This research highlights the multifunctional role of uPA in pancreatic cancer as a regulator of stemness and chemoresistance. uPA is found to be physically associated with the transcription factors Lhx2 and HOXA5 in the nucleus and regulated expression of p53.
Abstract
Pancreatic ductal adenocarcinoma (PDAC) is almost always lethal. One of the underlying reasons for this lethality is believed to be the presence of cancer stem cells (CSC), which impart chemoresistance and promote recurrence, but the mechanisms responsible are unclear. Recently the poor prognosis of PDAC has been correlated with increased expression of urokinase plasminogen activator (uPA). In the present study we examine the role of uPA in the generation of PDAC CSC. We observe a subset of cells identifiable as a side population (SP) when sorted by flow cytometry of MIA PaCa-2 and PANC-1 pancreatic cancer cells that possess the properties of CSC. A large fraction of these SP cells are CD44 and CD24 positive, are gemcitabine resistant, possess sphere-forming ability, and exhibit increased tumorigenicity, known characteristics of cancer stemness. Increased tumorigenicity and gemcitabine resistance decrease after suppression of uPA. We observe that uPA interacts directly with transcription factors LIM homeobox-2 (Lhx2), homeobox transcription factor A5 (HOXA5), and Hey to possibly promote cancer stemness. uPA regulates Lhx2 expression by suppressing expression of miR-124 and p53 expression by repressing its promoter by inactivating HOXA5. These results demonstrate that regulation of gene transcription by uPA contributes to cancer stemness and clinical lethality.
INTRODUCTION
Pancreatic adenocarcinoma is the fourth-most-common cause of cancer deaths in the United States. Despite new insights into the molecular profile of pancreatic cancer and its precursor lesions and advances in diagnosis and therapy, survival rates have changed little over the past 40 yr. Major hallmarks of pancreatic cancer are extensive local tumor invasion, early systemic dissemination, and extremely poor response to chemotherapy and radiation treatment. The basis for these adverse features is not well understood. Emerging evidence suggests that the capability of tumors to grow, propagate, and recur may depend on an initially small subset of cells within a tumor, called cancer stem cells (CSC) or cancer-initiating cells. CSC, like normal stem cells, can both self-renew and produce differentiated progeny. The stem cell phenotype is associated with “en bloc” silencing of cell cycle–inhibitor genes (Nguyen et al., 2012). The resistance of pancreatic cancer to treatment and the high rate of recurrence have been attributed to a highly tumorigenic CSC subpopulation expressing cell surface CD44, CD24, CD133, and epithelial-specific antigen. Du et al. (2011) demonstrated that the chemoresistance of pancreatic cancer cells correlates with the expression of cell surface markers similar to those present on CSC that undergo epithelial–mesenchymal transition (EMT; Lonardo et al., 2010; Moriyama et al., 2010; Rausch et al., 2010). Urokinase plasminogen activator (uPA) expression correlates with increased number of EMTs in cancer cells, linking uPA to emergence of CSC and the resultant chemoresistance (Chen et al., 2009). Down-regulating uPA expression by silencing Ets-1 transcription factors sensitizes pancreatic cancer cells to gemcitabine-induced apoptosis (Khanna et al., 2011). Furthermore, uPA and uPA receptor (uPAR) are strong independent prognostic indicators of cancer relapse after primary therapy and indicative of metastatic potential, advanced stage, and poor prognosis (Watabe et al., 1998; Kato et al., 2012). Serum levels of uPA are elevated in patients with pancreaticobiliary cancer (Harvey et al., 2003), and overexpression of uPA in tumors is associated with shorter survival times (Gibbs et al., 2009). We previously demonstrated that silencing uPA and uPAR inhibits the growth of pancreatic tumors in animal models (Gorantla et al., 2011). Inhibitors of uPA's catalytic activity and antagonists of its binding to uPAR have shown only partial success, however, in animal model studies and clinical trials (Mekkawy et al., 2009; Lund et al., 2011; Mazar et al., 2011). This outcome suggests that approaches targeting only the proteolytic or receptor-binding functions of uPA are suboptimal and critical functions of uPA in cancer progression have been overlooked. We also previously reported that uPA is internalized by proliferating cells and rapidly transported to cell nuclei (Stepanova et al., 2008; Gorantla et al., 2011). Within the nucleus of pancreatic cancer cells, uPA binds to a LIM homeobox-2 (Lhx2) transcription factor (Gorantla et al., 2011). Lhx2 helps to maintain stem/progenitor cell phenotype and EMT in vitro and in vivo (Pinto do et al., 2001; Richter et al., 2003; Tiede and Paus, 2006; Williams et al., 2006; Kim et al., 2012; Nadal et al., 2012). Lhx2 is up-regulated in several cancer transcriptome databases in mouse models of breast cancer (Chou and Yang, 2006) and neuroendocrine carcinoma (Zhao et al., 2010; Perez et al., 2012).
Together these data led us to investigate whether nuclear uPA contributes to maintenance of stemness in pancreatic cancer cells by binding and regulating Lhx2. Here we provide evidence that nuclear uPA promotes pancreatic cancer cell stemness by binding directly to specific homeobox transcription factors. We studied the mechanism of interplay between uPA and tumor-suppressive microRNA (miRNA) miR124 in regulating Lhx2 expression, which is linked to maintenance of pancreatic cancer cell stemness and chemoresistance. We demonstrate that uPA also down-regulates expression of the tumor suppressor p53 by binding to and interfering with the function of homeobox transcription factor A5 (HOXA5), which further contributes to acquisition and maintenance of stemness in pancreatic cancer cells. Our data suggest that targeting nuclear uPA and its binding to the homeobox transcription factors may sensitize pancreatic cancer cells to chemotherapy-induced apoptosis and therefore has the potential to significantly improve treatment outcomes.
RESULTS
Side population of pancreatic cancer cells shows enhanced stem cell–like properties and uPA expression
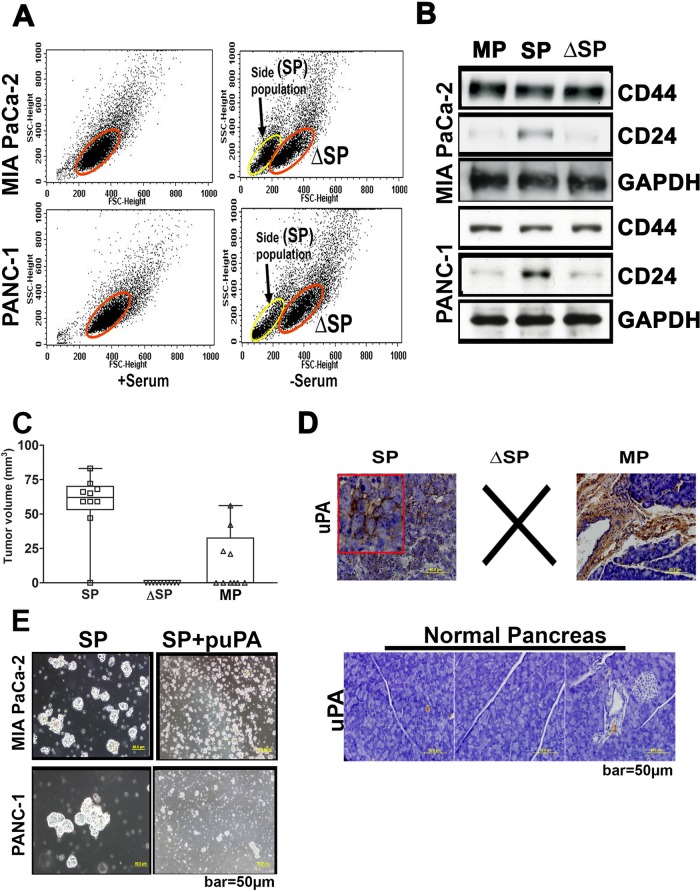
Side-population (SP) cells play a crucial role in tumorigenesis and cancer recurrence (Zhang et al., 2013). We first determined whether the side populations of pancreatic cancer cells we studied contain subpopulations of stem-like cells. Because culturing cancer cells under serum-free condition promotes the growth of cancer stem cells (Gou et al., 2007), we cultured MIA PaCa-2 and PANC-1 cells in complete or serum-free media with appropriate growth factors. We then detached the cells with trypsin and sorted them for density and size by standard flow cytometry. Cells cultured under serum-free conditions showed a side population of cells (25–36%) with lower density and size (Figure 1A) that characterize the CSC phenotype (Gou et al., 2007). To confirm this inference, protein extracts from the sorted populations of MIA PaCa-2 and PANC-1 cells grown under serum-free conditions were immunoprobed for the known cancer stem cell markers CD44 and CD24 (Lonardo et al., 2010; Moriyama et al., 2010; Rausch et al., 2010). The SP cells were positive for both CD44 and CD24, whereas the “residual” cells were positive only for CD44 (Figure 1B). These data indicate that the SP cells possess the cancer stem cell surface phenotype (Supplemental Figure S1). To further validate the stem cell character of MIA PaCa-2 SP cells and SP–depleted cells (ΔSP), we implanted these cells subcutaneously in nude mice (10,000 cells per mouse). The inoculates were allowed to grow for 40 d and then scored for the presence or absence of measurable (>1 mm in size) tumors. We observed that in 9 of 10 mice implanted with SP cells, tumors became visually evident within 40 d, whereas none of the mice implanted with CD24-negative cells (10,000 ΔSP cells) formed tumors over that time. When implanted with mixed population (MP) of MIA PaCa-2 cells (10,000 cells/mouse), 4 of 10 mice developed visually evident tumors (Figure 1C). Thus these in vivo studies indicate that the SP cells or cancer stem–like cells have a greater tumorigenicity potential than ΔSP or unseparated cancer cells. To obtain the orthotopic tumors derived from these subcutaneous tumors, we implanted naive nude mice orthotopically in the pancreas with fragments of these subcutaneous tumors as described previously (Fu et al., 1992) and allowed the tumors to develop for an additional 40 d. Forty days after implantation, pancreatic tissues were harvested and processed for paraffin sectioning and immunohistochemical analysis. Because increased expression of uPA is associated with higher “aggressiveness” for multiple tumor types, including pancreatic adenocarcinoma (Ceccarelli et al., 2010; Markl et al., 2010; Bekes et al., 2011; Provost et al., 2012), we studied expression levels of uPA in these orthotopic tumors using immunohistochemistry. We observed that orthotopic tumors grown from the implanted SP cell–derived subcutaneous tumors expressed uPA at much higher levels than those grown from implanted MP cells (Figure 1D). In contrast, normal pancreatic tissue expressed moderate-to-low levels of uPA (Figure 1D). To further assess the role of uPA in establishing the cancer stem cell phenotype, we overexpressed uPA in both SP and ΔSP MIA PaCa-2 cells (uPAOE-SP and uPAOE-ΔSP, respectively) and compared their proliferation and growth patterns using the sphere formation assay. We observed that SP cells possessed greater sphere-forming ability (p < 0.001) than ΔSP cells. Overexpression of uPA induced sphere formation in ΔSP cells (Supplemental Figure S2). The sphere-forming ability of SP cells was attenuated when uPA expression was suppressed with uPA-specific short hairpin RNA (shRNA; Mia PaCa-2(uPA-) and PANC-1(uPA-) cells), which led to significant disintegration of the pancreatospheres (Figure 1E). Fluorescence-activated cell sorting analysis of the mixed populations of MIA PaCa-2 and PANC-1 cells revealed that uPA overexpression (uPAOE) increased the proportion of SP cells (Supplemental Figure S3). Together these data indicate that uPA promotes pancreatic cancer cell stemness.
