Gene targeting was used to ablate Cep135, a component of the centriolar cartwheel, in chicken DT40 cells. Cep135-deficient cells showed no major defects in centrosome composition or function, although centrosome amplification after HU treatment increased significantly. EM revealed an unusual structure in the lumen of Cep135-null centrioles.
Abstract
Centrosomes are key microtubule-organizing centers that contain a pair of centrioles, conserved cylindrical, microtubule-based structures. Centrosome duplication occurs once per cell cycle and relies on templated centriole assembly. In many animal cells this process starts with the formation of a radially symmetrical cartwheel structure. The centrosomal protein Cep135 localizes to this cartwheel, but its role in vertebrates is not well understood. Here we examine the involvement of Cep135 in centriole function by disrupting the Cep135 gene in the DT40 chicken B-cell line. DT40 cells that lack Cep135 are viable and show no major defects in centrosome composition or function, although we note a small decrease in centriole numbers and a concomitant increase in the frequency of monopolar spindles. Furthermore, electron microscopy reveals an atypical structure in the lumen of Cep135-deficient centrioles. Centrosome amplification after hydroxyurea treatment increases significantly in Cep135-deficient cells, suggesting an inhibitory role for the protein in centrosome reduplication during S-phase delay. We propose that Cep135 is required for the structural integrity of centrioles in proliferating vertebrate cells, a role that also limits centrosome amplification in S-phase–arrested cells.
INTRODUCTION
The centrosome is the primary site of microtubule nucleation in animal somatic cells. Centrosomes control a number of processes, including mitotic spindle formation, cell polarity, motility, and intracellular trafficking. Defects in centrosome number or function compromise stem cell division, DNA-damage responses, and faithful chromosome segregation, leading to a variety of diseases, such as microcephaly, dwarfism, ciliopathies, and cancer. Centrosome numbers are therefore under strict control, with centrosome duplication limited to once per cell cycle.
The centrosome consists of a pair of centrioles—cylindrical structures of 0.2–0.5 μm composed of nine microtubule triplets. Centrioles are surrounded by electron-dense, pericentriolar material (PCM), which serves as the main microtubule nucleation site within the centrosome, and nearby lie the centriolar satellites, electron-dense granules that contribute to centrosome function, possibly by mediating protein transport to the centrosome (Dammermann and Merdes, 2002). Centriole duplication is initiated in late G1/early S phase. Each centriole, the so-called mother centriole, templates the assembly of a procentriole. The wall of procentrioles is initially composed of nine singlet microtubules, but these are converted to doublets and/or triplets as the procentrioles elongate to form a new centriole, the daughter centriole (Hinchcliffe and Sluder, 2001; Doxsey et al., 2005; Bettencourt-Dias and Glover, 2007; Nigg, 2007). Procentriole assembly depends on a structural scaffold. In Caenorhabditis elegans, procentrioles form around a central tube comprising the conserved proteins Sas-5 and Sas-6 (Dammermann et al., 2004; Delattre and Gonczy, 2004; Leidel et al., 2005). In other organisms, such as Chlamydomonas, Drosophila, and vertebrates, the procentriole forms around a cartwheel-like structure (Cavalier-Smith, 1974). The cartwheel consists of a central hub surrounded by nine radial spokes. This ninefold-symmetrical structure is believed to dictate the structure of the nascent centriole. Sas-6 is a highly conserved protein that localizes to the proximal end of the mother centriole at the position of the cartwheel (Kleylein-Sohn et al., 2007; Strnad et al., 2007). Recent reports show that Sas-6 oligomerizes into a ring-like structure with nine coiled-coil domains projecting outward, thus closely resembling the central hub and the nine radial spokes that constitute the cartwheel (Kitagawa et al., 2011; van Breugel et al., 2011).
