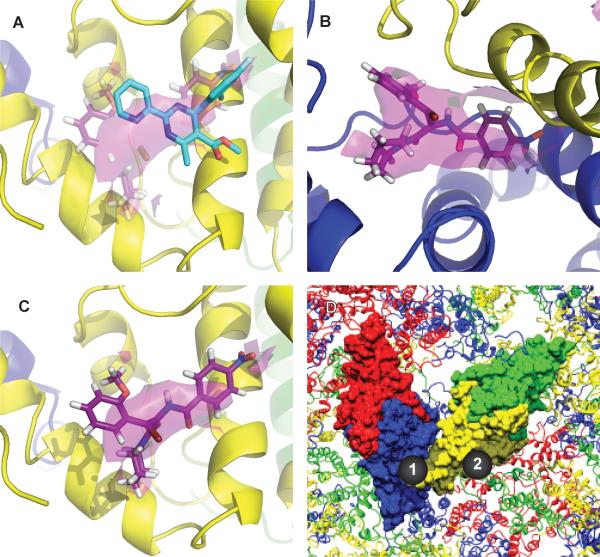
The binding sites of AT-130 were identified by electron density (particularly evident in the sharpened map) that could not be attributed to protein. In a hydrophobic pocket located at the contact between B and C dimers, the same pocket where the HAP compounds were proposed to bind (Figure 4 and S3B), density of comparable detail to that of the protein chains was visible. It was strongest in the pocket of the B-monomer, visible above 3σ (Figure 4B and S4A). While still clear and comparable in strength to that of the protein, the density in the pocket of the C-monomer was somewhat weaker; only a small mass of density was still visible at a 3σ cutoff in the sharpened density map (Figure 4C and S4B). No density was visible in the pockets of the A- or D-monomer. This suggests that AT-130 binds to both B and C pockets but favors that of the B-monomer; in the C-monomer, the weaker density may correspond to only partial occupancy. This implies relatively weak binding of phenylpropenamide compounds to C-sites despite crystallization conditions where there were two AT-130 molecules for every Cp dimer, implying that AT-130 binds the C-site with a dissociation constant on the order of 300 μM.
Figure 4. AT-130 Binding in Promiscuous Pocket at Base of the Dimer.
(A) AT-130 cis-isomer (purple) and HAP1 (cyan) molecular models shown overlapping in the AT-130 density in the C-subunit, the preferred location for HAP1 (Bourne et al., 2006). The AT-130 density (purple) is contoured at 1.5σ (Figure S4C-D).
(B) AT-130 density is strongest at the base of the B-subunit (Figure S4A).
(C) Weaker but clearly visible AT-130 density in the equivalent pocket in C.
(D) An overview of AT-130 binding sites. AT-130 binds in both the B/C quasi-equivalent binding pocket (Site 1) and the C/D pocket (Site 2), but favors Site 1. See also Figure S4 and Movie S3.
The observed density in our crystal structure is of sufficient resolution to confirm that the cis-isomer is the form of AT-130 that binds to the HBV capsid protein (Figure 4B and S4A,C-D). The stronger density in the B-pocket is unambiguous, and even the weaker density in the C-pocket, when observed at lower contour, can be identified as the cis-isomer of AT-130 (Figure 4C and S4B). Note that in a typical phenylpropenamide sample, the (Z)-olefin isomer (cis-isomer) makes up 70-90% of the total mass (Figure S4C) (King et al., 1998; Perni et al., 2000; Wang, 2011); this is known to be the active form of the compound with an EC50 of 1.2 μM, compared to over 100 μM for the trans-isomer (Figure S4D) (Wang, 2011). The relatively narrow hydrophobic pocket cannot accommodate the bulkier trans-isomer, whereas the cis-isomer fills the pocket neatly (Figure S4A-D).
Despite having entirely different structures and substantially different effects on assembly, AT-130 and HAP1 share the same binding pocket, indicating that this binding site is promiscuous and able to accommodate multiple structures (Figure 4A). However, AT-130 favored a different quasi-equivalent site than HAP1 (Figure 4B and S4A). The HAP1-bound structure showed significant extra density in the pocket of the C-monomer, and while there was additional density present in the B-pocket, it was much weaker (Bourne et al., 2006). The observed behavior of AT-130 was the opposite: the strongest observable density attributed to the compound was in the B-pocket, and while weaker, the additional density in the C-pocket was still as strong as that of the protein density.