FIGURE 1:
Stem cell–like properties of the SP cells derived from pancreatic cancer cells. (A) Mixed populations of MIA PaCa-2 and PANC-1 cells (2 × 106) were sorted by density-based flow cytometry (10,000 cells sorted per treatment condition, with three replications) to separate SP and ΔSP cells. Acquisition was performed on a FACSCalibur flow cytometer, and viable cells were analyzed with CellQuest software. (B) Cell lysates prepare from the sorted SP and ΔSP cells were immunoblotted for CD24 and CD44 to elucidate expression of cancer stem cell markers. (C) SP, ΔSP, and MP cells were implanted subcutaneously in nude mice (10,000 cells/mouse), and the tumor volumes in treated groups were quantified and represented graphically (mean ± SD; n = 5 and p < 0.001). (D) Subcutaneous tumors grown as in C were implanted orthotopically in the pancreas of nude mice as described in Materials and Methods and allowed to grow for 40 d. At the end of this period, pancreatic tissues were harvested and processed for paraffin sectioning. Expression levels of uPA were determined by immunohistochemistry using anti-uPA and control immunoglobulin G. Brown color denotes uPA-antibody–positive reaction. Normal pancreatic tissue was also sectioned and immunoprobed for uPA. (E) Proliferation and formation of the neurospheres by untreated SP cells derived from MIA-PA Ca-2 and PANC-1 cells (left). Right, disintegration of the neurospheres after exposure to shRNA specific for uPA (puPA).
Suppression of uPA expression sensitizes pancreatic CSC to gemcitabine
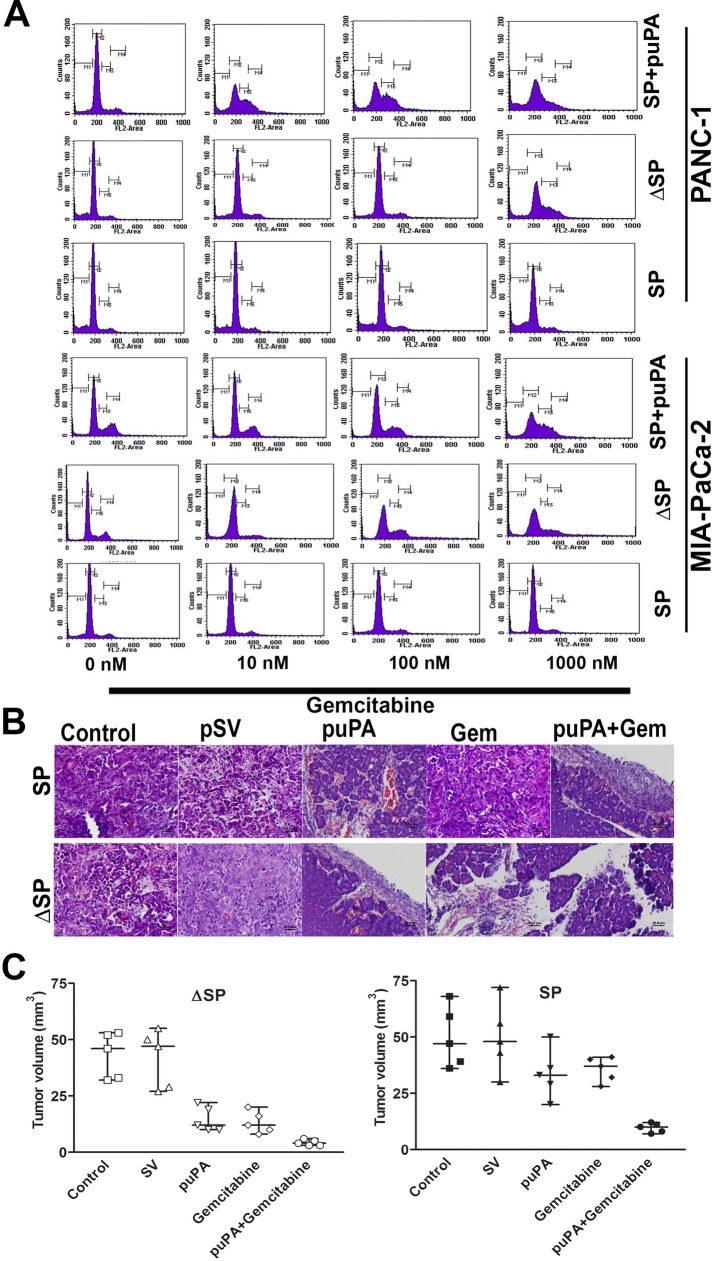
Human pancreatic CSC are highly tumorigenic and highly resistant to standard chemotherapy (Hermann et al., 2007), including gemcitabine (Hong et al., 2009). On the basis of our finding that uPA confers a stem-like phenotype to pancreatic cancer cells, we asked whether uPA contributes to chemoresistance and whether inhibition of uPA expression would sensitize these cells to gemcitabine. To do so, we expressed uPA-targeting or scrambled shRNA in SP and ΔSP MIA PaCa-2 and PANC-1 cells and added varying concentrations of gemcitabine (0–1000 nM). We observed that 1) SP cells are resistant to gemcitabine treatment and 2) silencing uPA expression sensitizes chemoresistant SP cells to gemcitabine, exemplified by a decrease in the proportion of cells in G0/G1 phase indicative of cell cycle arrest (Figure 2A). These data further suggest that uPA plays a critical role in the chemoresistance and survival of pancreatic CSCs.
FIGURE 2:
uPA controls chemoresistance of pancreatic cancer cells to gemcitabine. (A) SP cells derived from MIA PaCa-2 and PANC-1 cells treated with shRNA specific for uPA (SP + puPA), untreated SP, and ΔSP cells derived from MIA PaCa-2 and PANC-1 cells were subjected to various concentrations of gemcitabine (0, 10, 100, 1000 nM), and cell cycle analysis was performed by fluorescence-activated cell sorting. (B) H&E staining of the paraffin sections of the tumors obtained from nude mice in which SP and ΔSP cells derived from MIA PaCa-2 cells that had been exposed to either puPA or gemcitabine or both as described in Materials and Methods were implanted orthotopically (100,000 cells/mouse). (C) Graphic representation of the tumor sizes in nude mice after orthotopic implantation of SP and ΔSP cells derived from MIA PaCa-2 cells (100,000 cells/mouse) that had been exposed to either puPA or gemcitabine (mean ± SD; n = 5; ΔSP, p = 0.01, and SP, p = 0.008).
Suppression of uPA retards development of pancreatic cancer in nude mice and increases sensitization to gemcitabine
To determine whether tumorigenicity of MIA PaCa-2 cells was reduced after suppression of uPA expression, we orthotopically implanted nude mice with MIA PaCa-2 SP and ΔSP cells (100,000 cells initially) that had been pretreated with small interfering RNA (siRNA) specific for uPA (puPA), gemcitabine, or both, as described in Materials and Methods. Mice given puPA showed smaller tumor burdens than controls (p = 0.24). Mice implanted with ΔSP cells treated with gemcitabine alone showed the greatest reduction in tumor burden, whereas mice implanted with SP tumors did not respond to gemcitabine. The greatest reduction in tumor burden was seen in mice implanted with SP and ΔSP treated with both puPA and gemcitabine (Figure 2, B and C; p = 0.012 and 0.008, respectively).
uPA positively regulates Lhx2 expression in MIA PaCa-2 and PANC-1 pancreatic cancer cells and in human pancreatic tissues
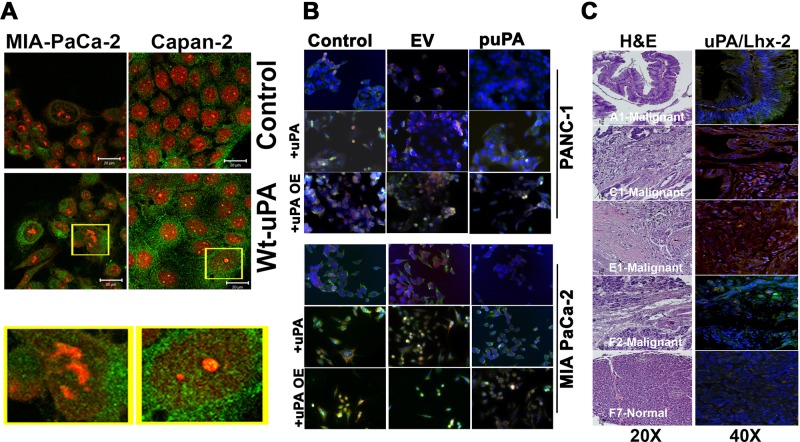
We demonstrated previously that uPA is found within the nuclei of various types of proliferating cells (Stepanova et al., 2008). We therefore asked whether uPA localizes to the nuclei in pancreatic cancer cells. Immunocytochemical analysis of MIA Pa Ca-2, Capan-2, and PANC-1 cells revealed partial nuclear localization of uPA, which is significantly (p = 0.40) increased when recombinant uPA protein is added exogenously (Figure 3A and Supplemental Figure S4A). More recently, we reported that uPA binds to the transcription factor Lhx2 within the nuclei of pancreatic cancer cells and knockdown of uPA suppresses Lhx2 expression (Gorantla et al., 2011). Because Lhx2 is known to be involved in maintenance of stem/progenitor cell phenotype (Dahl et al., 2008; Tornqvist et al., 2010; Mardaryev et al., 2011; Nadal et al., 2012), we next investigated whether uPA–Lhx cross-talk regulates the maintenance of stem/progenitor cell phenotype in pancreatic cancer cells. To further decipher the role of nuclear uPA in the regulation of Lhx2 expression, we knocked down endogenous uPA expression in MIA PaCa-2 and PANC-1 cells using the puPA plasmid or uPA siRNA (MIA PaCa-2(uPA-) and PANC-1(uPA-) cells, respectively) and added wild-type (WT) uPA exogenously. We observed that addition of exogenous WT-uPA and overexpression of uPA (uPAOE) in MIA PaCa-2(uPA-) and PANC-1(uPA-) cells induced the expression of Lhx2 (Figure 3B). To determine whether uPA positively regulates Lhx2 expression in human tissues, we immunoprobed a human pancreatic tissue array for uPA and Lhx2. In malignant tissues, high levels of expression of both uPA with Lhx2 were observed. In contrast, normal pancreatic tissues showed no detectable expression of uPA and low expression levels of Lhx2 throughout (Figure 3C). Our previous studies indicated that uPAR is not essential for translocation of uPA to the nucleus. A uPA mutant lacking the uPAR-binding domain (ΔGFD-uPA) also translocates to cell nuclei, whereas a kringle-deficient uPA mutant (ΔK-uPA) did not, despite its ability to bind to uPAR (Stepanova et al., 2008). We found identical requirements for translocation of uPA in pancreatic cancer cells (Supplemental Figure S4, A and B). To address the role of uPAR in the regulation of Lhx2 expression by uPA specifically, we then studied the effect of these uPA deletion mutants on expression levels of Lhx2. PANC-1(uPA-) cells overexpressed Lhx2 in response to exogenously added WT-uPA and ΔGFD-uPA, whereas levels were unchanged after addition of identical concentrations of ΔK-uPA (Supplemental Figure S4C).
