The biogenesis of Cox4 is unknown. Cox4, mtHsp70, and Mge1 form a complex that promotes the assembly of cytochrome c oxidase. In the absence of the mature cytochrome c oxidase, Cox4 arrests at the chaperone complex. This complex delivers Cox4 into the assembly line of complex IV when needed.
Abstract
The formation of the mature cytochrome c oxidase (complex IV) involves the association of nuclear- and mitochondria-encoded subunits. The assembly of nuclear-encoded subunits like cytochrome c oxidase subunit 4 (Cox4) into the mature complex is poorly understood. Cox4 is crucial for the stability of complex IV. To find specific biogenesis factors, we analyze interaction partners of Cox4 by affinity purification and mass spectroscopy. Surprisingly, we identify a complex of Cox4, the mitochondrial Hsp70 (mtHsp70), and its nucleotide-exchange factor mitochondrial GrpE (Mge1). We generate a yeast mutant of mtHsp70 specifically impaired in the formation of this novel mtHsp70-Mge1-Cox4 complex. Strikingly, the assembly of Cox4 is strongly decreased in these mutant mitochondria. Because Cox4 is a key factor for the biogenesis of complex IV, we conclude that the mtHsp70-Mge1-Cox4 complex plays an important role in the formation of cytochrome c oxidase. Cox4 arrests at this chaperone complex in the absence of mature complex IV. Thus the mtHsp70-Cox4 complex likely serves as a novel delivery system to channel Cox4 into the assembly line when needed.
INTRODUCTION
Hsp70 proteins play a fundamental role in the biogenesis of mitochondria. In the model organism baker's yeast (Saccharomyces cerevisiae), the major mitochondrial (mt) Hsp70 is encoded by the essential SSC1 gene and is required for import of preproteins and protein folding (Kampinga and Craig, 2010; Marom et al., 2011; Voos, 2013). The presequence translocase (translocase of the inner membrane, TIM23 complex) mediates the membrane potential–dependent import of preproteins across and into the inner membrane (Dolezal et al., 2006; Baker et al., 2007; Neupert and Herrmann, 2007; Chacinska et al., 2009; Endo and Yamano, 2009; Fox, 2012). Through cycles of ATP-dependent preprotein binding and release, mtHsp70 completes preprotein transfer across the inner membrane. MtHsp70 binds to the TIM23 complex via Tim44 and is regulated by the J-proteins Pam18 and Pam16 within the presequence translocase–associated motor (PAM). Mitochondrial GrpE (Mge1) acts as a nucleotide exchange factor to regenerate the ATP-bound form of mtHsp70 (Neupert and Herrmann, 2007; Chacinska et al., 2009; Endo and Yamano, 2009). mtHsp70 and Mge1 also associate with the matrix-localized J-protein mitochondrial DnaJ (Mdj1). This mtHsp70 pool stimulates the folding of imported as well as of some mitochondria-encoded proteins (Rowley et al., 1994; Herrmann et al., 1994; Westermann et al., 1996; Horst et al., 1997).
In yeast, cytochrome c oxidase (complex IV) consists of three mitochondria-encoded components that form the catalytic core and several nuclear-encoded subunits. Complex IV associates with cytochrome bc1 complex (complex III) in respiratory chain supercomplexes. The biogenesis of cytochrome c oxidase is a highly complicated process involving >20 assembly factors (Herrmann and Funes, 2005; Mick et al., 2011; Fox, 2012; Soto et al., 2012a; Winge, 2012). mtHsp70 plays a role in the maturation of the mitochondria-encoded cytochrome c oxidase subunit 1 (Cox1). It interacts with the mitochondrial splicing suppressor protein 51 (Mss51), which is a translational activator of Cox1 (Decoster et al., 1990; Perez-Martinez et al., 2003; Barrientos et al., 2004; Soto et al., 2012b). The Mss51-mtHsp70 complex binds to ribosomes synthesizing Cox1 and to a membrane-bound assembly intermediate of Cox1 containing additional biogenesis factors (Pierrel et al., 2007; Perez-Martinez et al., 2009; Ott and Herrmann, 2010; Fontanesi et al., 2010, 2011; Mick et al., 2010; Fox, 2012). The role of mtHsp70 at this step is unclear. It was suggested that the chaperone supports the folding of Cox1 (Fontanesi et al., 2010). In contrast, little is known about the assembly of nuclear-encoded complex IV subunits. It was reported that Cox5a and Cox6 associate with the Cox1-containing assembly intermediate (Horan et al., 2005; Mick et al., 2011; Soto et al., 2012a; McStay et al., 2013). Further assembly steps leading to the formation of mature complex IV are poorly understood. In particular, the association of Cox4 with assembly intermediates has not been analyzed in detail, although Cox4 is crucial for the stability of cytochrome c oxidase (Frazier et al., 2006; Coyne et al., 2007; Mick et al., 2007).
To identify factors specifically required for Cox4 assembly, we performed stable isotope labeling with amino acids in cell culture (SILAC) followed by affinity purification via histidine (His)-tagged Cox4 and mass spectrometric analyses. We found strong interaction of Cox4 with mtHsp70 and Mge1. Cox4 arrests at this chaperone complex in the absence of mature complex IV. We identified a novel temperature-sensitive mutant of mtHsp70 that is specifically impaired in binding to Cox4. Strikingly, the assembly of Cox4 into respiratory chain supercomplexes is selectively impaired in the mutant mitochondria. Because Cox4 is needed for assembly of other subunits of complex IV, we conclude that the mtHsp70-Mge1-Cox4 complex is important for the formation of the mature supercomplexes. Thus we found a novel function of mtHsp70 in the assembly of nuclear-encoded subunits of cytochrome c oxidase.