Bld10/Cep135 is another highly conserved centrosomal protein involved in cartwheel assembly (Carvalho-Santos et al., 2010). Analysis of Chlamydomonas mutants with defective flagella demonstrated a requirement for Bld10 in the assembly of basal bodies, which are analogous to centrioles (Matsuura et al., 2004). Expression of bld10-deletion mutants in Chlamydomonas caused the formation of centrioles with altered numbers of centriolar microtubules due to reduced cartwheel diameters (Hiraki et al., 2007). Bld10 is also required for basal body formation in Paramecium and, in both protists, it localizes to the tips of the cartwheel spokes (Matsuura et al., 2004; Hiraki et al., 2007; Jerka-Dziadosz et al., 2010). Its absence destabilized Sas-6p at the cartwheel hub, even though Bld10 was not required for the establishment of a structure with ninefold symmetry (Nakazawa et al., 2007; Jerka-Dziadosz et al., 2010). In Tetrahymena, Bld10 shows a similar localization to the outer cartwheel and is also required for basal body assembly and maintenance (Bayless et al., 2012). In Drosophila, Bld10 depletion led to defects in centriole duplication and PCM maturation (Dobbelaere et al., 2008), and bld10 mutants exhibited centrioles and basal bodies that were shorter than controls (Mottier-Pavie and Megraw, 2009). Recent data indicate that cartwheels can assemble in the absence of Drosophila Bld10, although they appear to lack stability (Roque et al., 2012).
The mammalian orthologue of Bld10, Cep135, localizes to the proximal lumen and exterior of centrioles, as determined by immuno–electron microscopy analyses in hamster (Ohta et al., 2002) and human (Kleylein-Sohn et al., 2007) cells. Recent work using superresolution three-dimensional structured illumination microscopy in human cells localized the principal Cep135 signal to the proximal end of mother centrioles, with a weaker signal on nascent daughter centrioles (Sonnen et al., 2012). Depletion of Cep135 prevented PLK4-induced centriole overduplication in human cells (Kleylein-Sohn et al., 2007). Cep135 deficiency caused the delocalization of other centrosomal proteins from the proximal end of the centrioles (Uetake et al., 2004; Kim et al., 2008), and recent data demonstrated significant centriole abnormalities resulting from Cep135 depletion in human cells (Lin et al., 2013). Furthermore, a mutation in Cep135 has been reported to cause primary microcephaly (Hussain, 2012), highlighting the potential importance of Cep135 in centrosome function. Despite these observations, the precise function of Cep135 in the vertebrate centrosome is not well understood.
In this study, we use gene targeting in the genetically tractable DT40 cell line to disrupt Cep135 function. Cep135-knockout cells are viable and capable of centriole duplication. A small increase in monopolar spindles is observed, with a concomitant reduction in centriole numbers, indicative of minor perturbations of centriole biogenesis. Although the centrosomes appear generally intact, we note an atypical electron-dense structure within centrioles. Furthermore, centriole amplification is significantly elevated during prolonged S-phase in cells that lack Cep135. These data indicate a role for Cep135 in centriole integrity and the control of centrosome number during S-phase delay.
RESULTS AND DISCUSSION
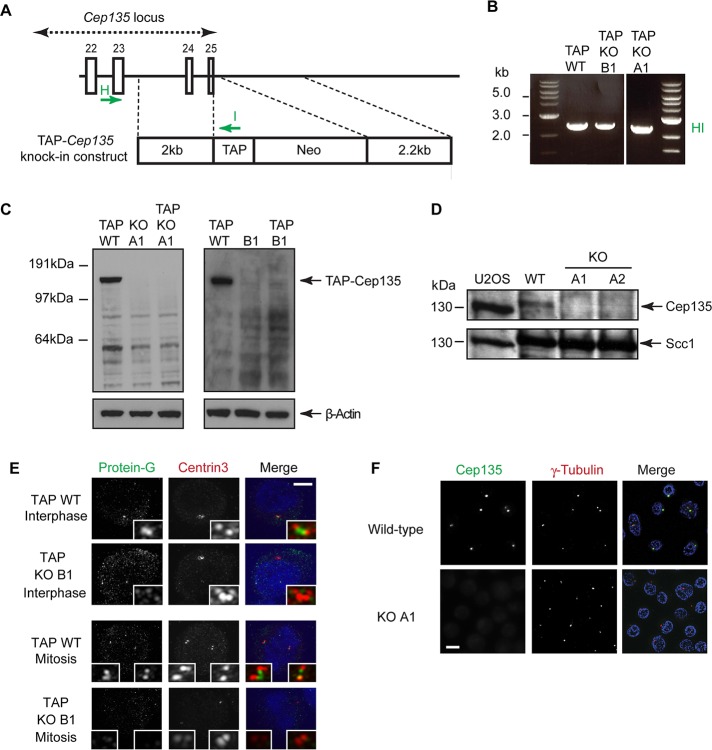
To investigate the effect of Cep135 deficiency on centrosome function and structure, we used targeted gene disruption in the hyperrecombinogenic chicken B-cell line, DT40. We identified the chicken Cep135 orthologue from the National Center for Biotechnology Information database and confirmed its sequence by 5′ rapid amplification of cDNA ends and reverse transcription (RT)-PCR analysis of DT40 RNA. The large size of the Cep135 locus precluded simple disruption of the entire gene. Therefore, as shown in Figure 1A, two knockout strategies were chosen to disrupt the Cep135 locus: the first, using construct A, deleted the genomic region encoding exons 1–4, and the second, using construct B, deleted the region encoding exons 5–9 (Figure 1A). Sequential gene targeting with either of these constructs yielded several clones in which the Cep135 locus was disrupted (Figure 1, B and C). To indicate which strategy was used, clones generated with construct A are described as Cep135 knockout (KO) A1 and A2 and those with construct B as Cep135 KO B1 and B2.