FIGURE 3:
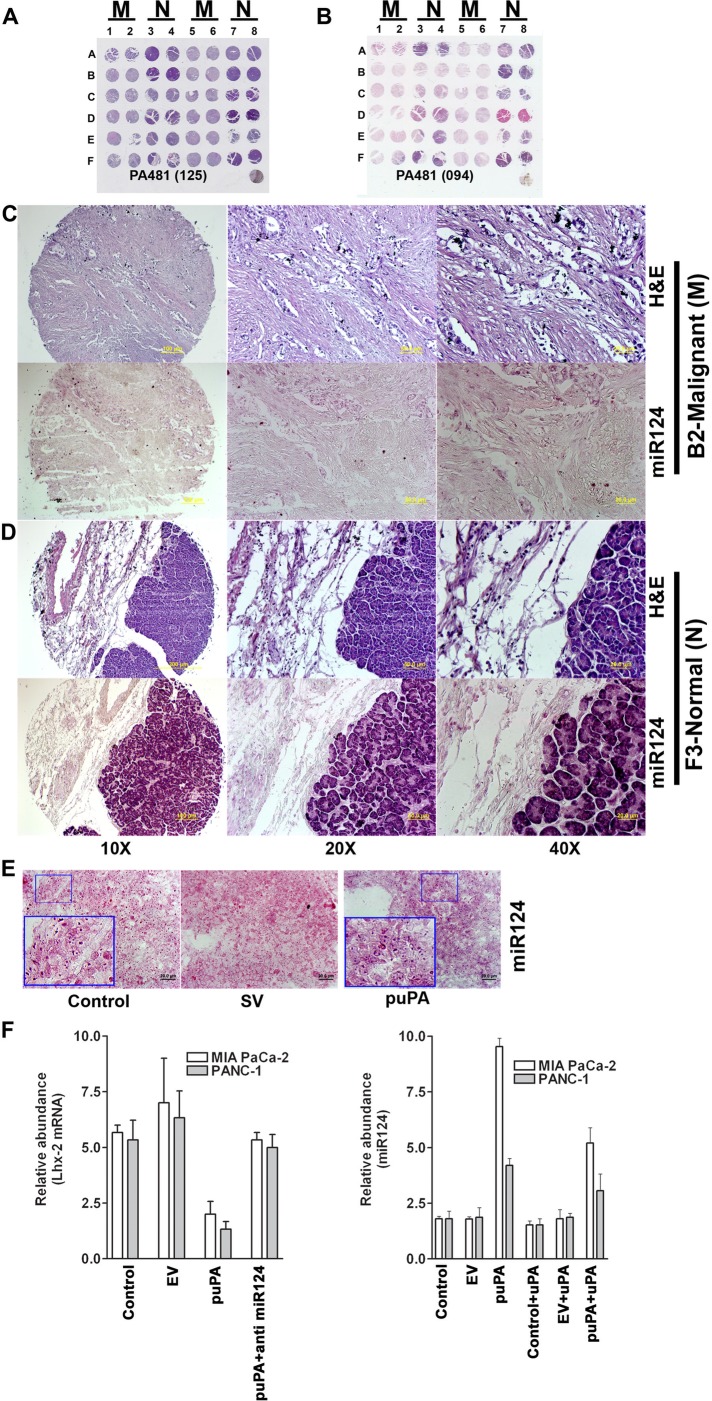
Nuclear uPA regulates expression of Lhx2 in pancreatic cancer cells. (A) MIA-PaCa-2 and Capan-2 cells were left untreated or incubated with 20 nM of recombinant WT-uPA for 1 h, fixed in MeOH, and stained with anti-uPA rabbit polyclonal Abs and Alexa 488–conjugated anti-rabbit secondary Abs. Nuclei were counterstained with propidium iodide (red). Green staining denotes cytoplasmic and nuclear localization of uPA. (B) MIA PaCa-2 and PANC-1 cells plated and grown on chamber slides were transfected with pSV (scrambled vector) or puPA to lower uPA or uPA-encoding plasmid for uPA overexpression (pUPAOE). Nontransfected cells were also incubated with exogenously added WT-uPA protein. Cells were immunoprobed for uPA (green) and Lhx2 (red) and mounted with DAPI-containing mounting medium, and fluorescent photomicrographs were obtained as described (Stepanova et al., 2008). (C) Human pancreatic cancer tissue array (± cancer) was stained with H&E or immunoprobed for uPA or Lhx2 (A1, C1, E1, and F2 are malignant pancreatic adenocarcinoma tissues, and F7 is normal pancreatic tissue).
miR-124 targets Lhx2 and is negatively regulated by uPA
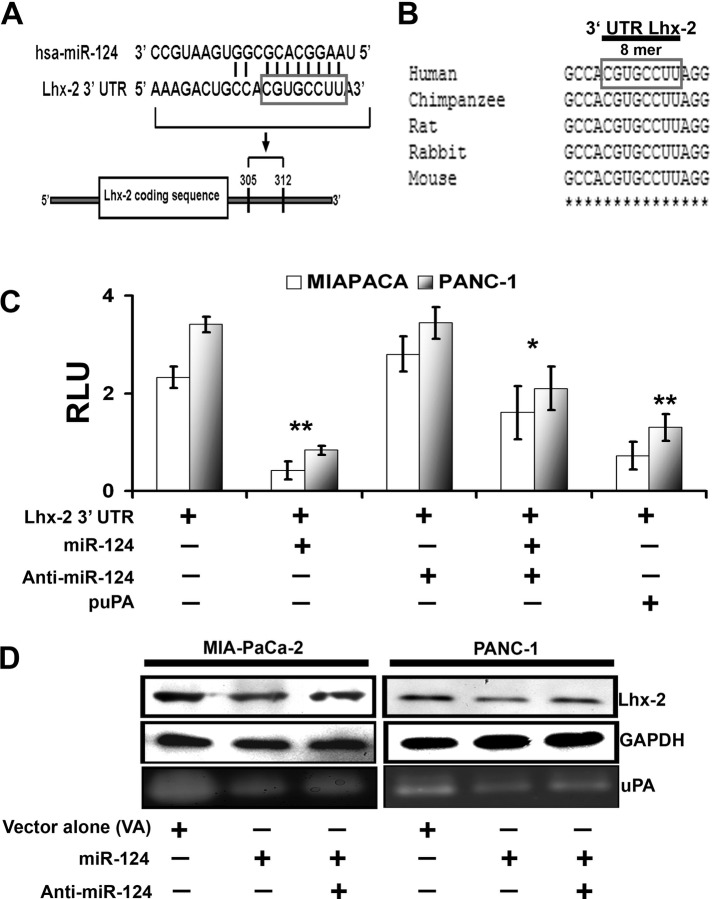
We next investigated the mechanism by which uPA up-regulates the expression of Lhx2. We first performed miRNA target prediction analysis using the MiRanda (Enright et al., 2003) and PITA (Kertesz et al., 2007) algorithms. We found that Lhx2 mRNA is a potential target of hsa-mir-124 (miR-124) (Figure 4A). Specifically, we identified an 8-mer (CGUGCCUU) motif in the 3′ untranslated region (UTR) of Lhx2 that is highly conserved in multiple mammalian species as a potential binding site for miR-124 (Figure 4B). To validate this in silico prediction, we developed a reporter construct in which luciferase expression is controlled by the human Lhx2 3′-UTR DNA fragment containing the putative miR-124 interaction sequence. The Lhx2 3′-UTR reporter construct was transiently transfected into MIA PaCa-2 and PANC-1 cells alone or together with hsa-miR-124 again alone or in combination with the miR-124 inhibitor (anti–miR-124). Overexpression of miR-124 repressed the Lhx2 3′-UTR reporter (Figure 4C), whereas anti–miR-124 prevented the miR-124–mediated repression of Lhx2 3′-UTR luciferase activity in both MIA PaCa-2 and PANC-1 cells, confirming the specificity of miR-124 toward the 3′-UTR region of Lhx2. We then examined expression levels of miR-124 in human pancreatic cancer tissues versus normal (unaffected) pancreas using human pancreatic cancer tissue arrays. Figure 5A shows hematoxylin and eosin (H&E) staining of normal and pancreatic ductal adenocarcinoma (PDAC). We also performed in situ hybridization (Figure 5B) to examine the expression of miR-124 in human pancreatic cancer tissue arrays. We observed that tumor tissues did not express miR-124 (Figure 5C), in contrast to its expression in normal pancreatic tissues (Figure 5D). This suggests that overexpression of Lhx2 in pancreatic cancer tissues (Figure 3) might be due to suppression of miR-124. Because 1) enhanced miR-124 expression has inhibitory effects on cancer stem–like traits and invasiveness (Xia et al., 2012), 2) miR-124 targets Lhx2 transcript, and 3) uPA up-regulates Lhx2 in pancreatic cancer cells, we examined the relationship of uPA and miR-124. To determine whether uPA affects the expression of miR-124 and thereby regulates Lhx2, we used miRNA-specific stem loop PCR to examine expression of miR-124 in control MIA PaCa-2 and PANC-1, MIA PaCa-2(uPA-), and PANC-1(uPA-) cells cultured in absence or presence of exogenously added WT-uPA. We observed that down-regulation of uPA expression (MIA PaCa-2(uPA-) and PANC-1(uPA-) cells) induced expression of miR-124 compared with control MIA PaCa-2 and PANC-1 cells, whereas addition of WT-uPA inhibited miR-124 expression (Figure 5F). Furthermore, cells treated with shRNA specific for uPA (MIA PaCa-2(uPA-) and PANC-1(uPA-) cells) and transfected with the Lhx2 3′-UTR luciferase reporter construct showed significant decrease in luciferase activity, suggesting that uPA up-regulates Lhx2 by suppressing expression of miR-124. To determine whether down-regulation of uPA induces expression of miR-124 in vivo, we orthotopically implanted MIA PaCa-2 SP cells that exhibit cancer stem cell–like characteristics (Figure 1A and Supplemental Figures S2 and S4) into the pancreas of nude mice, which were then injected intraperitoneally with shRNA targeting uPA (puPA; plasmid expressing shRNA targeting uPA). In vivo suppression of uPA resulted in significant (p = 0.02) increase in expression of miR-124 in tumor tissue after 40 d but not in normal tissue (Figure 5E). Of interest, hsa-miR-124 also suppressed expression of both Lhx2 and uPA in MIA PaCa-2 and PANC-1 cells, whereas transfection of these cells with anti–miR-124 enhanced expression of Lhx2 and uPA (Figure 4D). Together these data suggest the existence of a negative feedback loop between uPA and miR-124, which may regulate expression of Lhx2 and pancreatic cancer cell stemness.