RESULTS
mtHsp70 and Mge1 interact with the Cox4 subunit of cytochrome c oxidase
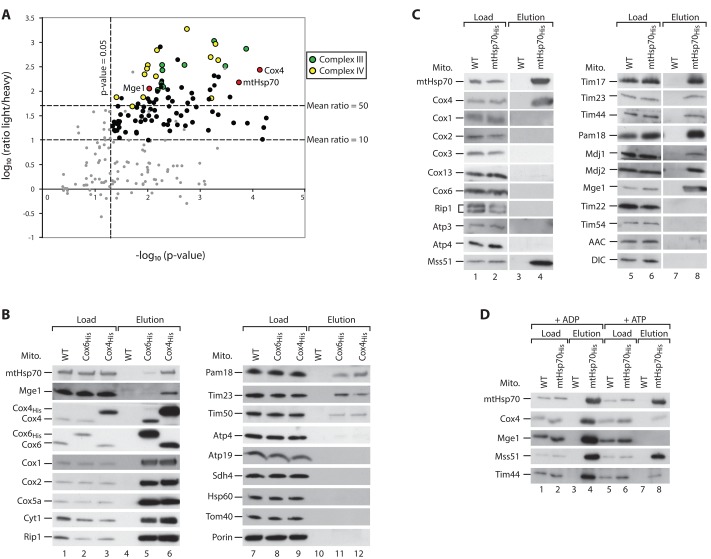
To identify factors specifically required for Cox4 biogenesis, we generated a yeast strain expressing His-tagged Cox4 (Cox4His) for affinity purification. We grew the cells in minimal medium containing either “heavy,” stable isotope-labeled (wild-type) or “light” forms (Cox4His) of arginine and lysine (Ong et al., 2002). Mitochondria were isolated and lysed with digitonin, and Cox4-interacting proteins were purified by Ni2+–nitriloacetic acid (NTA) chromatography. The light-over-heavy peptide ratios were determined by mass spectrometry and used to assess the specific enrichment of proteins in the Cox4His elution fraction. Strikingly, mtHsp70 and its cochaperone Mge1 were enriched to a similar extent as several subunits of complex IV and the cytochrome bc1 complex (complex III; Figure 1A and Supplemental Table S1). These interactions were verified by immunodetection of mtHsp70 and Mge1 in the elution fraction of Cox4His (Figure 1B, lane 6). The high yield of mtHsp70-Mge1 in the Cox4His elution sample was unexpected because both proteins were not coisolated with respiratory chain supercomplexes using TAP-tagged Cor1 of complex III (van der Laan et al., 2006; Wiedemann et al., 2007). To test whether the chaperone binds to mature complex IV, we also performed affinity purification with His-tagged Cox6. Here only small fractions of mtHsp70 and Mge1 were coeluted with Cox6His (Figure 1B, lane 5), whereas other subunits of complexes III and IV and TIM23 subunits were copurified to a similar extent with both tagged Cox4 and Cox6 (Figure 1B, lanes 5, 6, 11, and 12). The binding of TIM23 components to both tagged complex IV subunits confirms the reported association of the translocase with the respiratory chain supercomplexes (van der Laan et al., 2006; Wiedemann et al., 2007). Control proteins of both mitochondrial membranes and the matrix did not bind to the tagged proteins, demonstrating the specificity of the pull-down experiment (Figure 1B, lanes 11 and 12). The His tagging leads to a moderate overexpression of Cox4 (Figure 1B, lanes 1–3). To rule out that binding of mtHsp70 to Cox4 is due to the increased Cox4 content, we performed affinity purification via His-tagged mtHsp70 (Figure 1C). Cox4 was copurified with high efficiency, whereas other subunits of the respiratory chain complexes did not coelute with mtHsp70His or did so in low amounts, (Figure 1C, lane 4). Known interaction partners and cochaperones of mtHsp70, including components of the TIM23 complex, the complex IV biogenesis factor Mss51, the J-proteins Mdj1 and Mdj2, and Mge1 were specifically copurified with the tagged mtHsp70 (Figure 1C, lanes 4 and 8; Kampinga and Craig, 2010; Fontanesi et al., 2010; Mapa et al., 2010). A number of inner membrane control proteins did not bind to mtHsp70His (Figure 1C, lane 8). MtHsp70 binds to several partner proteins, like Tim44, in an ATP-sensitive manner (Figure 1D, lanes 4 and 8; Schneider et al., 1996; Liu et al., 2003). Similarly, the association of mtHsp70 to Cox4 was weakened upon incubation with ATP (Figure 1D, lanes 4 and 8). In contrast, the binding to Mss51 was only mildly affected in the presence of ATP, as reported (Figure 1D, lanes 4 and 8; Fontanesi et al., 2010). Thus we conclude that mtHsp70, Mge1, and Cox4 specifically interact with each other in an ATP-sensitive manner.
FIGURE 1:
Cox4 interacts with mtHsp70 and Mge1 in an ATP-sensitive manner. (A) Yeast cells were labeled with heavy (H) isotopes [15N213C6]lysine/[15N413C6]arginine (wild type) or their corresponding light (L) [14N12C]-containing variants (Cox4His strain). Mitochondria were isolated, lysed, and subjected to affinity purification. Elution fractions were mixed and analyzed by quantitative mass spectrometry. The mean log10(L/H) ratio and −log10 p values of three independent experiments were determined and plotted against each other. Red dots, Cox4, mtHsp70, and Mge1; yellow, subunits of complex IV; green, subunits of complex III; black, other proteins; gray, unspecific proteins (see Supplemental Table S1). (B) Isolated mitochondria from wild-type (WT), Cox6His, and Cox4His yeast strains were lysed in digitonin and incubated with Ni2+-NTA agarose. Bound complexes were eluted with imidazole, and samples were analyzed by SDS–PAGE and Western blotting. Load, 3%; elution, 100%. (C, D) Wild-type (WT) and mtHsp70His mitochondria were analyzed as described for B. Where indicated, lysis and purification of the samples were performed in the presence of 5 mM MgCl2 and 5 mM ADP or ATP. Load, 3%; elution, 100%. AAC, ADP/ATP carrier; DIC, dicarboxylate carrier.
Cox4 arrests at mtHsp70-Mge1 in the absence of complex IV
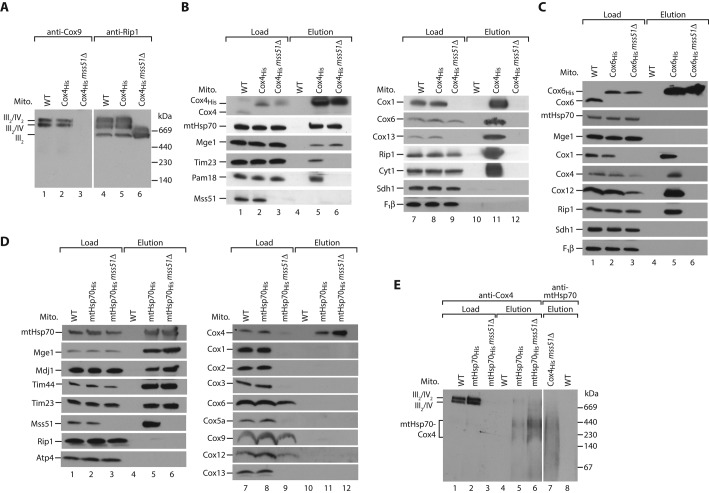
In contrast to the other major cytochrome c oxidase subunits, Cox4 was shown to be a prominent interaction partner of mtHsp70-Mge1. We asked whether this interaction occurs in the absence of mature complex IV. We fused chromosomally encoded Cox4 to a His tag in an mss51Δ strain. Loss of Mss51 leads to impaired synthesis of Cox1 and lack of assembled complex IV (Figure 2A; Perez-Martinez et al., 2003; Barrientos et al., 2004). Consequently, subunits of complex III and IV did not bind to Cox4His in mss51Δ mitochondria (Figure 2B, lane 12). In contrast, the association of mtHsp70-Mge1 to Cox4 was unaffected (Figure 2B, lane 6). Because mtHsp70 is the central component of the TIM23/PAM machinery, we wondered whether the Cox4-chaperone complex is associated with the translocase. Subunits of TIM23 were recovered with Cox4His expressed in wild-type but not in mss51Δ cells (Figure 2B, lanes 5 and 6). We conclude that Cox4 binds to mtHsp70-Mge1 independently of complex IV and the presequence translocase. Because Cox4 binds to the chaperone complex in the absence of Mss51, we conclude that this pool is distinct from the previously reported Mss51-mtHsp70 association (Fontanesi et al., 2010). Thus mtHsp70-Mge1-Cox4 represents a novel complex of mtHsp70. As control, His-tagged Cox6, the other matrix-exposed peripheral subunit of complex IV, did not bind in considerable amounts to mtHsp70-Mge1 in the absence of Mss51 (Figure 2C, lane 6). Moreover, we could not detect any binding of other complex IV subunits except Cox4 to His-tagged mtHsp70 in the mss51Δ background (Figure 2D, lane 12). Thus we conclude that the binding to mtHsp70-Mge1 in the absence of mature complex IV is specific for Cox4. To determine the size of the mtHsp70-Mge1-Cox4 complex, we analyzed the proteins bound to His-tagged mtHsp70 in either wild-type or mss51Δ mitochondria by blue native electrophoresis (Figure 2E). With antibodies against Cox4 we detected two protein complexes between 200 and 400 kDa (Figure 2E, lanes 5 and 6). Complexes of similar size were detected with antibodies against mtHsp70 after purification via Cox4His in mss51Δ mitochondria (Figure 2E, lane 7), confirming that both protein complexes contain mtHsp70 and Cox4.