A HAP-resistant mutant confirms the phenylpropenamide binding site
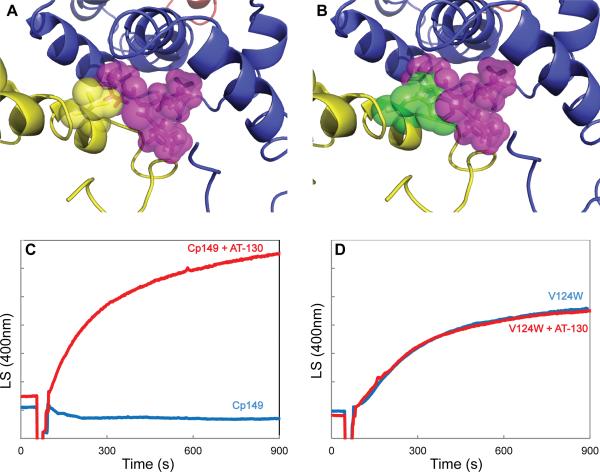
The phenylpropenamide and HAP binding site is an interfacial hydrophobic pocket. A mutant capsid protein was constructed, V124W (Figure 5A), that partially fills the pocket resulting in resistance to HAPs (Figure 5B) (Tan et al., 2013). Tryptophan occupies about half the volume of HAP1 or AT-130. Furthermore, the Cp149-V124W mutant is assembly hyperactive, consistent with a filled HAP site. HAP-12, the most effective of the previously-tested HAP compounds (Bourne, 2008), had negligible effect on Cp149-V124W assembly (Tan et al., 2013).
Figure 5. HAP-resistant Capsid Protein Mutant is Insensitive to AT-130.
(A) A valine residue (yellow sticks/spheres) makes up part of the assigned AT-130 (purple sticks/spheres) binding site.
(B) The V124W mutation (green sticks/spheres) partially occludes the proposed binding site for AT-130.
(C) Light scattering (in arbitrary units) of Cp149 assembly in 150 mM NaCl with AT-130.
(D) Light scattering of Cp149-V124W assembly in 50 mM NaCl with AT-130.
To test the assignment of the putative AT-130 density, we examined the effect of AT-130 on Cp149-V124W assembly. We used 90° light scattering to examine the effect of AT-130 on Cp149-V124W assembly kinetics. Wild type protein showed increased assembly in the presence of AT-130 (Figure 5C) (Katen, 2010) whereas Cp149-V124W showed no significant change in assembly rate or extent (Figure 5D). The valine-to-tryptophan mutation nullified the effects of AT-130.
DISCUSSION
AT-130 binds in the same putative location as the HAP compounds correlating with its properties as an assembly effector. In the context of a monomer, this pocket is located at the juncture of all four Cp subdomains (Figure S3). The binding of the hydrophobic phenylpropenamides molecule could serve as a molecular “glue,” bringing the surrounding flexible subdomains and the central chassis into a more compact, assembly-active state (Packianathan, 2010). Indeed, filling this pocket with a tryptophan in the V124W mutant leads to a more compact dimer, observed by size exclusion chromatography (Tan et al., 2013). This structure more readily forms the nucleation complex needed to initiate assembly (Zlotnick et al., 1999a). The kinetic trapping of incomplete intermediates observed with the related phenylpropenamide B-21 (Figure S4E) supports the conclusion that nucleation is the affected step of assembly (Katen, 2010).
The hydrophobic pocket was put forward as the binding site for the HAP compounds based on a 5 Å resolution structure. Our higher resolution structure enabled us see the density with sufficient detail to confirm this binding site and the published biochemical data identifying the phenylpropenamide cis-isomer as the active form. Our model of AT-130 in its site is consistent with phenylpropenamide structure-activity relationship studies (Wang, 2011). Substitutions on the piperidine ring abolished activity (Wang, 2011); in our structure we observe that this ring is buried in the pocket so that additions would likely abrogate binding. The o-position of the A-ring and p-position of the B-ring are oriented toward the capsid interior and exterior, respectively, and the phenylpropenamides are tolerant of additions and substitutions to these positions. The other positions of the ring structures are buried within the hydrophobic interior, and the loss of activity observed with substitutions at these positions suggest a loss of binding due to a poor fit within the binding site (Wang, 2011). Lastly, the HAP-resistant V124W mutant, which partially occludes the binding pocket with a tryptophan residue, is also resistant to AT-130. These results lead us to confirm that that this site is a promiscuous binding pocket and can accommodate multiple compounds, including both the HAPs and the phenylpropenamides.