FIGURE 4:
Lhx2 is the predicted target for miR-124, which negatively regulates Lhx2. (A) Sequence alignment of miR-124 and predicted sequence pairing with a region of Lhx2 mRNA 3′-UTR. The nucleotides within the Lhx2 3′-UTR region that may interact with miR-124 are framed. (B) Alignment of nucleotide sequences of Lhx2 3′-UTR corresponding to the targets for miR-124 from several mammalian species. A high level of conservation suggests a functional role for these sequences. (C) Luciferase reporter assay. Interaction of miR-124 with Lhx2 3′-UTR luciferase reporter vector was transfected in the control and puPA-treated MIA PaCa-2 and PANC-1 cells alone and/or in combination with hsa-miR-124 and miR-124 inhibitor (anti–miR-124). Luciferase activity, which reflects extent of inhibition of Lhx2 3′-UTR reporter by miR-124, was quantified and normalized as described in Materials and Methods. The y-axis denotes relative luciferase units (RLU; mean ± SD; n = 3; *p < 0.05; **p < 0.01). (D) Western blot analysis of cell lysates (40 μg of total protein) obtained from control and hsa-miR-124– and/or anti–miR-124–treated MIA PaCa-2 and PANC-1 cells. Separated proteins were probed with anti-Lhx2 antibodies, followed by HRP-conjugated anti-rabbit secondary Abs and the bands were visualized with the chemiluminescent substrate. Anti-GAPDH Abs were used as a loading control. Fibrin zymography (bottom) was performed to determine uPA activity in the conditioned media of the cultured cells as described in Materials and Methods.
FIGURE 5:
Down-regulation of uPA causes overexpression of miR-124 in pancreatic cancer cells. Human pancreatic cancer tissue arrays were subjected to (A) H&E staining or (B) miR-124 in situ hybridization. miR-124 expression was assessed in both PDAC (C) and normal pancreatic tissues (D). (E) Mice implanted subcutaneously with MIA PaCa-2 cells were treated with puPA as described in Materials and Methods, and expression levels of miR-124 in paraffin sections were assessed by in situ hybridization. (F) Total RNA was isolated from MIA PaCa-2 and PANC-1 cells treated with puPA, and expression levels of Lhx2 mRNA and miR-124 were determined.
uPA complexes with HOXA5 and inhibits p53 promoter activity
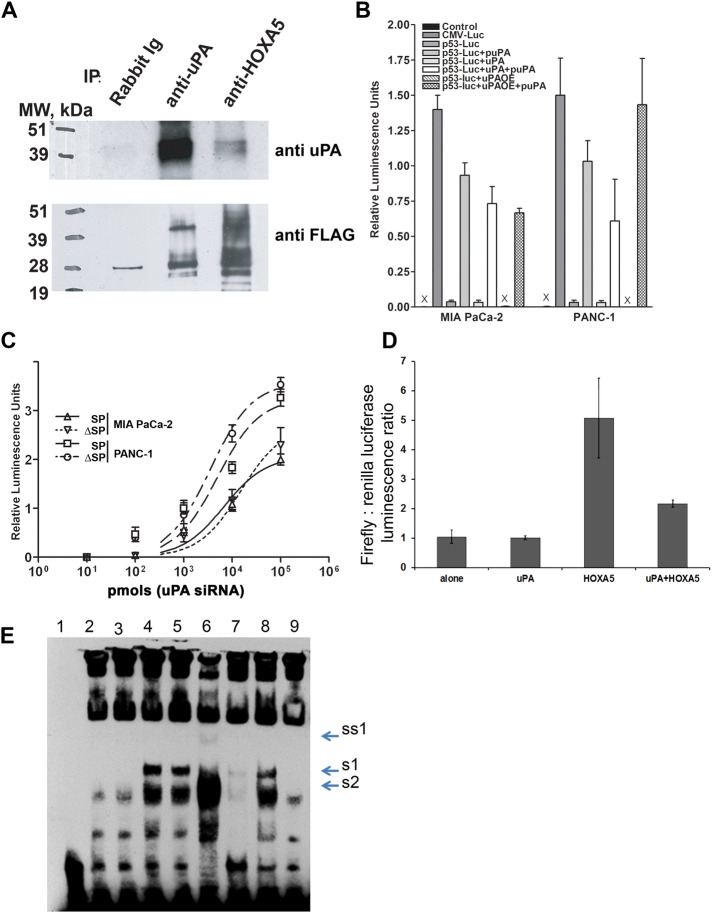
To further elucidate the functions of uPA in the nucleus, we examined whether uPA binds directly to transcription factors (TFs), using a protein TF array. Supplemental Figure S5A shows that uPA binds to homeobox transcription factors Lhx2, HOXA5, and Hey. Binding of uPA to Lhx2 (Gorantla et al., 2011) and HOXA5 (Figure 6A) was confirmed by coimmunoprecipitation pull-down assays. HOXA5 up-regulates expression of the p53 tumor suppressor gene in breast cancer cells (Raman et al., 2000). It has been suggested that down-regulation of HOXA5 expression or loss of its function results in inhibition of p53 expression (Raman et al., 2000), which enhances tumorigenicity, EMT, and acquisition of stem-like phenotype by breast cancer cells (Mizuno et al., 2010). Loss of p53 function has also been linked to the induction of EMT and stemness in pancreatic cells (Keck and Brabletz, 2011; Pasi and Pelicci, 2011). Therefore we hypothesized that uPA further promotes stemness of pancreatic cancer cells by down-regulating p53 through interference with HOXA5. To assess whether uPA regulates activity of the p53 promoter, we stably transfected MIA PaCa-2 and PANC-1 cells with the human p53 promoter-driven luciferase reporter construct (Mia PaCa-2(p53-luc) and PANC-1(p53-luc) cells, respectively). Suppression of uPA expression in Mia PaCa-2(p53-luc) and PANC-1(p53-luc) cells caused activation of p53 promoter-driven luciferase expression, whereas uPA overexpression (uPAOE) inhibited p53 promoter-controlled luciferase activity (Figure 6B). Similar results were obtained using SP or ΔSP cells (Figure 6C). To determine whether uPA suppresses p53 promoter activity by interfering with the function of HOXA5, we used human embryonic kidney 293 cells (HEK293 cells), which express low levels of endogenous HOXA5 and uPA. HEK293 cells were cotransfected with the p53-luc construct and HOXA5- and/or uPA-encoding vectors. Figure 6D shows that coexpression of p53-luc and HOXA5 leads to activation of p53 promoter activity, whereas concomitant coexpression of uPA attenuates HOXA5-mediated activation of p53 promoter.
FIGURE 6:
uPA suppresses p53 promoter activity via binding to and attenuation of the Hoxa-5 function. (A) HEK293 cells were transfected with HOXA5-FLAG in pcDNA3.1 and uPA/pcDNA3.1 vector. Two days after transfection, cells were harvested, and nuclear extracts were prepared using the Novagen NucBuster Protein Extraction Kit. uPA and/or HOXA5 were immunoprecipitated using anti-uPA mouse monoclonal Abs (IMTEK, Moscow, Russia) and rabbit polyclonal anti-HOXA5 Abs (Santa Cruz Biotechnology). Immunoprecipitated proteins were subjected to Western blot analysis. Immunoprecipitated and coimmunoprecipitated uPA and/or HOXA5-FLAG were detected using anti-uPA rabbit polyclonal Abs (389; American Diagnostica, Stamford, CT) and mouse monoclonal HRP-conjugated anti-FLAG M2 Abs (Sigma-Aldrich). (B) MIA PaCa-2 and PANC-1 cancer cells were stably transfected with p53 promoter luciferase reporter plasmid. uPA expression was suppressed with puPA or uPA was overexpressed (uPAOE) in these cells. In parallel, cells were incubated with WT-uPA. p53 promoter activity was determined by measurement of luciferase activity as described in Materials and Methods. (C) MIA PaCa-2 and PANC-1 cells were stably transfected with p53-luc plasmid and sorted to obtain SP and ΔSP cells. uPA was suppressed or overexpressed or added to the cells, and luciferase activity was measured as described. (D) HEK293 cells were cotransfected with p53-luc, uPA/pcDNA3.1+ plasmid encoding human WT-uPA, or HOXA5-FLAG/pcDNA3.1 plasmid encoding C-terminus–tagged HOXA5-FLAG. Empty pcDNA3.1 was used as the negative control, and pRL TK plasmid encoding constitutively expressed Renilla luciferase was cotransfected to normalize the data. Luciferase activity was determined using a Promega Dual Luciferase Reporter Assay Kit. (E) Effect of uPA on DNA-binding capacity of HOXA5. HEK293 cells were transfected either with empty pcDNA3.1 (mock transfected) or with HOXA5-FLAG in pcDNA3.1 alone or in combination with uPA/pcDNA3.1 vector. Two days after transfection, cells were harvested. Nuclear extracts were prepared using the NucBuster Protein Extraction Kit. EMSA reactions were performed using biotinylated, double-stranded, p53 promoter-derived Hoxa-5–specific oligonucleotides. 1, No NE; 2, + mock-transfected NE; 3, + uPA-transfected NE; 4, + HOXA5-transfected NE; 5, + HOXA5-transfected NE + BSA; 6, + HOXA5-transfected NE + scuPA (500 ng); 7, + HOXA5-transfected NE + specific “cold” oligo duplex; 8, + HOXA5-transfected NE + scuPA + anti-uPA Abs; 9, + mock-transfected NE + scuPA (500 ng). S1, Probe shift, caused by HOXA5 overexpression; S2, DNA–protein complex, formed in presence of NE from the mock-transfected cells; SS1, probe supershift caused by HOXA5-bound anti-HOXA5 Ab.