FIGURE 2:
Cox4 binds independently of the respiratory chain supercomplexes to mtHsp70-Mge1. (A) Mitochondria from wild-type (WT), Cox4His, and Cox4His mss51Δ cells were lysed in digitonin and analyzed by blue native electrophoresis and Western blotting. III, complex III; IV, complex IV of mitochondrial respiratory chain. (B) Wild-type (WT), Cox4His, and Cox4His mss51Δ mitochondria were lysed in digitonin and incubated with Ni2+-NTA agarose. Bound complexes were eluted with imidazole, and samples were analyzed by SDS–PAGE and Western blotting. Load, 3%; elution, 100%. F1β, β-subunit of F1Fo-ATP synthase. (C) Wild-type (WT), Cox6His, and Cox6His mss51Δ mitochondria were analyzed as described in B. (D) Wild-type (WT), mtHsp70His, and mtHsp70His mss51Δ mitochondria were analyzed as described in B. (E) Wild-type (WT), mtHps70His, mtHsp70His mss51Δ, and Cox4His mss51Δ mitochondria were lysed in digitonin and incubated with Ni2+-NTA agarose. Bound complexes were eluted with imidazole, and samples were analyzed by blue native electrophoresis and Western blotting. Load, 3%; elution, 100%.
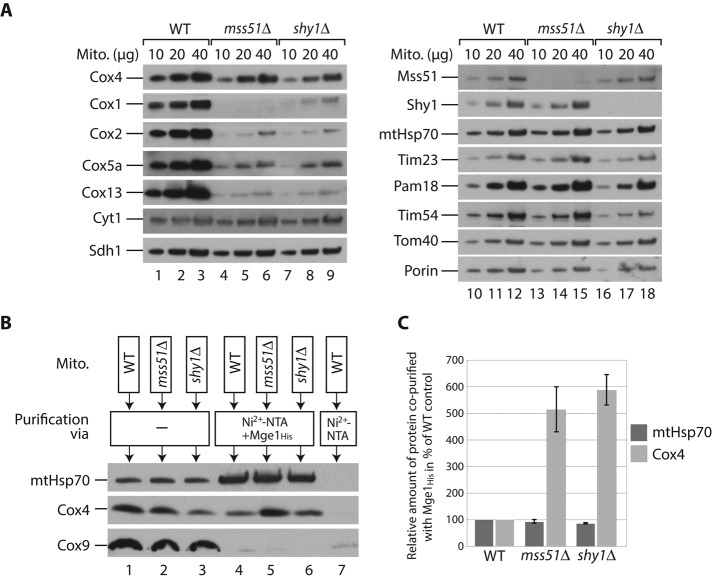
In mitochondria lacking the biogenesis factors Mss51 and Shy1, Cox4 protein levels are only mildly affected, whereas the levels of other subunits of complex IV are strongly reduced (Figure 3A). We asked whether unassembled Cox4 arrests at the chaperone complex in these mutants. Because the His tagging of Cox4 caused a moderate overproduction of the protein, we incubated solubilized wild-type, mss51Δ, or shy1Δ mitochondria with an Mge1 affinity matrix (Figure 3B; Schneider et al., 1996). MtHsp70 from wild-type and mutant mitochondria bound efficiently to the Mge1-coated matrix (Figure 3, B, lanes 4 and 6, and C). The amount of associated Cox4 relative to the load, however, was fivefold to sixfold increased in mss51Δ and shy1Δ compared with wild-type mitochondria (Figure 3C). Similarly, we observed a highly increased fraction of Cox4 bound to His-tagged mtHsp70 in the mss51Δ strain compared with the wild-type background (Figure 2D, lanes 11 and 12). Thus Cox4 arrests at mtHsp70-Mge1 when it cannot assemble into the mature complex IV.
FIGURE 3:
Cox4 arrests at mtHsp70-Mge1 in the absence of mature complex IV. (A) Wild-type (WT), mss51Δ, and shy1Δ mitochondria were lysed under denaturing conditions and analyzed by SDS–PAGE and Western blotting. (B) Wild-type (WT), mss51Δ, and shy1Δ mitochondria were lysed in digitonin and incubated with a noncoated or Mge1His-coated Ni2+-NTA matrix. Bound proteins were eluted with imidazole, and samples were analyzed by SDS–PAGE and Western blotting. Load (lanes 1–3), 3%; elution (lanes 4–7), 100%. (C) Quantification of three independent experiments like the one shown in B. Amount of coeluted Cox4 or mtHsp70, respectively, relative to the corresponding total mitochondrial protein content in the load fractions. Values for wild-type (WT) mitochondria were set to 100%. Error bars, SEM (n = 3).
A novel mutant of mtHsp70 is selectively impaired in Cox4 binding
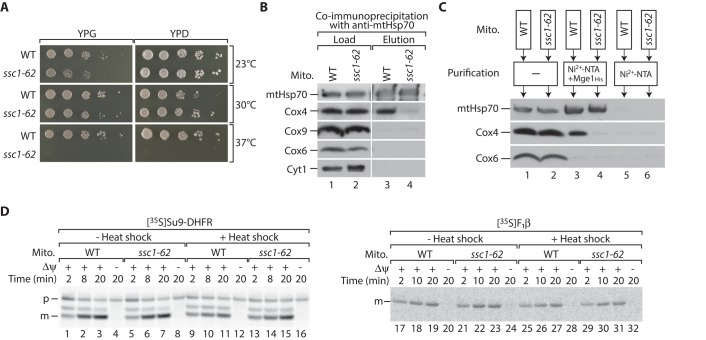
What is the function of the mtHsp70-Mge1-Cox4 association? To address this question, we generated novel temperature-sensitive mutants of the SSC1 gene (ssc1-ts) encoding mtHsp70 (Hutu et al., 2008). We screened this library of new ssc1-ts strains for a mutant that was selectively impaired in binding to Cox4. For this purpose, we isolated mitochondria of these strains grown under permissive conditions and tested the binding of the mtHsp70 variants to Cox4 by coimmunoprecipitation. We could identify one ssc1-ts mutant, termed ssc1-62, which shows mildly reduced growth on nonfermentable medium at 24 and 30°C and does not grow on either nonfermentable or fermentable media at 37°C (Figure 4A). In isolated mitochondria, association of Cox4 to the mutant Hsp70 is strongly reduced, as shown by coimmunoprecipitation with an mtHsp70-specific antibody (Figure 4B, lane 4). In contrast, binding of mtHsp70 to the Mge1 affinity matrix was only mildly affected (Figure 4C, lane 4). We tested the capability of the mutant mitochondria to import the 35S-labeled model precursor Su9-DHFR and the precursor of the β-subunit of F1Fo-ATP synthase (F1β). The import of both precursors was unaffected in the ssc1-62 mitochondria regardless of previous in vitro heat treatment (Figure 4D; in several temperature-sensitive mutants, an in vitro heat shock inactivates the gene product; Stojanovski et al., 2007). We conclude that the overall functionality of mtHsp70 was not compromised under these conditions, but the binding to Cox4 was specifically impaired in the ssc1-62 mutant mitochondria. Therefore we chose this strain for functional characterization of the mtHsp70-Mge1-Cox4 complex.
FIGURE 4:
A temperature-sensitive mutant of mtHsp70 (ssc1-62) is specifically defective in binding to Cox4. (A) Serial dilutions of wild-type (WT) and ssc1-62 yeast cells were spotted onto agar plates containing glucose (YPD) or glycerol (YPG) as carbon source. Plates were incubated at the indicated temperatures. (B) Isolated mitochondria from WT and ssc1-62 mutant cells were lysed in digitonin and incubated with anti-mtHsp70 antibodies coupled to protein A–Sepharose. Bound proteins were eluted with glycine, pH 2.5, and analyzed by SDS–PAGE and Western blotting. Load, 3%; elution, 100%. (C) Isolated mitochondria from WT and ssc1-62–mutant cells were lysed in digitonin and incubated with Mge1His-coated or uncoated Ni2+-NTA. Bound proteins were eluted with imidazole and analyzed by SDS–PAGE and Western blotting. Load, 3%; elution, 100%. (D) 35S-labeled Su9-DHFR (left) or F1β (right) was imported into WT and ssc1-62 mitochondria for the indicated time points with or without previous in vitro heat shock. In control reactions, the membrane potential (Δψ) was dissipated. Nonimported F1β precursor proteins were removed by treatment with proteinase K. Samples were subjected to SDS–PAGE and digital autoradiography. m, mature; p, precursor.