Given the same binding site and their similar accelerating and stabilizing effects on assembly in vivo, we had expected that the HAPs and phenylpropenamides would have the same effect on capsid structure: quaternary shifting of the capsid dimers as rigid bodies. With AT-130 the A/B dimer quaternary structure change is very similar to what was observed with HAP1. With the HAPs this shift can result in mis-assembly of the capsid protein into non-capsid polymers; however, AT-130 supports formation of morphologically normal capsids. The structural basis of this difference was evident in the C/D dimers which, unlike with the rigid body movement seen with the HAPs (Bourne et al., 2006), had altered tertiary structure. The C/D intradimer interface was distorted, the spike-tip broadening and splaying outward, to the point of unwinding the spike helix at the uppermost residues.
The observed combination of quaternary and tertiary structural changes led us to propose a mechanism by which AT-130-bound capsids maintain their structural integrity where HAP-bound capsids do not. When AT-130 is bound, the protein-protein interactions around five-folds are destabilized (as HAPs also do (Bourne et al., 2006)), but the C/D dimers adjust their conformation to accommodate the altered quaternary interactions, holding the A/B dimers in place. This has the observed effect of retaining the icosahedral quaternary geometry, eliminating the tendency to adopt misdirected aggregate structures (Figure 6A-B). The tertiary structure change of the C/D dimer may be a compromise to fit the bulk of the phenylpropenamide (compared to a relatively planar HAP) and still assemble a T=4 capsid. The cost of distorting the C/D dimer would be paid by the global capsid stability. Conversely, the HAP molecules do not impose C/D distortion and consequently lead to aberrant, non-capsid polymers. We note that HAPs stabilize protein-protein interaction in HBV much more than phenylpropenamides. We suggest that the phenylpropenamides pay a thermodynamic price for distorting the four helix bundles that makes their enhancement of capsid stability weaker than seen with the HAPs, which cause no such tertiary distortion.
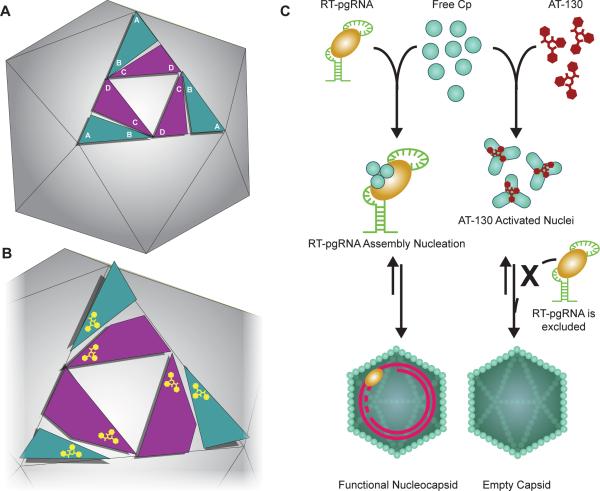
Figure 6. Model of AT-130 as a Kinetic Effector of HBV Assembly through Subtle Conformational Changes.
(A) Schematic of HBV capsid structure showing the arrangement of the asymmetric unit on an icosahedral face.
(B) Model of allosteric change with AT-130 binding; the quaternary upward motion of the A/B dimers is compensated for by tertiary conformational changes within the C/D dimers.
(C) Proposed AT-130 mechanism of action. Normal virus assembly involves the packaging of the RT-pgRNA complex (Nassal, 2008); AT-130 binds to the capsid protein and induces subtle conformational changes that cause the formation of nuclei without the RT-pgRNA complex, leading to the formation of empty particles. See also Figure S5.
Subtle conformational shifts in capsid subunits are reminiscent of molecular motions that have been proposed to be responsible for signal transduction in the infectious virion. Specifically, motion around the spike tip upon DNA synthesis and capsid maturation and also exposure of the C-terminus from the capsid exterior may be structural signals for virus secretion (Bottcher et al., 1998; Chen et al., 2011; Ning, 2011; Roseman et al., 2005; Wang et al., 2012a). Image reconstructions of HBV capsids with inserts in the spikes have revealed an additional degree of flexibility within dimers of the assembled capsid (Bottcher et al., 2006). Thus, all current evidence points to the HBV capsid being a highly dynamic molecule that undergoes structural changes as necessary steps in the virus lifecycle, but without disrupting the overall capsid geometry.