These data suggest that binding of uPA to HOXA5 attenuates its DNA-binding capacity. To test this hypothesis, we examined whether uPA affects binding of HOXA5 to its target DNA sequence. We performed the electrophoretic mobility shift assay (EMSA) using p53 promoter-derived, double-stranded oligonucleotides that possess a HOXA5 consensus region (see Materials and Methods). We found that uPA does not bind the HOXA5 DNA consensus sequence but instead forms a complex with HOXA5 that binds p53 promoter-derived oligonucleotide less well than free HOXA5 (Figure 6E). Finally, we used pancreatic cancer cell line Capan-2, which expresses wild-type p53, to assess the role of p53, HOXA5, and uPA in chemoresistance. Exogenously added WT-uPA down-regulated p53 mRNA in Capan-2 cells by 1.6 ± 0.2 times (p < 0.05), and silencing uPA expression in Capan-2 cells up-regulated p53 mRNA (Supplemental Figure S5B). Finally, WT-uPA and ΔGFD-uPA but not ΔK-uPA increased resistance of uPA-targeting, shRNA-treated Capan-2 cells to gemcitabine-induced apoptosis (Supplemental Figure S5C). Together these data suggest that uPA regulates pancreatic cancer cell survival by down-regulating expression of p53 in addition to up-regulating expression of Lhx2.
DISCUSSION
Pancreatic ductal adenocarcinoma has a high mortality rate, which is attributable in part to delay in diagnosis but also to lack of effective treatment options. Gemcitabine, the cornerstone of adjuvant and metastatic therapy, delays the development of recurrent disease after “complete resection” (Oettle et al., 2007), but the estimated disease-free survival at 3 and 5 yr remains 23.5 and 16.5%, respectively, compared with 7.5 and 5.5% in untreated patients. Thus the intrinsic and acquired resistance of pancreatic tumors to gemcitabine and other treatments continues to be a major clinical problem. The goal of our study was to better understand the molecular basis underlying the drug resistance of pancreatic cancer cells. The rapid progression of pancreatic cancer is characterized by molecular changes within the tumor and the surrounding stromal cells (Bailey and Leach, 2012; Feig et al., 2012; Tod et al., 2013; Whatcott et al., 2013) that emerge from a side population of cells indicated by EMT and a cancer stem cell phenotype associated with drug resistance (Kabashima et al., 2009; Haque et al., 2011; Kabashima-Niibe et al., 2013). Pancreatic cancer cell lines (MIA Pa Ca-2 and PANC-1) cultured under serum-free conditions expressed stem cell markers CD44 and CD24 (Figure 1 and Supplemental Figure S1), had an increased proportion of SP cancer stem cell–like cells, formed pancreatospheres (Figure 1E and Supplemental Figure S2), and showed gemcitabine resistance, in contrast to mixed or ΔSP populations (Figure 2A). Down-regulation of uPA expression attenuated the stem cell phenotype (Hamada and Shimosegawa, 2012), suppressed formation of pancreatospheres (Bao et al., 2011), and restored sensitivity to gemcitabine. In contrast, overexpression of uPA increased drug resistance and formation of pancreatospheres by cultured pancreatic cancer cells in vitro (Figure 1E) and promoted tumor growth in vivo (Figure 1C). These outcomes provide new insight into the mechanisms underlying the well-established correlation between elevated tumor uPA, propensity to metastasize, and poor prognosis of this disease (reviewed in Ischenko et al., 2010; Rasheed and Matsui, 2012).
The adverse outcome associated with uPA expression has generally been attributed to its proteolytic activity and uPAR-dependent signaling mechanisms (Gupta et al., 2011; Andres et al., 2012; He et al., 2012), but uPA's role in maintaining cancer cell stemness, chemoresistance, and survival has not been addressed. Our studies identify a novel uPA-mediated pathway that contributes to the stem-like phenotype. We found that uPA is localized in the nucleus of pancreatic cancer cells (Figure 2A and Supplemental Figure S2B; Gorantla et al., 2011) and associates with the transcription factor Lhx2, which has been implicated in the maintenance of stemness (Pinto do et al., 2001; Tiede and Paus, 2006). We show that uPA up-regulates expression of Lhx2 in pancreatic cancer cells. Lhx2 expression was also seen in tumor-associated cells in an in vivo mouse model, suggesting that uPA may participate in reprogramming of these associated cells to facilitate tumor growth via Lhx2 (Gorantla et al., 2011). Overexpression of Lhx2 in response to uPA was associated with concomitant uPA-dependent down-regulation of miR-124, which, as we found, targets Lhx2 3′-UTR. In support of this proposed pathway, expression of miR-124 was inversely related to expression of uPA. Pancreatic tissue array analysis revealed that normal pancreatic tissues expressed miR-124, whereas tumor tissues did not. Furthermore, overexpression of uPA in orthotopic tumors in nude mice paralleled suppression of miR-124, whereas tumor tissues in which uPA were down-regulated expressed elevated levels of miR-124. The mechanism of miR-124 suppression by uPA requires further study. Our data suggest that a negative feedback loop links uPA and miR-124, which in turn increases expression of Lhx2, leading to pancreatic cancer cell stemness. uPA can also interact with stromal cells, in which it may activate Lhx2 and promote stemness through paracrine pathways. This paracrine regulation mediated by uPA may be one of the reasons uPA-overexpressing tumors are more resilient to conventional chemotherapies (Khanna et al., 2011).
uPA may up-regulate Lhx2 through several intracellular mechanisms that eventuate in cell survival and chemoresistance. For example, we found that uPA binds directly to several homeobox transcription factors, including HOXA5, which up-regulates p53 expression through direct transactivation of the p53 promoter (Raman et al., 2000; Gendronneau et al., 2010). Using EMSA and p53 promoter-driven luciferase reporter assays, we observed that uPA inhibits binding of HOXA5 to its DNA consensus sequence, attenuates the ability of HOXA5 to activate the p53 promoter, and inhibits p53 expression in the pancreatic cancer cell line Capan-2 (Figure 6 and Supplemental Figure S5). This is accompanied by increased chemoresistance of Capan-2 cells (Supplemental Figure S5), suggesting that the effect of uPA and HOXA5 on p53 contributes to pancreatic cancer cell stemness. HOXA5-binding elements are present in Lhx2 and CD24 promoters (Qiagen, Valencia, CA; available at www.sabiosciences.com/chipqpcrsearch.php?app=TFBS). Whether and how HOXA5 regulates activity of Lhx2 and CD24 promoters in clinical disease and whether uPA regulates expression of these genes in a HOXA5-dependent manner remain to be determined.
uPAR has been implicated in the progression of pancreatic and other types of cancer through uPA-dependent and uPA-independent pathways (Adachi et al., 2002; Blasi and Carmeliet, 2002; Gondi et al., 2003; de Bock and Wang, 2004; Kondraganti et al., 2006; Gorantla et al., 2011; Asuthkar et al., 2012). It is likely that uPA binds to uPAR on the surface of pancreatic cancer cells and uPAR binds to other receptors and/or ligands that might contribute to survival and progression. On the other hand, the incomplete response of pancreatic cancer to inhibition of uPA-proteolytic activity and incapacitation of the uPA-uPAR axis (Mekkawy et al., 2009; Hildenbrand et al., 2010; Boonstra et al., 2011; Carriero and Stoppelli, 2011; Lund et al., 2011) provides compelling evidence that other pathways must be involved as well. Nucleolin, which we showed mediates nuclear translocation of uPA (Stepanova et al., 2008), associates with uPAR (Dumler et al., 1999) and participates in the nuclear translocation of endostatin (Song et al., 2012) and perhaps uPAR itself (Asuthkar et al., 2012). Whether nuclear translocation of uPAR in pancreatic cancer cells contributes to pancreatic cancer stemness and chemoresistance through its association with nucleolin remains to be determined. The uPA variant ΔGFD-uPA, however, which is unable to bind to uPAR but is capable of nuclear translocation, up-regulates Lhx2 expression in PANC-1 cells (Supplemental Figure S4) and increases chemoresistance of Capan-2 cells (Supplemental Figure S5) similar to WT-uPA, suggesting that uPAR is not an obligate carrier for uPA translocation to the nucleus and its effect on gene transcription. Thus, to our knowledge, this is the first evidence that links nuclear uPA to the maintenance of pancreatic cancer cell stemness and chemoresistance in an uPAR-independent manner. Further delineation of this cellular and genetic program might offer new therapeutic targets in this still devastating disease.
MATERIALS AND METHODS
Cell lines and culture conditions
Pancreatic cancer cells MIA PaCa-2 and PANC-1 were obtained from the American Type Culture Collection (Manassas, VA) and maintained in DMEM supplemented with 10% fetal bovine serum in a humidified atmosphere containing 5.0% CO2 at 37.2°C with media changes every 48 h. To facilitate growth of cancer stem cells, MIA PaCa-2 and PANC-1 cells were cultured in serum-free DMEM supplemented with basal fibroblast growth factor (10 ng/ml), 1× B27 supplement, and epidermal growth factor (50 ng/ml; all from Life Technologies, Frederick, MD).
shRNA construct
shRNA-expressing plasmids targeting uPA (puPA) were constructed as described previously (Gondi et al., 2007a, b). Lentivirus-delivered, pLKO-based, shRNA-targeting uPA mRNA in Capan-2 cells was obtained from Thermo Scientific (formerly Open Biosystems; West Palm Beach, FL). uPA siRNA was obtained from Santa Cruz Biotechnology (sc-36779; Santa Cruz, CA).
Chemotherapy compound
Gemcitabine (G6423) was purchased from Sigma-Aldrich (St. Louis, MO) and diluted in sterile water. MIA PaCa-2 and PANC-1 side-population cancer stem like cells were incubated with gemcitabine (0–1000 nM), and the cytotoxicity was determined after 48 h of incubation.
Side-population sorting, cell cycle analysis, and sphere formation
MIA PaCa-2 and PANC-1 cells (2 × 106) resuspended in 0.5 ml of phosphate-buffered saline (PBS)–bovine serum albumin (BSA) were analyzed, and the SP cells were sorted by density-based flow cytometry (10,000 cells sorted per treatment condition, with three replications for each treatment). Acquisition was performed on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA), and viable cells were analyzed with CellQuest software. In separate experiments, SP cells were labeled with anti-CD44 and anti-CD24 antibodies using a similar staining protocol. The control for each sample was prepared identically, except that an isotype-specific antibody was used. Progression through different cell cycle phases by MIA PaCa-2 and PANC-1 cells alone and by puPA and after exposure to gemcitabine for 48 h was monitored by flow cytometric analysis of DNA content of cell populations stained with 50 μg/ml propidium iodide (BioSure, Grass Valley, CA). CSC neurospheres were obtained from SP cells sorted from parental MIA PaCa-2 and PANC-1 cells as described previously (Velpula et al., 2011).
miRNA and anti-miRNA
The pcDNA3.2/V5 hsa-miR-124 (plasmid 26306) was purchased from Addgene (Cambridge, MA), and hsa-miR-124 miRCURY LNA microRNA inhibitor (412512-00) was purchased from Exiqon (Woburn, MA).