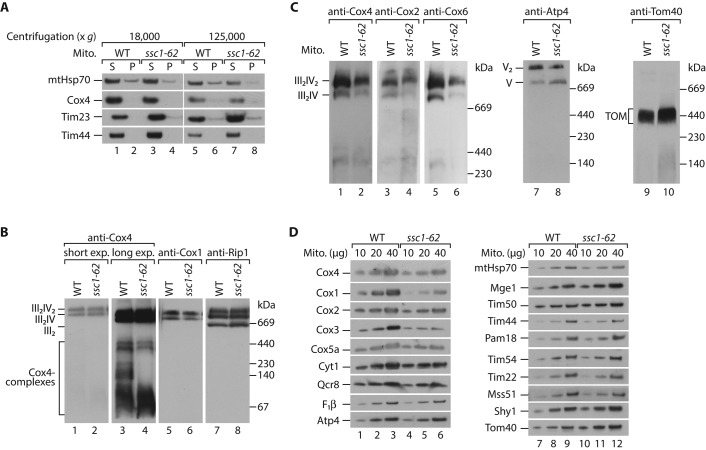
We used ssc1-62 mitochondria to address whether Cox4 aggregates upon loss of association to mtHsp70. To address this issue, we lysed mitochondria with digitonin and separated soluble and aggregated proteins by centrifugation. We could not detect any precipitated Cox4 in mitochondria isolated from the ssc1-62 mutant (Figure 5A). We asked whether the respiratory chain supercomplexes are affected in the mutant mitochondria. The levels of mitochondrial proteins, including Cox4 and other respiratory chain subunits, were not or only mildly altered in ssc1-62 mitochondria (Supplemental Figure S1). Respiratory chain supercomplexes containing a dimer of the cytochrome bc1 complex and one or two copies of the cytochrome c oxidase can be resolved by blue native electrophoresis upon mild solubilization with the nonionic detergent digitonin (Cruciat et al., 2000; Schägger and Pfeiffer, 2000; Zara et al., 2007; Böttinger et al., 2012). The formation of these complexes detected by antibodies against Cox4, Cox1, and Rip1 was similar to wild type in ssc1-62–mutant mitochondria (Figure 5B). Longer exposure revealed that Cox4 was also present in complexes of lower molecular weight, which is in line with earlier observations (Church et al., 2005). Some of these smaller Cox4-containing complexes of around 140–400 kDa were not detectable or less abundant in ssc1-62 mitochondria (Figure 5B, compare lanes 3 and 4) and might represent the identified mtHsp70-Mge1-Cox4 complex (Figure 2E). Thus under permissive conditions stable binding of Cox4 to the chaperone complex seems not to be critical for the formation of respiratory chain complexes. We speculated that the mtHsp70-Mge1-Cox4 complex may be required under stress conditions. To test this notion, we shifted ssc1-62 cells initially grown at low temperature to nonpermissive conditions (37°C) and isolated mitochondria for biochemical characterization. We performed the growth at restrictive conditions for a limited time to minimize secondary effects due to heat inactivation of the mutated mtHsp70. Under these conditions a small fraction of mtHsp70 and Mge1 aggregated in the mutant mitochondria (Supplemental Figure S2A). This aggregation of the essential chaperone system explains why the ssc1-62 strain did not grow on fermentable or nonfermentable medium when the cells were constantly incubated at 37°C (Figure 4A). The levels of the respiratory chain supercomplexes were considerably reduced, whereas the F1Fo-ATP synthase (complex V) and the TOM complex were unaffected (Figure 5C). The activity of complex IV was strongly impaired in the mutant mitochondria, whereas complex III activity was only moderately reduced (Supplemental Figure S2, B and C). The protein levels of several complex IV subunits, including Cox4, were substantially decreased in comparison to wild-type mitochondria (Figure 5D). In contrast, the levels of other mitochondrial proteins were less affected (Figure 5D). We conclude that under stringent conditions, the formation of an active complex IV is inhibited in ssc1-62 mitochondria.
FIGURE 5:
Respiratory chain supercomplexes are affected in ssc1-62 mitochondria. (A) Mitochondria from wild-type (WT) and ssc1-62 cells were lysed in digitonin and subjected to centrifugation at 18,000 × g or 125,000 × g, respectively. Supernatant (S) and pellet (P) fractions were separated and analyzed by SDS–PAGE and Western blotting. (B) Mitochondria isolated from WT and ssc1-62 cells grown under permissive conditions were lysed in digitonin and analyzed by blue native electrophoresis followed by Western blotting. III, complex III; IV, complex IV of mitochondrial respiratory chain. (C) WT and ssc1-62 cells were grown under permissive conditions and shifted to 37°C for 32 h. Mitochondria were isolated, and protein complexes were analyzed by blue native electrophoresis followed by Western blotting. TOM, translocase of outer membrane; V, complex V (F1Fo-ATP synthase). (D) Wild-type and ssc1-62 cells were grown under grown under permissive conditions and shifted to 37°C for 32 h. Isolated mitochondria were lysed under denaturing conditions and analyzed by SDS–PAGE and Western blotting.
mtHsp70-Mge1-Cox4 complex promotes assembly of respiratory chain supercomplexes
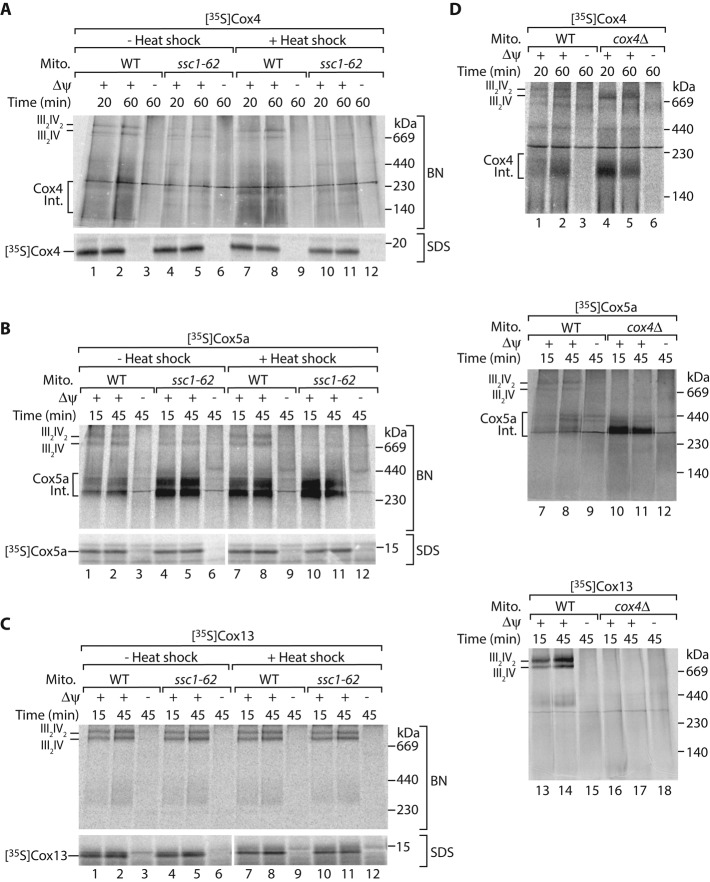
In mitochondria isolated from ssc1-62 cells grown under permissive conditions the interaction between mtHsp70 and Cox4 is specifically lost. We wondered whether the import or assembly of Cox4 is impaired in these mitochondria. We incubated 35S-labeled Cox4 precursors with isolated mitochondria and subsequently removed nonimported precursors by proteinase K treatment. We found that the uptake of the Cox4 precursor by ssc1-62 mitochondria was not affected (Figure 6A, bottom). To study the assembly of Cox4 into the mature complex IV, we imported the 35S-labeled Cox4 precursor into ssc1-62 mitochondria, solubilized the mitochondria with the mild detergent digitonin, and monitored Cox4 assembly on a blue native gel. Indeed the assembly of Cox4 into respiratory chain supercomplexes was impaired in ssc1-62 mitochondria (Figure 6A, top). We conclude that the association of mtHsp70 to Cox4 promotes the integration of imported Cox4 into the respiratory chain supercomplexes.