Changes in conformation are clearly transmitted across the entire structure. AT-130 was shown to bind only in the hydrophobic pockets of the B-and C-monomers, and yet the A-monomer showed the greatest vertical displacement, while the D-monomer spike had the largest RMSD from the native structure. Small local changes communicated across the capsid protein are the basis of the argument for the role of allostery in viral assembly, signaling, and disassembly (Bottcher et al., 2006; Tang J, 2006). Thus, the structure presented here demonstrates an important implication of the high degree of plasticity of the HBV capsid: quaternary changes are compensated for by tertiary structural changes to maintain capsid integrity. The plasticity we observe suggests that the HBV capsid (and capsids of other viruses) accommodate a broad continuum of asymmetric motion and conformations. While we observe icosahedrally ordered structural changes, the high degree of conformational plasticity suggests a freedom that may break icosahedral symmetry (Wang et al., 2012b).
AT-130 and HAP1 show different quasi-equivalent binding preferences. In the +HAP1 capsid structure, the strongest HAP density was located in the pocket of the C-subunit (Bourne et al., 2006). In the +AT-130 structure, the strongest density was in the B-subunit pocket, indicating a binding preference there. It seems that the breakdown of HBV quasi-equivalence extends to the binding pockets. An HBV capsid has 60 copies each of four unique binding pockets. This raises the possibility of designing antiviral compounds that target different quasi-equivalent sites for maximal disruption of assembly. In fact, this has already been observed to some degree; low concentrations of AT-130 and BAY41-4109 (HAP1) synergistically reduced virus production (Billioud, 2011). However, antagonism exists between the two compounds at higher concentrations. Our structure reveals that while AT-130 does prefer the B-pocket, it can also bind in the C-position, and vice versa for HAP1. It is possible that the high concentration antagonism stems from competition between the two compounds. Examination of the effects of both these molecules together on HBV capsid assembly may shed further light on the synergy and antagonism between different binding pockets. The observed effects may arise solely from competition for the same site, or filling one site may affect binding to the other. Thus, both compounds in concert may also change the thermodynamic and kinetic assembly properties of the capsid in ways different from the compounds alone.
The lifecycle of any virus is an exquisitely timed and regulated process; a focused disruption can be devastating to virus replication. The greater implication is that a molecule does not need to induce large structural changes or possess inhibitory properties in order to function as a successful antiviral. Phenylpropenamides are effective antiviral compounds even though they have relatively subtle effects on capsid structure and assembly, decreasing virus production to levels comparable with the superior members of the HAP family (Figure S5). Virion assembly initiated at the wrong point in time, which is what the phenylpropenamides appear to do, results in morphologically normal capsids that are empty and non-infectious (Figure 6C) (Feld, 2007). While most antiviral therapies tend to focus on inhibition or termination of a viral process, we propose that deregulation or mistiming of a viral process is an equally viable strategy for the design of antiviral compounds. Potential antiviral targets have been overlooked because their effects on virus replication were not classically inhibitory.
The HBV capsid protein and its assembly process have no human analogs, making HBV assembly a very attractive target for new antiviral therapies. We have determined the binding site of a representative of a second family of assembly effectors, a member of the phenylpropenamide family, AT-130, and in doing so have shown that the HAP binding pocket is promiscuous and can be considered a target for a wide variety of small molecules. Our results demonstrate that the quasi-equivalent nature of the capsid results in four subtly different targets. The capsid can accommodate a great range of motion and tertiary and quaternary conformational change while still maintaining its structural integrity. However, even small changes can significantly alter the assembly properties of the virus and significantly disrupt the normal lifecycle. That the modest effects of AT-130 can have such a significant decrease in virus production illustrates an alternative paradigm for antiviral compounds; while inhibitors of viral processes (e.g. nucleoside analogs) are potent antiviral compounds, “deregulators” of the viral processes may be equally effective antiviral therapies. This novel approach offers new opportunities for effective combination therapies.
EXPERIMENTAL PROCEDURES
Sample Preparation
Cp149 capsid protein dimer from Hepatitis B subtype adyw was expressed in E. coli and purified as described previously (Zlotnick et al., 2002). The V124W mutant was purified using a modified protocol (Tan et al., 2013). Frozen aliquots of Cp were dialyzed against assembly buffer (50 mM HEPES, pH 7.5) prior to light scattering experiments. For crystallization, a Cp149 mutant, 3CA-Cp150, was used, wherein three native cysteine residues were mutated to alanine and an additional cysteine was appended at position 150 (Bourne et al., 2006). T=4 3CA-Cp150 capsids were purified from free dimer and T=3 particles by sucrose gradient sedimentation as described previously and immediately dialyzed for crystallization (Zlotnick et al., 1999b). The phenylpropenamide compound AT-130 was synthesized as described previously (Katen, 2010).