3′-UTR reporter assay
3′-UTR mRNA sequences of Lhx2 (5′-GGTACCTTTCTAATGACTCGCAACC-3′; 5′-TAAACAAAAAACAACCTCACAAAGAAGATC-3′) were amplified and cloned into pISO (plasmid 12178) mammalian expression vector (Addgene). For the reporter gene assay, MIA PaCa-2 and PANC-1 cells were transiently transfected with the 200 ng/ml Lhx2 3′-UTR construct alone or in combination with either the 1 nmol hsa-miR-124 mimic or 5 nmol hsa-miR-124 inhibitor (anti-miR-124). After 48 h, cells were lysed to determine luciferase activity using a dual-luciferase reporter assay system (Promega, Madison, WI), and the relative luciferase units were measured in a luminometer (TD-20/20 DLReady).
Stem-loop PCR (reverse transcriptase PCR)
Total RNA was isolated using TRIZOL reagent (Invitrogen, Carlsbad, CA). For miRNA analysis, 100 ng of template RNA was reverse transcribed using universal first-strand cDNA synthesis kit and miR-124 specific reverse transcriptase (RT) primers from the miRCURY LNATM microRNA PCR system (Exiqon) following the supplied protocol. The miR-124 transcript levels were examined by RT-PCR using the CFX96 Real Time System (Bio-Rad, Hercules, CA) and the SYBR Green PCR Master Mix (Applied Biosystems, Framingham, MA). The following PCR conditions were used: 95°C for 10 min, followed by 50 cycles at 95°C/10 s and 60°C/30 s. The fold change was calculated using 2−ΔΔCT, where CT is the threshold cycle and ΔΔCT = ΔCT of treatment − ΔCT of control.
RT-PCR analysis
MIA PaCa-2 and PANC-1 cells were transfected with uPA-directed siRNA for 48 h. The cells were collected, and total cell RNA was isolated. RT-PCR was set up using primers specific for uPA (Table 1). The PCR cycle was 95°C/5 min, (95°C/30 s, 65°C/1 min, 72°C/1 min) × 30, 72°C/10 min. The PCR product was quantified and plotted relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression as arbitrary units.
TABLE 1:
Primers used for PCR and RT-PCR.
| Forward | Reverse | |
|---|---|---|
| uPA | TGCGTCCTGGTCGTGAGCGA | CAAGCGTGTCAGCGCTGTAG |
| GAPDH | CGGAGTCAACGGATTTGGTCGTAT | AGCCTTCTCCATGGTGGTGAAGAC |
| P53 | TCAACAAGATGTTTTGCCAACTG | ATGTGCTGTGACTGCTTGTAGATG |
Antibodies
The following antibodies were obtained from Santa Cruz Biotechnology: anti-uPA (sc-14019), anti-Lhx2 (sc-81311), anti–HOXA5 (sc-13199), anti-P53 (sc-126), anti–p-P53 (Ser-15; sc-101762), and anti-GAPDH (sc-59541).
Western blotting
MIA PaCa-2 and PANC-1 cells were transfected with uPA-targeted siRNA for 48 h. Cells were collected, and total cell lysates were prepared in standard RIPA extraction buffer containing aprotinin and phenylmethylsulfonyl fluoride. Protein samples (40 μg) were separated under nonreducing conditions by 12% SDS–PAGE and transferred to nitrocellulose membranes (Schleicher & Schuell, Keene, NH). The membranes were immunoprobed using 1:500 dilutions of primary antibodies and 1:1000 dilutions of species-specific, horseradish peroxidase (HRP)–conjugated secondary antibodies and then developed according to an enhanced chemiluminescence protocol (Amersham, Arlington Heights, IL).
Fibrin zymography
The enzymatic activity of electrophoretically separated forms of uPA in the conditioned media of MIA PaCa-2 and PANC-1 transfected with uPA-targeted siRNA for 48 h was determined by SDS–PAGE as described previously (Yamamoto et al., 1994; Mohanam et al., 1997). The acrylamide gels were enriched with purified plasminogen and fibrinogen before polymerization. Equal amounts of sample proteins were electrophoresed, and the gels were washed and stained to determine enzymatic activity as per standard protocols.
Subcutaneous tumor growth and surgical orthotopic implantation of MIA PaCa-2 tumors
We carried out subcutaneous implantation (10,000 or 100,000 cells) and orthotopic implantation of MIA PaCa-2 SP and ΔSP cells as described previously (Fu et al., 1992). Three days after implantation, mice were administered intraperitoneal injections of puPA five times at 150 μg/mouse every other day and or in combination with gemcitabine (0.33 mg/mouse; Koppe et al., 2006). Mice were monitored daily, and body weight was measured daily to ensure that weight loss did not exceed 20%. At 40 d after implantation, mice were killed, and pancreatic tissues were isolated and processed for paraffin embedding.
Immunohistochemistry analysis
Pancreatic tumors tissues from control (untreated) and pSV- and puPA-treated mice were cut into thin sections (5–6 μm thick), which were deparaffinized in xylene and rehydrated in graded ethanol solutions. Antigen retrieval was carried out with 10 mM citrate buffer (pH 6) at boiling temperature for 60 min, with permeabilization in 0.1% Triton-X-100. Permeabilized sections were blocked for 1 h using 3% BSA in PBS. Cells were incubated with primary antibody (uPA or Lhx2) and Alexa Fluor– or HRP-conjugated secondary antibody for 60 min at room temperature. Before mounting, the slides were washed with PBS and incubated for 5 min with a 1:100 dilution of 4′-6-diamidino-2-phenylindole (DAPI) for fluorescence nuclear staining and analyzed using confocal microscopy (BX61 FluoView; Olympus, Minneapolis, MN) at 40× magnification. If the HRP-conjugated secondary antibodies (Abs) were used, the sections were developed using diaminobenzidine substrate. Human pancreatic cancer tissue arrays were obtained from US Biomax (Rockville, MD). Tissue arrays were processed for immunohistochemistry using a standard protocol (Gondi et al., 2004). A control study was performed using a normal rabbit immunoglobulin fraction as the primary antibody (control Ab) in lieu of uPA or Lhx2. The tissues were counterstained with hematoxylin dye to visualize the nucleus.
In situ hybridization
The miRCURY LNA microRNA ISH Optimization Kit (FFPE) and the full-length miR-124 hybridization probe were purchased from Exiqon. The detailed procedure for in situ hybridization was performed following the supplied protocol. In brief, paraffin-embedded tumor tissue sections were subjected to deparaffinization, and the following steps were performed: proteinase K treatment at 37°C, prehybridization at 55°C for 15 min, hybridization with digoxigenin (DIG)-labeled LNA miR-124 probe (50 nM) at 55°C for 60 min, and stringent washes with saline–sodium citrate buffer at 55°C for more than a total of 33 min, followed by DIG blocking reagent (15 min at room temperature), alkaline phosphatase–conjugated anti-DIG at 1:1000 dilution (60 min at room temperature), alkaline phosphatase substrate enzymatic development (120 min at room temperature), and nuclear fast red counterstain (5 min). The slides were mounted and air dried, and the images were captured with an Olympus BX61 fluorescence microscope (Olympus, Tokyo, Japan) with an attached charge-coupled device camera.
Electrophoretic mobility shift assay
HEK293 cells were transfected with empty pcDNA3.1 (mock transfected) or HOXA5-FLAG in pcDNA3.1 alone or in combination with uPA/pcDNA3.1 vector. Two days after transfection, cells were harvested and nuclear extracts were prepared using the NucBuster Protein Extraction Kit (Novagen, Gibbstown, NJ). EMSA reactions were performed using biotinylated double-stranded oligonucleotide derived from p53 promoter, 5′-AATGCTATTTTTGAATTAAGAAAGGTGAGA3′, and the LightShift chemiluminescent EMSA kit (Pierce, Rockford, IL). uPA or BSA (500 ng/reaction) was added to the binding reaction, where indicated.
p53 promoter luciferase reporter activity assay
For the reporter gene assay, MIA PaCa-2 and PANC-1 cells were transiently transfected with 200 ng/ml p53 luciferase reporter vector constructs (Panomics, Redwood City, CA) alone or in combination with puPA and or exogenous wild-type uPA. After 48 h, luciferase activity was determined using a dual-luciferase reporter assay system (Promega) in a luminometer (TD-20/20 DLReady). In some experiments, HEK293 cells were transfected with the human p53 promoter (959 base pairs)-driven luciferase reporter pSGG_PROM (SwitchGear Genomics, Menlo Park, CA), which was cotransfected into HEK293T cells with uPA- and HOXA5-encoding vectors. Luciferase activity was measured at 24 h using a Dual Luciferase Reporter Assay Kit (Promega). Outcomes were normalized to the activity of cotransfected Renilla luciferase-encoding pRL-TK vector.
Transcription factor–binding array analysis
TF protein–protein binding array analysis was performed using the TF Protein Array kit from Panomics (MA3501-08) as per the manufacturer's instructions. Briefly, purified human wild-type uPA (American Diagnostics, Lexington, MA) was suspended in 1× blocking buffer at a concentration of 100 ng/ml. The TF membranes were incubated in 1× blocking buffer for 2 h at room temperature, followed by incubation with uPA in 1× blocking buffer for 2 h at room temperature. This was followed by washings and further incubation with anti-uPA antibody, followed by secondary HRP-conjugated antibody as per kit instructions. HRP was detected using the buffers provided, followed by exposure of membranes to x-ray film. Binding of uPA to TFs was observed as spots on the x-ray film.
Statistical analysis
All experiments were repeated in triplicate, with the exception of the human pancreatic tissue array.
Supplementary Material
Acknowledgments
We thank Peggy Mankin for assistance with flow cytometry analysis and Noorjahan Ali for histology processing. Support from the Department of Medicine, Department of Cancer Biology and Pharmacology, William E. McElroy Foundation, Springfield, IL (to C.S.G.), and the National Cancer Institute (1R21CA141228-01 to V.S.) for this research is acknowledged.
Abbreviations used:
- CSC
cancer stem sells
- HOXA5
homeobox transcription factor A5
- Lhx2 3′-UTR
LIM homeobox-2 3′-untranslated region
- p53
tumor suppressor p53
- PDAC
pancreatic ductal adenocarcinoma
- puPA
plasmid expressing shRNA tageting uPA
- SP
side-population cells
- ΔSP
side-population–depleted cells
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-04-0306) on July 17, 2013.
*These authors contributed equally to this work.