FIGURE 6:
mtHsp70-Mge1-Cox4 stimulates assembly of the respiratory supercomplexes. The 35S-labeled precursors of Cox4 (A), Cox5a (B), or Cox13 (C) were imported into wild-type (WT) or ssc1-62 mitochondria for the indicated time points with or without previous in vitro heat shock. In control reactions the membrane potential (Δψ) was dissipated. Samples were either lysed in digitonin and subjected to blue native electrophoresis (BN; top), or treated with proteinase K, denatured, and subjected to SDS–PAGE (bottom), followed by digital autoradiography. III, complex III; IV, complex IV of mitochondrial respiratory chain. (D) The 35S-labeled precursors of Cox4 (top), Cox5a (middle), or Cox13 (bottom) were imported into isolated mitochondria from WT and cox4Δ cells for the indicated time points in presence or absence of Δψ. Samples were lysed in digitonin and subjected to blue native electrophoresis followed by digital autoradiography. III, complex III; IV, complex IV of mitochondrial respiratory chain. Int., assembly intermediate.
Is mtHsp70 involved in the assembly of other nuclear-encoded complex IV subunits as well? To explore this possibility, we studied the assembly of Cox5a and Cox13 in ssc1-62 mutant mitochondria. Cox13 assembles directly into preformed respiratory supercomplexes, whereas imported Cox5a forms assembly intermediates, which can be detected by blue native electrophoresis (Brandner et al., 2005; Mick et al., 2007; Vukotic et al., 2012). Even though the import of both precursors into ssc1-62 mutant mitochondria occurred with wild-type efficiency (Figure 6, B and C, bottom), we observed delayed integration of Cox5a into the respiratory chain supercomplexes in the ssc1-62 mitochondria, whereas Cox13 assembly was not altered (Figure 6, B and C, top). Interestingly, we detected accumulation of Cox5a assembly intermediates in ssc1-62 (Figure 6B, lanes 1–6). Similar accumulation of imported Cox5a at an intermediate stage can be detected in mitochondria lacking Cox4 (Figure 6D, lanes 7–12; Mick et al., 2007). Formation of the Cox5a intermediates indicates that several complex IV subunits are already preassembled, and Cox4 is required to complete the biogenesis of respiratory chain complexes. To challenge this idea, we imported 35S-labeled precursors of Cox4 and Cox13 into cox4Δ mitochondria (Figure 6D). Imported Cox4 restored the assembly of the respiratory chain supercomplex consisting of one copy of complex IV and two copies of complex III (Figure 6D, lanes 4 and 5). In contrast, imported Cox13 did not restore the formation of respiratory chain supercomplexes in cox4Δ mitochondria (Figure 6D, lanes 13–18). These observations indicate that Cox4 promotes the assembly of mature complex IV from preassembled intermediates. Because the imported Cox5a accumulates at similar assembly intermediates in both cox4Δ and ssc1-62 mitochondria, we conclude that mtHsp70-Mge1 provides Cox4 for the formation of respiratory chain supercomplexes.
DISCUSSION
Assembly of the cytochrome c oxidase is a highly complicated process. Several biogenesis factors for mitochondria-encoded subunits have been identified (Mick et al., 2011). In contrast, little is known about the assembly of nuclear-encoded subunits. We show here that mtHsp70 promotes the assembly of the nuclear-encoded Cox4 and Cox5a proteins into the respiratory chain supercomplexes. mtHsp70 is also required for import of the Cox4 and Cox5a precursors (Scherer et al., 1990; Gärtner et al., 1995), indicating that the chaperone fulfills a dual function in the biogenesis of these complex IV subunits. Several reported observations indicate that also other components involved in protein translocation might play a role in the assembly of cytochrome c oxidase. The TIM23 translocase, as well as Pam16 and Pam18, associates with respiratory supercomplexes (van der Laan et al., 2006; Wiedemann et al., 2007). Recently an involvement of the human TIM23 translocase component Tim21 in the assembly of complex IV subunits was reported (Mick et al., 2012).
The mtHsp70 stimulates assembly of Cox4 by formation of an intermediate complex with Cox4 and Mge1. We identified a novel temperature-sensitive mutant of mtHsp70 that is selectively impaired in the formation of the Cox4-containing assembly intermediate but not in the translocation of the Cox4 precursor across the inner mitochondrial membrane. This observation indicates that the interaction of mtHsp70 and Cox4 within the intermediate complex represents a protein import–independent function of mtHsp70. It remains to be determined whether different domains of mtHsp70 mediate substrate binding during protein import and the association with Cox4 in the newly identified assembly intermediate. An ATP dependence of Hsp70 interactions, as shown here for Cox4 binding, has been found both for client proteins that bind to the canonical peptide-binding pocket (Kampinga and Craig, 2010; Marom et al., 2011; Voos, 2013) and for regulatory partner proteins, like Tim44 or Mge1, that bind to different regions of mtHsp70 (Figure 1D; Voos et al., 1994; Schneider et al., 1996; Strub et al., 2002).
Our assembly assay using isolated mitochondria reveals that the mtHsp70-Mge1-Cox4 complex promotes the efficient incorporation of both Cox4 and Cox5a into complex IV. This mtHsp70-Mge1-Cox4 interaction does not seem to be the limiting step in the formation of respiratory chain supercomplexes in vivo when cells are grown under standard laboratory conditions but likely contributes to efficient assembly of active complex IV under more stringent conditions such as heat stress (Figure 5C and Supplemental Figure S2). Because Cox4 assembly is critical for the formation of mature, active cytochrome c oxidase (Frazier et al., 2006; Coyne et al., 2007; Mick et al., 2007), we propose that the mtHsp70-Mge1-Cox4 complex serves as a backup system to allow fast delivery of Cox4 to the assembly line of the cytochrome c oxidase when needed. This idea is supported by the observation that Cox4 arrests at the chaperone complex in the absence of mature cytochrome c oxidase. Notably, in vitro import of Cox4 into mitochondria lacking Cox4 is sufficient to trigger the formation of a respiratory chain supercomplex from complex III and preassembled complex IV building blocks (Figure 6D). In the absence of Cox4, imported Cox5a accumulates in similar assembly intermediates as in mitochondria isolated from the temperature-sensitive mutant of mtHsp70 lacking the association of Cox4 to mtHsp70. Based on these findings, we conclude that the efficient channeling of Cox4 into the complex IV assembly line via the mtHsp70-Mge1-Cox4 complex prevents Cox4 supply from becoming limiting for complex IV biogenesis under stress conditions.
In addition, mtHsp70 acts in the biogenesis of the mitochondria-encoded subunit Cox1 (Herrmann et al., 1994; Fontanesi et al., 2010). Here the chaperone binds to Mss51, the translational activator of Cox1, and might stimulate the folding of Cox1 (Fontanesi et al., 2010). We show that the mtHsp70-Mge1-Cox4 complex forms independently of Mss51, demonstrating that mtHsp70 promotes the biogenesis of complex IV at two different steps. Thus mtHsp70 fulfills central functions in the formation of respiratory chain complexes. Defects of the human homologue of mtHsp70, mortalin, have been reported to play a role in carcinogenesis and Parkinson's disease (Czarnecka et al., 2006; Deocaris et al., 2008; Burbulla et al., 2010; Goswami et al., 2012). The function of mtHsp70 in the assembly of respiratory chain complexes might have important implications for its role in the pathogenesis of these severe human disorders.