FIGURE 1:
Gene targeting of chicken Cep135. (A) Schematic shows the chicken Cep135 genomic locus and the targeting constructs used to replace either exons 1–4 (construct A) or exons 5–9 (construct B) of Cep135 with the indicated drug resistance cassettes. The positions of the allele-specific and the resistance cassette–specific primers used for PCR-based genotypic analysis are shown as capital letters (A–G). (B, C) Agarose gel images showing the allele-specific PCR products from the wild-type (Cep135+/+), (B) Cep135−/− (KO) A1 and A2, and (C) Cep135−/− (KO) B1 and B2 cell lines. Capital letters correspond to primer pairs indicated in A. Bsr, blasticidin; Neo, neomycin; Puro, puromycin. (D) An agarose gel image showing the Cep135 and β-actin–specific RT-PCR products from the wild-type and Cep135 KO A1 and A2 DT40 cell lines. (E) qPCR analysis of several Cep135 exons in clones of the indicated genotype. Histogram indicates CT values referenced to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and normalized to wild-type values. Error bars indicate SEM from two triplicate experiments.
RT-PCR analysis showed that the full-length Cep135 transcript was absent from Cep135 KO A1 and A2 (Figure 1D), although a weak signal from the region 3′ of the deleted sequence was observed (data not shown), suggesting that some downstream transcript remains in cells. Similarly, quantitative RT-PCR on clones B1 and B2 showed that no transcript was detectable from the deleted exon 6, although sequences from the neighboring exons 4 and 13 were present at levels similar to those seen wild-type cells (Figure 1E). To investigate whether Cep135-targeted cells were indeed deficient in Cep135 protein expression, we placed a tandem affinity purification (TAP) tag (Burckstummer et al., 2006), comprising protein G and streptavidin-binding protein, in frame with the last exon of Cep135 by gene targeting in wild-type, A1, and B1 cells (Figure 2, A and B). These cell lines were called TAP KO A1 or TAP KO B1. Western blots with anti–protein G antibodies revealed a 150-kDa band in TAP-wild-type cells. This corresponds to full-length Cep135 plus the TAP tag, which has a molecular weight of 17 kDa. In TAP KO A1 and TAP KO B1 cells, however, no product was detected, suggesting that the Cep135 KO cell lines are protein null (Figure 2C). To further confirm that the cell lines were protein null, we examined Cep135 protein levels by immunoblot with an antibody raised against the C-terminal end of the human Cep135 coding sequence (Ohta et al., 2002). As shown in Figure 2D, this antibody detected no bands corresponding to Cep135 in Cep135 KO cells. Consistently, despite robust centrosomal signals in wild-type cells, immunofluorescence microscopy failed to detect a centrosomal signal in TAP KO A1 (data not shown) or TAP KO B1 cells when using antibodies directed against the TAP tag (Figure 2E). Similarly, antibodies to human Cep135 did not reveal a centrosomal signal in KO A1 cells (Figure 2F). Together these data clearly demonstrate that our targeting of Cep135 resulted in the loss of the full-length Cep135 protein in both KO A1 and KO B1 cell lines. We therefore used the two cell lines interchangeably to assay the loss-of-function phenotype of Cep135.