Light Scattering
Observation of kinetics by 90° light scattering was observed with a Photon Technology International fluorometer set to 400 nm for both excitation and emission (Katen, 2010). Light scattering was measured for 10 μM (final concentration) wild-type Cp149 reduced with 5% β-mercaptoethanol (Packianathan, 2010; Tan et al., 2013). Assembly was induced by addition of NaCl to a final concentration of 150 mM NaCl, with and without the addition of 20 μM AT-130. The V124W assembly experiment followed the same procedure, with the exception that assembly was initiated with 50 mM NaCl due to the mutant assembly hyperactivity (Tan et al., 2013).
Electron Microscopy
Samples from light scattering experiments were adsorbed to glow-discharged carbon over paralodian copper grids (EM sciences). Samples were stained with 2% uranyl acetate and visualized with a JEOL 1010 transmission electron microscope equipped with a 4K×4K Gatan CCD camera.
Crystallization
Crystallization was optimized from previously determined conditions for the adyw 3CA-Cp150 capsid (Bourne et al., 2006). Co-crystals were grown in the presence of half-molar, equimolar, and twofold excess molar concentration of AT-130 in DMSO (relative to the concentration of Cp in capsid form). Crystallization was carried out at room temperature with 4 μL sitting drops, initiated by a 1:1 mixture of protein solution with well solution. Protein solutions contained 10 mg/mL 3CA Cp150 capsid in 5 mM Tris buffer, pH 7.5, 150 mM NaCl, and 0.7-3% DMSO. Well solutions were composed of 5-10% polyethylene glycol 5000 monomethylether, 0-5% polyethylene glycol 8000 monomethylether, 6-28% 2,3-butanediol, 100 mM Tris pH 9.0, 150 mM NaCl, and 300 mM KCl.
Diffraction Data Collection
Crystals were cryoprotected as described previously (Bourne et al., 2006). Crystals were flash-frozen in a stream of gaseous -170°C nitrogen before transport to the Advanced Photon Source (APS) Beamline 14BMC for data collection. The final dataset was collected from a single cryo-cooled crystal containing a 2:1 molar ratio of AT-130 to Cp dimer; statistics are in Table 1.
Structure Solution and Refinement
Molecular replacement and averaging was used for phasing. The native adw-like capsid structure (Protein Data Bank Code 1QGT) was used as the phasing model (Wynne et al., 1999). Molecular replacement was carried out with the Phaser program in the CCP4 program suite (Collaborative Computational Project, 1994; McCoy, 2007). Phases calculated to 7 Å were subjected to 60-fold non-crystallographic symmetry (NCS) averaging and phase extension using the RAVE suite (Kleywegt et al., 2001). NCS averaging produced an averaging R factor of 26.0% and a correlation coefficient of 93.7%.
Refinement was carried out with CNS (Brunger et al., 1998) using strict icosahedral NCS, isotropic B-factor correction, and bulk solvent scaling. AT-130 refinement was carried out through positional refinement in conjunction with torsion molecular dynamics, with slow-cooling from 2,000 K in 50 K steps. Further refinement was iterated with 60-fold NCS averaging and manual rebuilding in Coot (Emsley, 2010). A test set of 5,000 reflections was for Rfree cross-validation, but the resulting Rfree was virtually identical to the crystallographic R factor due to NCS correlation of the test and working data sets. The molecular model was refined to yield a crystallographic R factor of 37% (Table 1), a value consistent with those of other heavily averaged structures at comparable resolutions.
Superpositions of models were carried out in Coot (Emsley, 2010). Figures were generated with PyMol (DeLano, 2009) and Chimera (Pettersen EF, 2004).
Supplementary Material
HIGHLIGHTS.
Phenylpropenamides induce tertiary and quaternary structural changes in HBV capsids
AT-130 binds to a promiscuous pocket at the dimer-dimer interface
AT-130 favors a unique quasi-equivalent binding site in the capsid
Small molecules that do not disrupt capsid structure are still effective antivirals
ACKNOWLEDGEMENTS
This work was supported by grants from NIH (R01 AI067417 to AZ) and a Ruth L. Kirchstein Predoctoral Fellowship (F31 AI077323 to SPK). Time at APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W31-109-Eng-38
We thank the BioCARS beamline staff at APS. We also thank Drs. Jeff Speir and David Morgan for computational assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Protein structure Accession Numbers
The atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (www.rcsb.org/pdb) with the accession number 4G93.