REFERENCES
- Adachi Y, et al. Suppression of glioma invasion and growth by adenovirus-mediated delivery of a bicistronic construct containing antisense uPAR and sense p16 gene sequences. Oncogene. 2002;21:87–95. doi: 10.1038/sj.onc.1204999. [DOI] [PubMed] [Google Scholar]
- Andres SA, Edwards AB, Wittliff JL. Expression of urokinase-type plasminogen activator (uPA), its receptor (uPAR), and inhibitor (PAI-1) in human breast carcinomas and their clinical relevance. J Clin Lab Anal. 2012;26:93–103. doi: 10.1002/jcla.21488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asuthkar S, Gondi CS, Nalla AK, Velpula KK, Gorantla B, Rao JS. Urokinase-type plasminogen activator receptor (uPAR)-mediated regulation of WNT/beta-catenin signaling is enhanced in irradiated medulloblastoma cells. J Biol Chem. 2012;287:20576–20589. doi: 10.1074/jbc.M112.348888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JM, Leach SD. Signaling pathways mediating epithelial-mesenchymal crosstalk in pancreatic cancer: Hedgehog, Notch and TGFbeta. In: Grippo PJ, Munshi HG, editors. Pancreatic Cancer and Tumor Microenvironment. Chapter 7. Trivandrum, India: Transworld Research Network; 2012. pp. 123–138. [PubMed] [Google Scholar]
- Bao B, Wang Z, Ali S, Kong D, Li Y, Ahmad A, Banerjee S, Azmi AS, Miele L, Sarkar FH. Notch-1 induces epithelial-mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells. Cancer Lett. 2011;307:26–36. doi: 10.1016/j.canlet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bekes EM, Deryugina EI, Kupriyanova TA, Zajac E, Botkjaer KA, Andreasen PA, Quigley JP. Activation of pro-uPA is critical for initial escape from the primary tumor and hematogenous dissemination of human carcinoma cells. Neoplasia. 2011;13:806–821. doi: 10.1593/neo.11704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi F, Carmeliet P. uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol. 2002;3:932–943. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- Boonstra MC, Verspaget HW, Ganesh S, Kubben FJ, Vahrmeijer AL, van de Velde CJ, Kuppen PJ, Quax PH, Sier CF. Clinical applications of the urokinase receptor (uPAR) for cancer patients. Curr Pharm Des. 2011;17:1890–1910. doi: 10.2174/138161211796718233. [DOI] [PubMed] [Google Scholar]
- Carriero MV, Stoppelli MP. The urokinase-type plasminogen activator and the generation of inhibitors of urokinase activity and signaling. Curr Pharm Des. 2011;17:1944–1961. doi: 10.2174/138161211796718143. [DOI] [PubMed] [Google Scholar]
- Ceccarelli F, Fuso A, Civitelli L, Ranieri E, Caprio G, Pagni P, Rengo M, Scarpa S. Urokinase expression in course of benign and malignant mammary lesions: comparison between nodular and healthy tissues. J Cancer Res Clin Oncol. 2010;136:157–163. doi: 10.1007/s00432-009-0694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Hao J, Wang L, Li Y. Coexpression of invasive markers (uPA, CD44) and multiple drug-resistance proteins (MDR1, MRP2) is correlated with epithelial ovarian cancer progression. Br J Cancer. 2009;101:432–440. doi: 10.1038/sj.bjc.6605185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YT, Yang YC. Post-transcriptional control of Cited2 by transforming growth factor beta. Regulation via Smads and Cited2 coding region. J Biol Chem. 2006;281:18451–18462. doi: 10.1074/jbc.M601720200. [DOI] [PubMed] [Google Scholar]
- Dahl L, Richter K, Hagglund AC, Carlsson L. Lhx2 expression promotes self-renewal of a distinct multipotential hematopoietic progenitor cell in embryonic stem cell-derived embryoid bodies. PLoS One. 2008;4:e2025. doi: 10.1371/journal.pone.0002025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bock CE, Wang Y. Clinical significance of urokinase-type plasminogen activator receptor (uPAR) expression in cancer. Med Res Rev. 2004;24:13–39. doi: 10.1002/med.10054. [DOI] [PubMed] [Google Scholar]
- Du Z, Qin R, Wei C, Wang M, Shi C, Tian R, Peng C. Pancreatic cancer cells resistant to chemoradiotherapy rich in “stem-cell-like” tumor cells. Dig Dis Sci. 2011;56:741–750. doi: 10.1007/s10620-010-1340-0. [DOI] [PubMed] [Google Scholar]
- Dumler I, Stepanova V, Jerke U, Mayboroda OA, Vogel F, Bouvet P, Tkachuk V, Haller H, Gulba DC. Urokinase-induced mitogenesis is mediated by casein kinase 2 and nucleolin. Curr. Biol. 1999;9:1468–1476. doi: 10.1016/s0960-9822(00)80116-5. [DOI] [PubMed] [Google Scholar]
- Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;1:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18:4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Guadagni F, Hoffman RM. A metastatic nude-mouse model of human pancreatic cancer constructed orthotopically with histologically intact patient specimens. Proc Natl Acad Sci USA. 1992;89:5645–5649. doi: 10.1073/pnas.89.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendronneau G, Lemieux M, Morneau M, Paradis J, Tetu B, Frenette N, Aubin J, Jeannotte L. Influence of Hoxa5 on p53 tumorigenic outcome in mice. Am J Pathol. 2010;176:995–1005. doi: 10.2353/ajpath.2010.090499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JF, Schlieman M, Singh P, Saxena R, Martinick M, Hutson AD, Corasanti J. A pilot study of urokinase-type plasminogen activator (uPA) overexpression in the brush cytology of patients with malignant pancreatic or biliary strictures. HPB Surg 2009, 805971. 2009 doi: 10.1155/2009/805971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondi CS, Kandhukuri N, Dinh DH, Gujrati M, Rao JS. Down-regulation of uPAR and uPA activates caspase-mediated apoptosis and inhibits the PI3K/AKT pathway. Int J Oncol. 2007a;31:19–27. [PMC free article] [PubMed] [Google Scholar]
- Gondi CS, Lakka SS, Dinh DH, Olivero WC, Gujrati M, Rao JS. Downregulation of uPA, uPAR and MMP-9 using small, interfering, hairpin RNA (siRNA) inhibits glioma cell invasion, angiogenesis and tumor growth. Neuron Glia Biol. 2004;1:165–176. doi: 10.1017/s1740925x04000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondi CS, Lakka SS, Dinh DH, Olivero WC, Gujrati M, Rao JS. Intraperitoneal injection of an hpRNA-expressing plasmid targeting uPAR and uPA retards angiogenesis and inhibits intracranial tumor growth in nude mice. Clin Cancer Res. 2007b;13:4051–4060. doi: 10.1158/1078-0432.CCR-06-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondi CS, Lakka SS, Yanamandra N, Siddique K, Dinh DH, Olivero WC, Gujrati M, Rao JS. Expression of antisense uPAR and antisense uPA from a bicistronic adenoviral construct inhibits glioma cell invasion, tumor growth, and angiogenesis. Oncogene. 2003;22:5967–5975. doi: 10.1038/sj.onc.1206535. [DOI] [PubMed] [Google Scholar]
- Gorantla B, Asuthkar S, Rao JS, Patel J, Gondi CS. Suppression of the uPAR-uPA system retards angiogenesis, invasion, and in vivo tumor development in pancreatic cancer cells. Mol Cancer Res. 2011;9:377–389. doi: 10.1158/1541-7786.MCR-10-0452. [DOI] [PubMed] [Google Scholar]
- Gou S, Liu T, Wang C, Yin T, Li K, Yang M, Zhou J. Establishment of clonal colony-forming assay for propagation of pancreatic cancer cells with stem cell properties. Pancreas. 2007;34:429–435. doi: 10.1097/MPA.0b013e318033f9f4. [DOI] [PubMed] [Google Scholar]
- Gupta R, Chetty C, Bhoopathi P, Lakka S, Mohanam S, Rao JS, Dinh DE. Downregulation of uPA/uPAR inhibits intermittent hypoxia-induced epithelial-mesenchymal transition (EMT) in DAOY and D283 medulloblastoma cells. Int J Oncol. 2011;38:733–744. doi: 10.3892/ijo.2010.883. [DOI] [PubMed] [Google Scholar]
- Hamada S, Shimosegawa T. Pancreatic cancer stem cell and mesenchymal stem cell. In: Grippo PJ, Munshi HG, editors. Pancreatic Cancer and Tumor Microenvironment. Chapter 6. Trivandrum, India: Transworld Research Network; 2012. pp. 111–122. [PubMed] [Google Scholar]
- Haque I, Mehta S, Majumder M, Dhar K, De A, McGregor D, Van Veldhuizen PJ, Banerjee SK, Banerjee S. Cyr61/CCN1 signaling is critical for epithelial-mesenchymal transition and stemness and promotes pancreatic carcinogenesis. Mol Cancer. 2011 doi: 10.1186/1476-4598-10-8. DOI: 10.1186/1476-4598-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SR, et al. Evaluation of urinary plasminogen activator, its receptor, matrix metalloproteinase-9, and von Willebrand factor in pancreatic cancer. Clin Cancer Res. 2003;9:4935–4943. [PubMed] [Google Scholar]
- He X, et al. DJ-1 promotes invasion and metastasis of pancreatic cancer cells by activating SRC/ERK/uPA. Carcinogenesis. 2012;33:555–562. doi: 10.1093/carcin/bgs002. [DOI] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Hildenbrand R, Allgayer H, Marx A, Stroebel P. Modulators of the urokinase-type plasminogen activation system for cancer. Expert Opin Investig Drugs. 2010;19:641–652. doi: 10.1517/13543781003767400. [DOI] [PubMed] [Google Scholar]
- Hong SP, Wen J, Bang S, Park S, Song SY. CD44-positive cells are responsible for gemcitabine resistance in pancreatic cancer cells. Int J Cancer. 2009;125:2323–2331. doi: 10.1002/ijc.24573. [DOI] [PubMed] [Google Scholar]
- Ischenko I, Seeliger H, Kleespies A, Angele MK, Eichhorn ME, Jauch KW, Bruns CJ. Pancreatic cancer stem cells: new understanding of tumorigenesis, clinical implications. Langenbecks Arch Surg. 2010;395:1–10. doi: 10.1007/s00423-009-0502-z. [DOI] [PubMed] [Google Scholar]
- Kabashima A, et al. Side population of pancreatic cancer cells predominates in TGF-beta-mediated epithelial to mesenchymal transition and invasion. Int J Cancer. 2009;124:2771–2779. doi: 10.1002/ijc.24349. [DOI] [PubMed] [Google Scholar]