MATERIALS AND METHODS
Yeast strains, growth conditions, and isolation of mitochondria
The yeast strains mtHsp70His, cox4Δ, and YPH499 arg4Δ have been described before (Truscott et al., 2003; Frazier et al., 2006; von der Malsburg et al., 2011). The strains mss51Δ and shy1Δ and the corresponding BY4741 wild type were obtained from the European Saccharomyces cerevisiae Archive for Functional Analysis (Frankfurt, Germany). Yeast strains expressing Cox4His and Cox6His were generated by chromosomal integration of a HIS3MX6 cassette encoding for a deca-histidine tag using gene-specific primers (Meisinger et al., 2001). For SILAC analysis, a kanMX4 cassette was integrated into the ARG4 locus of a Cox4His strain by homologous recombination. For the generation of the temperature-sensitive strain ssc1-62, the SSC1 locus was disrupted with an ADE2 marker in the presence of the plasmid pYEp352 encoding for wild-type SSC1 and a URA3 selection marker. This strain was transformed with a second plasmid carrying the mutant SSC1-62 allele and the TRP selection marker. The SSC1-62 allele was produced by error-prone PCR. Subsequently yeast cells were grown on 5-fluoroorotic acid–containing medium to select cells that lost the plasmid encoding for the wild-type version of SSC1 and the URA3 marker (Hutu et al., 2008). The generation of the corresponding wild-type strain followed the same procedure, except that the remaining plasmid contains a wild-type copy of SSC1. The strains were grown to logarithmic growth phase at 23 or 30°C on YP medium (1% [wt/vol] yeast extract, 2% [wt/vol] bacto peptone) containing 3% (vol/vol) glycerol or 2% (wt/vol) galactose. For in vivo heat treatment of ssc1-62 cells, yeast cultures grown at permissive conditions were shifted for 32 h to 37°C. Mitochondria were isolated by differential centrifugation, resuspended in SEM buffer (10 mM 3-(N-morpholino)propanesulfonic acid [MOPS]/KOH, pH 7.2, 1 mM EDTA, 250 mM sucrose) at a protein concentration of 10 mg/ml, aliquoted, shock frozen in liquid nitrogen, and stored at −80°C until use (Meisinger et al., 2006).
Protein import into isolated mitochondria
Precursor proteins were synthesized in the presence of [35S]methionine in rabbit reticulocyte lysate (Promega, Madison, WI). For import reactions, 35S-labeled precursors (5–10% of the reaction volume) were incubated with mitochondria at 25°C in import buffer (3% [wt/vol] bovine serum albumin [BSA], 250 mM sucrose, 5 mM methionine, 80 mM KCl, 5 mM MgCl2, 10 mM MOPS/KOH, pH 7.2, 2 mM KH2PO4) in the presence of 2 mM ATP, 2 mM NADH, 5 mM creatine phosphate, and 0.1 mg/ml creatine kinase for different time points. Import reactions were stopped by dissipation of the membrane potential through the addition of an AVO mixture (8 μM antimycin A, 1 μM valinomycin, 20 μM oligomycin final concentrations; Stojanovski et al., 2007). In control reactions, the membrane potential was dissipated before import by the addition of AVO mix. Where indicated, samples were treated with 50 μg/ml proteinase K to remove nonimported precursor proteins. For analysis by blue native electrophoresis, mitochondria were washed with SEM buffer and solubilized in lysis buffer (20 mM Tris/HCl, pH 7.4, 50 mM NaCl, 10% [vol/vol] glycerol, 0.1 mM EDTA) containing 1% (wt/vol) digitonin.
Affinity purification assays
Isolated mitochondria were solubilized in lysis buffer containing 1% (wt/vol) digitonin and 10 mM imidazole at a final protein concentration of 1 mg/ml and incubated with Ni2+-NTA agarose (Qiagen, Hilden, Germany) under constant rotation at 4°C. The Ni2+-NTA agarose beads were reisolated and washed intensively with lysis buffer containing 0.1% (wt/vol) digitonin and 40 mM imidazole. Bound proteins were eluted with 250 mM imidazole and 0.1% (wt/vol) digitonin in lysis buffer. For coimmunoprecipitation, anti-mtHsp70 antibodies were coupled covalently to protein A–Sepharose (GE Healthcare, Little Chalfront, UK) with dimethylpimelidate. Mitochondria were solubilized in lysis buffer containing 1% (wt/vol) digitonin at a final protein concentration of 1 mg/ml and incubated with the anti-mtHsp70 beads under constant rotation at 4°C. After excessive washing of the beads with lysis buffer containing 0.1% (wt/vol) digitonin, bound proteins were eluted with 0.1 M glycine, pH 2.5. Samples were analyzed on blue native electrophoresis or denatured and analyzed on SDS–PAGE.
SILAC-based affinity purification and analysis by mass spectrometry
For SILAC analysis, YPH499 arg4Δ (wild type) and Cox4His arg4Δ were grown in defined synthetic medium (0.67% [wt/vol] bacto-yeast nitrogen base, amino acid mix, 3% [vol/vol] glycerol, 0.2% [wt/vol] glucose), supplemented with either [15N213C6]lysine and [15N413C6]arginine (Euriso-Top, Saint-Aubin, France) for wild-type or [14N212C6]lysine and [14N412C6]arginine for Cox4His arg4Δ cells (von der Malsburg et al., 2011). Mitochondria were isolated, proteins were purified as described, and the elution fractions were pooled.
The elution samples were acetone precipitated and tryptically digested in 20 mM NH4HCO3 solved in 60% (vol/vol) methanol. Resulting peptide mixtures were separated by ultra high-performance liquid chromatography (HPLC) on an UltiMate 3000 RSLCnano HPLC system (Thermo Scientific Dionex, Idstein, Germany), which was directly coupled to an LTQ-Orbitrap XL instrument (Thermo Scientific, Bremen, Germany) for electrospray ionization tandem mass spectrometry (MS/MS) analyses. Liquid chromatography MS/MS experiments of three independent replicates were performed as described (Niemann et al., 2013), with slight modifications. Peptides were concentrated and washed on a C18 precolumn (Acclaim PepMap μ-Precolumn Cartridge; 0.3 mm × 5 mm; particle size, 5 μm; Thermo Scientific Dionex) using 0.1% (vol/vol) trifluoroacetic acid at a flow rate of 30 μl/min. Subsequently the peptides were separated on a 50 cm × 75 μm C18-column (Acclaim PepMap RSLC column; particle size, 2 μm; pore size, 100 Å; Thermo Scientific Dionex) applying a 125-min gradient of 4–26% (vol/vol) acetonitrile (ACN) in 0.1% (vol/vol) formic acid (FA) followed by 26–43% ACN/0.1% FA in 40 min (flow rate, 300 nl/min). High-resolution MS spectra ranging from m/z = 370 to 1700 were acquired in the Orbitrap (resolution, 60,000 at m/z = 400; automatic gain control, 5 × 105 ions; maximum fill time, 500 ms), and up to six of the most intense peptide ions (charge state ≥+2) were further fragmented by collision-induced dissociation in the linear ion trap (normalized collision energy, 35%; automatic gain control, 10,000 ions; maximum fill time, 400 ms). Previously selected precursor ions were dynamically excluded from fragmentation for 65 s.