REFERENCES
- Billioud G, Pichoud Christian, Puerstinger Gerhard, Neyts Johan, Zoulim Fabien. The main Hepatitis B virus (HBV) mutants resistant to nucleoside analogs are susceptible in vitro to non-nucleoside inhibitors of HBV replication. Antiviral Res. 2011;92:271–276. doi: 10.1016/j.antiviral.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Bothner B, Dong XF, Bibbs L, Johnson JE, Siuzdak G. Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. J Biol Chem. 1998;273:673–676. doi: 10.1074/jbc.273.2.673. [DOI] [PubMed] [Google Scholar]
- Bottcher B, Tsuji N, Takahashi H, Dyson MR, Zhao S, Crowther RA, Murray K. Peptides that block hepatitis B virus assembly: analysis by cryomicroscopy, mutagenesis and transfection. EMBO J. 1998;17:6839–6845. doi: 10.1093/emboj/17.23.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher B, Vogel M, Ploss M, Nassal M. High plasticity of the hepatitis B virus capsid revealed by conformational stress. J Mol Biol. 2006;356:812–822. doi: 10.1016/j.jmb.2005.11.053. [DOI] [PubMed] [Google Scholar]
- Bourne C, Finn MG, Zlotnick A. Global structural changes in hepatitis B capsids induced by the assembly effector HAP1. J Virol. 2006;80:11055–11061. doi: 10.1128/JVI.00933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne CR, Lee S, Venkataiah B, Lee A, Korba B, Finn MG, Zlotnick A. Small-Molecule effectors of Hepatitis B Virus Capsid Assembly Give Insight into Virus Life Cycle. J Virol. 2008;82:10262–10270. doi: 10.1128/JVI.01360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Ceres P, a.Z.A. Weak Protein-Protein Interactions Are Sufficent to Drive Assembly of Hepatitis B Virus Capsids. Biochemistry. 2002;41:11525–11531. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- Chen C, Wang JC, Zlotnick A. A kinase chaperones hepatitis B virus capsid assembly and captures capsid dynamics in vitro. PLoS pathogens. 2011;7:e1002388. doi: 10.1371/journal.ppat.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, N. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Delaney W.E.t., Edwards R, Colledge D, Shaw T, Furman P, Painter G, Locarnini S. Phenylpropenamide derivatives AT-61 and AT-130 inhibit replication of wild-type and lamivudine-resistant strains of hepatitis B virus in vitro. Antimicrob Agents Chemother. 2002;46:3057–3060. doi: 10.1128/AAC.46.9.3057-3060.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system, version 1.21r1. Delano Scientific. San Carlos, CA: 2009. [Google Scholar]
- Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, Kramer T, Niewohner U, Pleiss U, Stoltefuss J, et al. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science. 2003;299:893–896. doi: 10.1126/science.1077215. [DOI] [PubMed] [Google Scholar]
- DiMattia MA WN, Stahl SJ, Grimes JM, Steven AC. Antigenic Switching of Hepatitis B Virus by Alternative Dimerization of the Capsid Protein. Structure. 2013;21:133–142. doi: 10.1016/j.str.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink D, Berkhout B, Sanders RW. Inhibition of HIV-1 by fusion inhibitors. Curr Pharm Des. 2010;16:3716–3728. doi: 10.2174/138161210794079218. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and Development of Coot. Acta Cryst. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feld JJ, Colledge C, Sozzi V, Edwards R, Littlejohn M, Locarnini SA. The phenylpropenamide derivative AT-130 blocks HBV replication at the level of viral RNA packaging. Antiviral Res. 2007;76:168–177. doi: 10.1016/j.antiviral.2007.06.014. [DOI] [PubMed] [Google Scholar]
- Ganem D, Schneider RJ. Hepadnaviridae: The Viruses and Their Replication. 4 edn Lippincott Williams & Wilkins; Philadelphia: 2001. [Google Scholar]
- Gertsman I, Komives EA, Johnson JE. HK97 maturation studied by crystallography and H/2H exchange reveals the structural basis for exothermic particle transitions. Journal of molecular biology. 2010;397:560–574. doi: 10.1016/j.jmb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilmer JK, Zlotnick A, Bothner B. Conformational Equilibria and Rates of Localized Motion within Hepatitis B Capsids. J Mol Biol. 2007;375:581–594. doi: 10.1016/j.jmb.2007.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Cooper KL, Tappenden P, Rees A, Simpson EL, Read RC, Nicholson KG. Oseltamivir, zanamivir, and amantadine in the prevention of influenza: a systematic review. J Infect. 2011;62:14–25. doi: 10.1016/j.jinf.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Johnson JE. Virus particle dynamics. Adv Protein Chem. 2003;64:197–218. doi: 10.1016/s0065-3233(03)01005-2. [DOI] [PubMed] [Google Scholar]