- Kabashima-Niibe A, et al. Mesenchymal stem cells regulate epithelial-mesenchymal transition and tumor progression of pancreatic cancer cells. Cancer Sci. 2013;104:157–164. doi: 10.1111/cas.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Fujita Y, Nakane K, Kojima T, Nozawa Y, Deguchi T, Ito M. ETS1 promotes chemoresistance and invasion of paclitaxel-resistant, hormone-refractory PC3 prostate cancer cells by up-regulating MDR1 and MMP9 expression. Biochem Biophys Res Commun. 2012;417:966–971. doi: 10.1016/j.bbrc.2011.12.047. [DOI] [PubMed] [Google Scholar]
- Keck T, Brabletz T. Under stress: p53 controls EMT and stemness in pancreatic epithelial cells. Cell Cycle. 2011;10:1715. doi: 10.4161/cc.10.11.15645. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Khanna A, Mahalingam K, Chakrabarti D, Periyasamy G. Ets-1 expression and gemcitabine chemoresistance in pancreatic cancer cells. Cell Mol Biol Lett. 2011;16:101–113. doi: 10.2478/s11658-010-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Lee J, Oh T, Moon Y, Chang E, Seo KS, Hoehn BD, An S, Lee JH. Genome-wide identification of OTP gene as a novel methylation marker of breast cancer. Oncol Rep. 2012;27:1681–1688. doi: 10.3892/or.2012.1691. [DOI] [PubMed] [Google Scholar]
- Kondraganti S, Gondi CS, McCutcheon I, Dinh DH, Gujrati M, Rao JS, Olivero WC. RNAi-mediated downregulation of urokinase plasminogen activator and its receptor in human meningioma cells inhibits tumor invasion and growth. Int J Oncol. 2006;28:1353–1360. [PMC free article] [PubMed] [Google Scholar]
- Koppe MJ, Oyen WJ, Bleichrodt RP, Verhofstad AA, Goldenberg DM, Boerman OC. Combination therapy using gemcitabine and radioimmunotherapy in nude mice with small peritoneal metastases of colonic origin. Cancer Biother Radiopharm. 2006;21:506–514. doi: 10.1089/cbr.2006.21.506. [DOI] [PubMed] [Google Scholar]
- Lonardo E, Hermann PC, Heeschen C. Pancreatic cancer stem cells—update and future perspectives. Mol Oncol. 2010;4:431–442. doi: 10.1016/j.molonc.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund IK, Illemann M, Thurison T, Christensen IJ, Hoyer-Hansen G. uPAR as anti-cancer target: evaluation of biomarker potential, histological localization, and antibody-based therapy. Curr Drug Targets. 2011;12:1744–1760. doi: 10.2174/138945011797635902. [DOI] [PubMed] [Google Scholar]
- Mardaryev AN, et al. Lhx2 differentially regulates Sox9, Tcf4 and Lgr5 in hair follicle stem cells to promote epidermal regeneration after injury. Development. 2011;138:4843–4852. doi: 10.1242/dev.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markl B, Renk I, Oruzio DV, Jahnig H, Schenkirsch G, Scholer C, Ehret W, Arnholdt HM, Anthuber M, Spatz H. Tumour budding, uPA and PAI-1 are associated with aggressive behaviour in colon cancer. J Surg Oncol. 2010;102:235–241. doi: 10.1002/jso.21611. [DOI] [PubMed] [Google Scholar]
- Mazar AP, Ahn RW, O'Halloran TV. Development of novel therapeutics targeting the urokinase plasminogen activator receptor (uPAR) and their translation toward the clinic. Curr Pharm Des. 2011;17:1970–1978. doi: 10.2174/138161211796718152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekkawy AH, Morris DL, Pourgholami MH. Urokinase plasminogen activator system as a potential target for cancer therapy. Future Oncol. 2009;5:1487–1499. doi: 10.2217/fon.09.108. [DOI] [PubMed] [Google Scholar]
- Mizuno H, Spike BT, Wahl GM, Levine AJ. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc Natl Acad Sci USA. 2010;107:22745–22750. doi: 10.1073/pnas.1017001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanam S, Chintala SK, Go Y, Bhattacharya A, Venkaiah B, Boyd D, Gokaslan ZL, Sawaya R, Rao JS. In vitro inhibition of human glioblastoma cell line invasiveness by antisense uPA receptor. Oncogene. 1997;14:1351–1359. doi: 10.1038/sj.onc.1200963. [DOI] [PubMed] [Google Scholar]
- Moriyama T, Ohuchida K, Mizumoto K, Cui L, Ikenaga N, Sato N, Tanaka M. Enhanced cell migration and invasion of CD133+ pancreatic cancer cells cocultured with pancreatic stromal cells. Cancer. 2010;116:3357–3368. doi: 10.1002/cncr.25121. [DOI] [PubMed] [Google Scholar]
- Nadal N, Chapiro E, Flandrin-Gresta P, Thouvenin S, Vasselon C, Beldjord K, Fenneteau O, Bernard O, Campos L, Nguyen-Khac F. LHX2 deregulation by juxtaposition with the IGH locus in a pediatric case of chronic myeloid leukemia in B-cell lymphoid blast crisis. Leuk Res. 2012;36:e195–e198. doi: 10.1016/j.leukres.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- Oettle H, et al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. J Am Med Assoc. 2007;297:267–277. doi: 10.1001/jama.297.3.267. [DOI] [PubMed] [Google Scholar]
- Pasi CE, Pelicci PG. Inhibition of epithelial-to-mesenchimal transition: a novel tumor suppressor function of p53? Cell Cycle. 2011;10:2616–2617. doi: 10.4161/cc.10.16.16543. [DOI] [PubMed] [Google Scholar]
- Perez C, Dastot-Le Moal F, Collot N, Legendre M, Abadie I, Bertrand AM, Amselem S, Sobrier ML. Screening of LHX2 in patients presenting growth retardation with posterior pituitary and ocular abnormalities. Eur J Endocrinol. 2012;167:85–91. doi: 10.1530/EJE-12-0026. [DOI] [PubMed] [Google Scholar]
- Pinto do OP, Wandzioch E, Kolterud A, Carlsson L. Multipotent hematopoietic progenitor cells immortalized by Lhx2 self-renew by a cell nonautonomous mechanism. Exp Hematol. 2001;29:1019–1028. doi: 10.1016/s0301-472x(01)00666-x. [DOI] [PubMed] [Google Scholar]
- Provost JJ, et al. Urokinase plasminogen activator receptor induced non-small cell lung cancer invasion and metastasis requires NHE1 transporter expression and transport activity. Cell Oncol (Dordr) 2012;35:95–110. doi: 10.1007/s13402-011-0068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman V, Tamori A, Vali M, Zeller K, Korz D, Sukumar S. HOXA5 regulates expression of the progesterone receptor. J Biol Chem. 2000;275:26551–26555. doi: 10.1074/jbc.C000324200. [DOI] [PubMed] [Google Scholar]
- Rasheed ZA, Matsui W. Biological and clinical relevance of stem cells in pancreatic adenocarcinoma. J Gastroenterol Hepatol. 2012;27 (Suppl 2):15–18. doi: 10.1111/j.1440-1746.2011.07015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch V, et al. Synergistic activity of sorafenib and sulforaphane abolishes pancreatic cancer stem cell characteristics. Cancer Res. 2010;70:5004–5013. doi: 10.1158/0008-5472.CAN-10-0066. [DOI] [PubMed] [Google Scholar]
- Richter K, Pinto do OP, Hagglund AC, Wahlin A, Carlsson L. Lhx2 expression in hematopoietic progenitor/stem cells in vivo causes a chronic myeloproliferative disorder and altered globin expression. Haematologica. 2003;88:1336–1347. [PubMed] [Google Scholar]
- Song N, Ding Y, Zhuo W, He T, Fu Z, Chen Y, Song X, Fu Y, Luo Y. The nuclear translocation of endostatin is mediated by its receptor nucleolin in endothelial cells. Angiogenesis. 2012;15:697–711. doi: 10.1007/s10456-012-9284-y. [DOI] [PubMed] [Google Scholar]
- Stepanova V, et al. Nuclear translocation of urokinase-type plasminogen activator. Blood. 2008;112:100–110. doi: 10.1182/blood-2007-07-104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiede S, Paus R. Lhx2—decisive role in epithelial stem cell maintenance, or just the “tip of the iceberg”? Bioessays. 2006;28:1157–1160. doi: 10.1002/bies.20506. [DOI] [PubMed] [Google Scholar]
- Tod J, Jenei V, Thomas G, Fine D. Tumor-stromal interactions in pancreatic cancer. Pancreatology. 2013;13:1–7. doi: 10.1016/j.pan.2012.11.311. [DOI] [PubMed] [Google Scholar]
- Tornqvist G, Sandberg A, Hagglund AC, Carlsson L. Cyclic expression of lhx2 regulates hair formation. PLoS Genet. 2010;6:e1000904. doi: 10.1371/journal.pgen.1000904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velpula KK, Dasari VR, Tsung AJ, Dinh DH, Rao JS. hUCBSC reverts EMT to MET in glioma stem cells by downregulating synergistic transcriptional activation of Sox2 and Twist1. Oncotarget 2,1028–1042. 2011 doi: 10.18632/oncotarget.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe T, Yoshida K, Shindoh M, Kaya M, Fujikawa K, Sato H, Seiki M, Ishii S, Fujinaga K. The Ets-1 and Ets-2 transcription factors activate the promoters for invasion-associated urokinase and collagenase genes in response to epidermal growth factor. Int J Cancer. 1998;77:128–137. doi: 10.1002/(sici)1097-0215(19980703)77:1<128::aid-ijc20>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Whatcott C, Han H, Posner RG, Von Hoff DD. Tumor-stromal interactions in pancreatic cancer. Crit Rev Oncog. 2013;18:135–151. doi: 10.1615/critrevoncog.v18.i1-2.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Wirta V, Meletis K, Wikstrom L, Carlsson L, Frisen J, Lundeberg J. Catalog of gene expression in adult neural stem cells and their in vivo microenvironment. Exp Cell Res. 2006;312:1798–1812. doi: 10.1016/j.yexcr.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Xia H, et al. Loss of brain-enriched miR-124 microRNA enhances stem-like traits and invasiveness of glioma cells. J Biol Chem. 2012;287:9962–9971. doi: 10.1074/jbc.M111.332627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Sawaya R, Mohanam S, Rao VH, Bruner JM, Nicolson GL, Rao JS. Expression and localization of urokinase-type plasminogen activator receptor in human gliomas. Cancer Res. 1994;54:5016–5020. [PubMed] [Google Scholar]
- Zhang HH, et al. Characterization of cancer stem-like cells in the side population cells of human gastric cancer cell line MKN-45. J Zhejiang Univ Sci B. 2013;14:216–223. doi: 10.1631/jzus.B1200102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao Y, Mailloux CM, Hermesz E, Palkovits M, Westphal H. A role of the LIM-homeobox gene Lhx2 in the regulation of pituitary development. Dev Biol. 2010;337:313–323. doi: 10.1016/j.ydbio.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.