Mass spectrometric data analysis, including protein identification and SILAC-based relative protein quantification, was performed using MaxQuant, version 1.2.0.18 (Cox and Mann, 2008), and its integrated search engine Andromeda (Cox et al., 2011). For peptide and protein identification, MS/MS data were correlated with the Saccharomyces cerevisiae genome database (www.yeastgenome.org). The database search was performed with the following settings: mass tolerances of 7 ppm for precursor and 0.5 D for fragment ions; tryptic specificity with a maximum of two missed cleavages; oxidation of methionine and acetylation of protein N-termini as variable modifications; [15N213C6]lysine and [15N413C6]arginine as heavy labels; at least one unique peptide with a minimum of six amino acids; and false discovery rate of <1% on both peptide and protein levels. The determination of protein abundance ratios (light/heavy [L/H]) was based on unique peptides with ≥1 ratio count. Low-scoring variants of identified peptides were excluded from quantification. L/H protein ratios were log10 transformed, and the mean log10 ratio across all replicates, as well as the p value (calculated using a one-sided t test) of all proteins quantified in ≥2 replicates, was determined. Proteins exhibiting a sequence coverage of ≥4%, a posterior error probability (PEP) of <0.01, and a mean L/H ratio of >10 were considered potential Cox4 interaction partners.
Mge1 binding assay
Mge1 lacking the putative presequence was fused to a His tag, recombinantly expressed, and purified as described (Dekker and Pfanner, 1997). Purified Mge1His was coupled to Ni2+-NTA agarose (Qiagen). Mitochondria were lysed in 1% (wt/vol) digitonin and 0.1% (wt/vol) BSA in binding buffer (30 mM Tris/HCl, pH 7.4, 80 mM KCl, 5% [vol/vol] glycerol, 5 mM MgCl2) at a final protein concentration of 1 mg/ml and incubated with the Mge1 affinity matrix for 1 h under constant rotation at 4°C. After excessive washing of the beads with binding buffer containing 0.1% (wt/vol) digitonin and 40 mM imidazole, bound proteins were eluted with 250 mM imidazole and 0.1% (wt/vol) digitonin in binding buffer. Samples were denatured and analyzed on SDS–PAGE.
Protein aggregation assay
Mitochondria were solubilized in lysis buffer containing 1% (wt/vol) digitonin and incubated on ice for 30 min, followed by centrifugation for 45 min at 4°C at 18,000 × g or 125,000 × g, respectively. The precipitated supernatant and pellet fractions were denatured and subjected to SDS–PAGE and Western blotting.
Determination of mitochondrial enzyme activities
Activities of cytochrome c oxidase and cytochrome bc1 complex were assayed spectrophotometrically as described previously (Vukotic et al., 2012). For cytochrome c oxidase activity measurements, the assay system contained 10 mM Tris/HCl, pH 7.4, 120 mM KCl, 0.01 mM reduced cytochrome c, and 20 μg of mitochondria lysed in 0.5% (vol/vol) Triton X-100. For cytochrome bc1 complex activity measurements, the assay system contained 10 mM Tris/HCl, pH 7.4, 120 mM KCl, 0.01 mM oxidized cytochrome c, 0.5 mM NADH, 0.1 mM KCN, and 20 μg of mitochondria.
Miscellaneous
The 35S-labeled proteins and protein complexes were detected by digital autoradiography (Storm imaging system; GE Healthcare) and analyzed using ImageQuant 5.2 software (GE Healthcare). After Western blotting and immunodecoration with the indicated antibodies, the proteins were detected using a chemiluminescence kit (ECL; GE Healthcare). Quantification of Western blot signals was performed with ImageQuant 5.2 software.
Supplementary Material
Acknowledgments
We thank Nikolaus Pfanner and Peter Rehling for discussion and Conny Schütze for material. We thank Inge Perschil and Kurt Lobenwein for expert technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 746, the Excellence Initiative of the German Federal and State Governments (EXC 294 BIOSS), and the Alexander von Humboldt Stiftung.
Abbreviations used:
- Cox4
cytochrome c oxidase subunit 4
- Mdj1
mitochondrial DnaJ
- Mge1
mitochondrial GrpE
- mtHsp70
mitochondrial Hsp70
- PAM
presequence translocase–associated motor
- TIM
translocase of the inner membrane
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-02-0106) on July 17, 2013.
REFERENCES
- Baker MJ, Frazier AE, Gulbis JM, Ryan MT. Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol. 2007;17:456–464. doi: 10.1016/j.tcb.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttinger L, Horvath SE, Kleinschroth T, Hunte C, Daum G, Pfanner N, Becker T. Phosphatidylethanolamine and cardiolipin differentially affect the stability of mitochondrial respiratory chain supercomplexes. J Mol Biol. 2012;423:677–686. doi: 10.1016/j.jmb.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner K, Mick DU, Frazier AE, Taylor RD, Meisinger C, Rehling P. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth syndrome. Mol Biol Cell. 2005;16:5202–5214. doi: 10.1091/mbc.E05-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulla LF, et al. Dissecting the role of the mitochondrial chaperone mortalin in Parkinson's disease: functional impact of disease-related variants on mitochondrial homeostasis. Hum Mol Genet. 2010;19:4437–4452. doi: 10.1093/hmg/ddq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church C, Goehring B, Forsha D, Wazny P, Poyton RO. A role for Pet100p in the assembly of yeast cytochrome c oxidase: interaction with a subassembly that accumulates in a pet100 mutant. J Biol Chem. 2005;280:1854–1863. doi: 10.1074/jbc.M410726200. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Coyne HJ III, Ciofi-Baffoni S, Banci L, Bertini I, Zhang L, George GN, Winge DR. The characterization and role of zinc binding in yeast Cox4. J Biol Chem. 2007;282:8926–8934. doi: 10.1074/jbc.M610303200. [DOI] [PubMed] [Google Scholar]
- Cruciat C-M, Brunner S, Baumann F, Neupert W, Stuart RA. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J Biol Chem. 2000;275:18093–18098. doi: 10.1074/jbc.M001901200. [DOI] [PubMed] [Google Scholar]
- Czarnecka AM, Campanella C, Zummo G, Cappello F. Mitochondrial chaperones in cancer: from molecular biology to clinical diagnostics. Cancer Biol Ther. 2006;5:714–720. doi: 10.4161/cbt.5.7.2975. [DOI] [PubMed] [Google Scholar]
- Decoster E, Simon M, Hatat D, Faye G. The MSS51 gene product is required for the translation of the COX1 mRNA in yeast mitochondria. Mol Gen Genet. 1990;224:111–118. doi: 10.1007/BF00259457. [DOI] [PubMed] [Google Scholar]
- Dekker PJ, Pfanner N. Role of mitochondrial GrpE and phosphate in the ATPase cycle of matrix Hsp70. J Mol Biol. 1997;270:321–327. doi: 10.1006/jmbi.1997.1131. [DOI] [PubMed] [Google Scholar]
- Deocaris CC, Kaul SC, Wadhwa R. From proliferative to neurological role of an hsp70 stress chaperone, mortalin. Biogerontology. 2008;9:391–403. doi: 10.1007/s10522-008-9174-2. [DOI] [PubMed] [Google Scholar]
- Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K. Multiple pathways for mitochondrial protein traffic. Biol Chem. 2009;390:723–730. doi: 10.1515/BC.2009.087. [DOI] [PubMed] [Google Scholar]
- Fontanesi F, Clemente P, Barrientos A. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J Biol Chem. 2011;286:555–566. doi: 10.1074/jbc.M110.188805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F, Soto IC, Horn D, Barrientos A. Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol Cell Biol. 2010;30:245–259. doi: 10.1128/MCB.00983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox TD. Mitochondrial protein synthesis, import and assembly. Genetics. 2012;192:1203–1234. doi: 10.1534/genetics.112.141267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier AE, Taylor RD, Mick DU, Warscheid B, Stoepel N, Meyer HE, Ryan MT, Guiard B, Rehling P. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J Cell Biol. 2006;172:553–564. doi: 10.1083/jcb.200505060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner F, Voos W, Querol A, Miller BR, Craig EA, Cumsky MG, Pfanner N. Mitochondrial import of subunit Va of cytochrome c oxidase characterized with yeast mutants. J Biol Chem. 1995;270:3788–3795. doi: 10.1074/jbc.270.8.3788. [DOI] [PubMed] [Google Scholar]