- Katen SP, Chirapu SR, Finn MG, Zlotnick A. Trapping of Hepatitus B Virus Capsid Assembly Intermediates by Phenylpropenamide Assembly Accelerators. Chem Biol. 2010;5:1125–1136. doi: 10.1021/cb100275b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Ladner SK, Miller TJ, Zaifert K, Perni RB, Conway SC, Otto MJ. Inhibition of human hepatitis B virus replication by AT-61, a phenylpropenamide derivative, alone and in combination with (-)beta-L-2',3'-dideoxy-3'-thiacytidine. Antimicrob Agents Chemother. 1998;42:3179–3186. doi: 10.1128/aac.42.12.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt GJ, Zou JY, Kjeldgaard M, Jones TA. Around O. In International Tables for Crystallography. 2001:353–356, 366-367. [Google Scholar]
- Lambert C, Döring T, Prange R. Hepatitis B virus matruation is sensitive to functional inhibition of ESCRT-III, Vps4,and gamma 2-adaptin. J Virol. 2007;81:9050–9060. doi: 10.1128/JVI.00479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan YT, Li J, Liao W, Ou J. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology. 1999;259:342–348. doi: 10.1006/viro.1999.9798. [DOI] [PubMed] [Google Scholar]
- Lewis JK, Bothner B, Smith TJ, Siuzdak G. Antiviral agent blocks breathing of the common cold virus. Proc Natl Acad Sci U S A. 1998;95:6774–6778. doi: 10.1073/pnas.95.12.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott L, Beames B, Notvall L, Lanford RE. Interaction between hepatitis B virus core protein and reverse transcriptase. J Virol. 2000;74:11479–11489. doi: 10.1128/jvi.74.24.11479-11489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser Crystallographic Software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menédez-Arias L. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 2008;134:124–146. doi: 10.1016/j.virusres.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Monroe EB, Kang S, Kyere SK, Li R, Prevelige PE., Jr. Hydrogen/deuterium exchange analysis of HIV-1 capsid assembly and maturation. Structure. 2010;18:1483–1491. doi: 10.1016/j.str.2010.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235–249. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Ning X, Nguyen D, Mentzer L, Adams C, Lee H, Ashley R, Havenstein S, Hu J. Secretion of genome-free hepatitis B virus--single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog. 2011;7:e1002255. doi: 10.1371/journal.ppat.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packianathan C, Katen SP, Dann CE, 3rd, Zlotnick A. Conformational changes in the Hepatitis B virus core protein are consistent with a role for allostery in virus assembly. J Virol. 2010;84:1607–1615. doi: 10.1128/JVI.02033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packianathan C, Katen SP, Dann CE, 3rd, Zlotnick A. Conformational Changes in the Hepatitis B Virus Core Protein are Consistent with a Role for Allostery in Virus Assembly. J Virol. 2010;84:1607–1615. doi: 10.1128/JVI.02033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perni RB, Conway SC, Ladner SK, Zaifert K, Otto MJ, King RW. Phenylpropenamide derivatives as inhibitors of hepatitis B virus replication. Bioorg Med Chem Lett. 2000;10:2687–2690. doi: 10.1016/s0960-894x(00)00544-8. [DOI] [PubMed] [Google Scholar]
- Pettersen EF GT, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Phelps DK, Post CB. A novel basis of capsid stabilization by antiviral compounds. J Mol Biol. 1995;254:544–551. doi: 10.1006/jmbi.1995.0637. [DOI] [PubMed] [Google Scholar]
- Porterfield J, Dhason MS, Loeb DD, Nassal M, Stray SJ, Zlotnick A. Full-length hepatitis B virus core protein packages viral and heterologus RNA with similarly high levels of cooperativity. J Virol. 2010;84:7174–7184. doi: 10.1128/JVI.00586-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe B, Vlachou A, Pante N, Helenius A, Kann M. Nuclear import of hepatitis B virus capsids and release of the viral genome. PNAS. 2003;100:9849–9854. doi: 10.1073/pnas.1730940100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razonable RR. Antiviral drugs for viruses other than human immunodeficiency virus. Mayo Clin Proc. 2011;86:1009–1026. doi: 10.4065/mcp.2011.0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman AM, Berriman JA, Wynne SA, Butler PJ, Crowther RA. A structural model for maturation of the hepatitis B virus core. Proc Natl Acad Sci U S A. 2005;102:15821–15826. doi: 10.1073/pnas.0504874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoeck R. Antiviral therapy of herpes simplex. Int J Antimicrob Agents. 2000;16:157–159. doi: 10.1016/s0924-8579(00)00233-8. [DOI] [PubMed] [Google Scholar]
- Stray SJ, Bourne CR, Punna S, Lewis WG, Finn MG, Zlotnick A. A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. Proc Natl Acad Sci U S A. 2005;102:8138–8143. doi: 10.1073/pnas.0409732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stray SJ, Zlotnick A. BAY 41-4109 has multiple effects on Hepatitis B virus capsid assembly. J Mol Recognit. 2006;19:542–548. doi: 10.1002/jmr.801. [DOI] [PubMed] [Google Scholar]
- Tan Z, Maguire ML, Loeb DD, Zlotnick A. Genetically altering the thermo and kinetics of Hepatitis B Virus capsid assembly has profound effects on virus replication in cell culture. Journal of virology. 2013 doi: 10.1128/JVI.03014-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J JJ, Dryden KA, Young MJ, Zlotnick A, Johnson JE. The role of subunit hinges and molecular “switches” in the control of viral capsid polymorphism. Structural Biol. 2006;154:59–67. doi: 10.1016/j.jsb.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Toniutto P, Fabris C, Pirisi M. Antiviral treatment of hepatitis C. Expert Opin Pharmacother. 2006;7:2025–2035. doi: 10.1517/14656566.7.15.2025. [DOI] [PubMed] [Google Scholar]
- Tuma R, Tsuruta H, French KH, Prevelige PE. Detection of intermediates and kinetic control during assembly of bacteriophage P22 procapsid. J Mol Biol. 2008;381:1395–1406. doi: 10.1016/j.jmb.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC, Dhason MS, Zlotnick A. Structural organization of pregenomic RNA and the carboxy-terminal domain of the capsid protein of hepatitis B virus. PLoS pathogens. 2012a;8:e1002919. doi: 10.1371/journal.ppat.1002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Nadhthambi Devan, Mosley Ralph T., Niu Congrong, Furman Phillip A., Otto Michael J., Sofia Michael J. Phenylpropenamide derivatives: Anti-hepatitis B virus activity of the Z-isomer, SAR and the search for novel analogs. Bioorg Med Chem Lett. 2011;21:4642–4647. doi: 10.1016/j.bmcl.2011.05.077. [DOI] [PubMed] [Google Scholar]
- Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, et al. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nature structural & molecular biology. 2012b;19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber O, Schlemmer KH, Hartmann E, Hagelschuer I, Paessens A, Graef E, Deres K, Goldmann S, Niewoehner U, Stoltefuss J, et al. Inhibition of human hepatitis B virus (HBV) by a novel non-nucleosidic compound in a transgenic mouse model. Antiviral Res. 2002;54:69–78. doi: 10.1016/s0166-3542(01)00216-9. [DOI] [PubMed] [Google Scholar]
- Wensing AM, van Maarseveen NM, Nijhuis M. Fifteen years of HIV Protease Inhibitors: raising the barrier to resistance. Antiviral Res. 2010;85:59–74. doi: 10.1016/j.antiviral.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- Yeh CT, Liaw YF, Ou JH. The arginine-rich domain of hepatitis B virus precore and core proteins contains a signal for nuclear transport. JVirol. 1990;64:6141–6147. doi: 10.1128/jvi.64.12.6141-6147.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen MF, Lai CL. Treatment of chronic hepatitis B: Evolution over two decades. J Gastroenterol Hepatol. 2011;26:138–143. doi: 10.1111/j.1440-1746.2010.06545.x. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Ceres P, Singh S, Johnson JM. A small molecule inhibits and misdirects assembly of hepatitis B virus capsids. J Virol. 2002;76:4848–4854. doi: 10.1128/JVI.76.10.4848-4854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A, Cheng N, Stahl SJ, Conway JF, Steven AC, Wingfield PT. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. ProcNatlAcadSciUSA. 1997;94:9556–9561. doi: 10.1073/pnas.94.18.9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A, Johnson JM, Wingfield PW, Stahl SJ, Endres D. A theoretical model successfully identifies features of hepatitis B virus capsid assembly. Biochemistry. 1999a;38:14644–14652. doi: 10.1021/bi991611a. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Palmer I, Stahl SJ, Steven AC, Wingfield PT. Separation and Crystallization of T=3 and T=4 Icosahedral Complexes of the Hepatitis B Virus Core Protein. Acta Cryst D. 1999b;55:717–720. doi: 10.1107/s090744499801350x. [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Stray SJ. How does your virus grow? Understanding and interfering with virus assembly. Trends Biotechnol. 2003;21:536–542. doi: 10.1016/j.tibtech.2003.09.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.