- Goswami AV, Samaddar M, Sinha D, Purushotham J, D´Silva P. Enhanced J-protein interaction and compromised protein stability of mtHsp70 variants lead to mitochondrial dysfunction in Parkinson´s disease. Hum Mol Genet. 2012;21:3317–3332. doi: 10.1093/hmg/dds162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Funes S. Biogenesis of cytochrome oxidase—sophisticated assembly lines in the mitochondrial inner membrane. Gene. 2005;354:43–52. doi: 10.1016/j.gene.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Stuart RA, Craig EA, Neupert W. Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J Cell Biol. 1994;127:893–902. doi: 10.1083/jcb.127.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan S, Bourges I, Taanman J-W, Meunier B. Analysis of Cox2 mutants reveals cytochrome oxidase subassemblies in yeast. Biochem J. 2005;390:703–708. doi: 10.1042/BJ20050598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst M, Oppliger W, Rospert S, Schönfeld HJ, Schatz G, Azem A. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J. 1997;16:1842–1849. doi: 10.1093/emboj/16.8.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutu DP, Guiard B, Chacinska A, Becker D, Pfanner N, Rehling P, van der Laan M. Mitochondrial protein import motor: differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase. Mol Biol Cell. 2008;19:2642–2649. doi: 10.1091/mbc.E07-12-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, D´Silva P, Walter W, Marszalek J, Craig EA. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science. 2003;300:139–141. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
- Mapa K, Sikor M, Kudryavtsev V, Waegemann K, Kalinin S, Seidel CAM, Neupert W, Lamb DC, Mokranjac D. The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol Cell. 2010;38:89–100. doi: 10.1016/j.molcel.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Marom M, Azem A, Mokranjac D. Understanding the molecular mechanism of protein translocation across the mitochondrial inner membrane: still a long way to go. Biochim Biophys Acta. 2011;1808:990–1001. doi: 10.1016/j.bbamem.2010.07.011. [DOI] [PubMed] [Google Scholar]
- McStay GP, Su CH, Tzagoloff A. Modular assembly of yeast cytochrome oxidase. Mol Biol Cell. 2013;24:440–452. doi: 10.1091/mbc.E12-10-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN. Isolation of yeast mitochondria. Methods Mol Biol. 2006;313:33–39. doi: 10.1385/1-59259-958-3:033. [DOI] [PubMed] [Google Scholar]
- Meisinger C, Ryan MT, Hill K, Model K, Lim JH, Sickmann A, Müller H, Meyer HE, Wagner R, Pfanner N. Protein import channel of the outer mitochondrial membrane: a highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small tom proteins, and import receptors. Mol Cell Biol. 2001;21:2337–2348. doi: 10.1128/MCB.21.7.2337-2348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Fox TD, Rehling P. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat Rev Mol Cell Biol. 2011;12:14–20. doi: 10.1038/nrm3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Vukotic M, Piechura H, Meyer HE, Warscheid B, Deckers M, Rehling P. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J Cell Biol. 2010;191:141–154. doi: 10.1083/jcb.201007026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, Wagner K, van der Laan M, Frazier AE, Perschil I, Pawlas M, Meyer HE, Warscheid B, Rehling P. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 2007;26:4347–4358. doi: 10.1038/sj.emboj.7601862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick DU, et al. MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell. 2012;151:1528–1541. doi: 10.1016/j.cell.2012.11.053. [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Niemann M, Wiese S, Mani J, Chanfon A, Jackson C, Meisinger C, Warscheid B, Schneider A. Mitochondrial outer membrane proteome of Trypanosoma brucei reveals novel factors required to maintain mitochondrial morphology. Mol Cell Proteomics. 2013;12:515–528. doi: 10.1074/mcp.M112.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Ott M, Herrmann JM. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta. 2010;1803:767–775. doi: 10.1016/j.bbamcr.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Perez-Martinez X, Broadley SA, Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez X, Butler CA, Shingu-Vazquez M, Fox TD. Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol Biol Cell. 2009;20:4371–4380. doi: 10.1091/mbc.E09-06-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel F, Bestwick ML, Cobine PA, Khalimonchuk O, Cricco JA, Winge DR. Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 2007;26:4335–4346. doi: 10.1038/sj.emboj.7601861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W. Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell. 1994;77:249–259. doi: 10.1016/0092-8674(94)90317-4. [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer PE, Krieg UC, Hwang ST, Vestweber D, Schatz G. A precursor protein partially translocated into yeast mitochondria is bound to a 70 kd mitochondrial stress protein. EMBO J. 1990;9:4315–4322. doi: 10.1002/j.1460-2075.1990.tb07880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H-C, Westermann B, Neupert W, Brunner M. The nucleotide exchange factor MGE exerts a key function in the ATP-dependent cycle of mt-Hsp70-Tim44 interaction driving mitochondrial protein import. EMBO J. 1996;15:5796–5803. [PMC free article] [PubMed] [Google Scholar]
- Soto IC, Fontanesi F, Liu J, Barrientos A. Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim Biophys Acta. 2012a;1817:883–897. doi: 10.1016/j.bbabio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto IC, Fontanesi F, Myera RS, Hamel P, Barrientos A. A heme-sensing mechanism in the translational regulation of mitochondrial cytochrome c oxidase biogenesis. Cell Metabol. 2012b;16:801–813. doi: 10.1016/j.cmet.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D, Pfanner N, Wiedemann N. Import of proteins into mitochondria. Methods Cell Biol. 2007;80:783–806. doi: 10.1016/S0091-679X(06)80036-1. [DOI] [PubMed] [Google Scholar]
- Strub A, Röttgers K, Voos W. The Hsp70 peptide-binding domain determines the interaction of the ATPase domain with Tim44 in mitochondria. EMBO J. 2002;21:2626–2635. doi: 10.1093/emboj/21.11.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott KN, et al. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J Cell Biol. 2003;163:707–713. doi: 10.1083/jcb.200308004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr Biol. 2006;16:2271–2276. doi: 10.1016/j.cub.2006.10.025. [DOI] [PubMed] [Google Scholar]
- von der Malsburg K, et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Voos W. Chaperone-protease networks in mitochondrial protein homeostasis. Biochim Biophys Acta. 2013;1833:388–399. doi: 10.1016/j.bbamcr.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Voos W, Gambill BD, Laloraya S, Ang D, Craig EA, Pfanner N. Mitochondrial GrpE is present in a complex with hsp70 and preproteins in transit across membranes. Mol Cell Biol. 1994;14:6627–6634. doi: 10.1128/mcb.14.10.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukotic M, et al. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15:336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Westermann B, Gaume B, Herrmann JM, Neupert W, Schwarz E. Role of the mitochondrial DnaJ homolog Mdj1p as a chaperone for mitochondrially synthesized and imported proteins. Mol Cell Biol. 1996;16:7063–7071. doi: 10.1128/mcb.16.12.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann N, van der Laan M, Hutu DP, Rehling P, Pfanner N. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J Cell Biol. 2007;179:115–1122. doi: 10.1083/jcb.200709087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winge DR. Sealing the mitochondrial respirasome. Mol Cell Biol. 2012;32:2647–2652. doi: 10.1128/MCB.00573-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zara V, Conte L, Trumpower BL. Identification and characterization of cytochrome bc1 subcomplexes in mitochondria from yeast with single and double deletions of genes encoding cytochrome bc1 subunits. FEBS J. 2007;274:4526–4539. doi: 10.1111/j.1742-4658.2007.05982.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.