FIGURE 2:
Cep135−/− cells lack Cep135 protein. (A) TAP-Cep135 knock-in construct designed to introduce a C-terminal TAP tag to Cep135. Targeting of this construct into the Cep135 locus removes the endogenous stop codon and allows expression of the TAP sequence as a fusion with Cep135. (B) PCR using the primers HI shown in A to assess correct integration of the TAP tag in the indicated clones. (C) Western blots of heterozygous Cep135-TAP–tagged clones of the indicated genotype using antibodies to protein G, a component of the TAP tag. We used β-actin as a loading control. (D) Western blots of human osteosarcoma U2OS and DT40 wild-type and Cep135 KO A1 and A2 total protein extracts with anti-Cep135 antibody. Scc1 serves as a loading control. (E) Localization of TAP-Cep135 protein products in the indicated clones visualized with structured illumination microscopy. Centrin3 (red) was used as a distal centriolar marker, and DNA was stained with Hoechst 33258 (blue). TAP-Cep135 was detected by antibodies against protein G (green). Scale bar, 3 μm. (F) Immunofluorescence microscopy analysis of clones of the indicated genotype. Cells were stained with antibodies against Cep135 (green) and γ-tubulin (red) as a reference marker. Scale bar, 10 μm.
We next analyzed the proliferative properties of Cep135 KO cells. As shown in Figure 3A, Cep135-deficient cells proliferated at a rate indistinguishable from that of wild-type cells. Flow cytometry analysis of the Cep135-targeted cells showed no difference from wild type with respect to the proportions of the population undergoing DNA replication (Figure 3B) or mitosis (Figure 3C). To extend our analysis of mitosis in Cep135-deficient cells, we measured the duration of mitosis by live cell imaging using wild-type and Cep135-deficient cells that stably expressed histone H2B-RFP (Dodson et al., 2007). The mean time taken from chromosome condensation to decondensation was found to be 42.9 min in both wild-type cells and Cep135-deficient cells (Figure 3D), indicating that mitotic progression is normal in cells that lack Cep135. Although the data presented in this figure were obtained from clones generated with construct A, those that resulted from targeting with construct B showed similar proliferative characteristics with no significant difference from wild-type cells (data not shown). These results indicate that full-length Cep135 is dispensable for cell viability and normal cell cycle progression in DT40 cells. Given the degree to which Cep135 is evolutionarily conserved, we had expected a stronger phenotype, so an obvious issue is that of potential redundancy. Extensive BLAST searching of the chicken genome and EST databases has not provided any evidence for a duplication of Cep135. In a detailed bioinformatic analysis of Cep135, Carvalho-Santos et al. (2010) found that testis-specific protein 10 (Tsga10) shared high levels of sequence similarity with Cep135 within a specific, conserved region. We confirmed that Tsga10 was expressed in DT40 cells (Supplemental Figure S1C). To test whether Tsga10 might serve redundant functions with Cep135, we disrupted Tsga10 in Cep135-knockout cells, using the strategy shown in Supplemental Figure S1, A and B, and confirmed the loss of Tsga10 expression (Supplemental Figure S1C). We found no significant alteration of spindle integrity in Cep135/Tsga10 knockouts (Supplemental Figure S1D) and no effect on centriole number in monastrol-induced monopoles (Supplemental Figure S1E). From these data, we conclude that Cep135 is nonredundant in DT40 cells.
FIGURE 3:

Cep135 null cells are viable and show no proliferative defect. (A) Proliferation analysis of Cep135 KO DT40 cells. Cells were seeded at 105/ml, and cell numbers were counted over 72 h. Data show mean ± SD of three separate experiments. (B) Quantitative cell cycle analysis of asynchronous cells of the indicated genotype stained for incorporated BrdU and total DNA levels. The G1 (bottom left), S (top), and G2/M (bottom right) gates are indicated in boxes, and the numbers refer to the percentage of cells detected in each of the gates averaged from three separate experiments. (C) Mitotic indices for wild type and two Cep135 KO clones were determined by M-phase marker phosphorylated histone H3 (PH3). The numbers refer to the percentage of cells in M phase averaged from three separate experiments. (D) Duration of mitosis in wild-type and Cep135 KO cells. Graph shows the mean of time in mitosis and the mean of time from the beginning of prometaphase to the end metaphase and the mean of time from the beginning of anaphase to the end of telophase. Each data point represents one cell. Data show mean ± SD for the 50 individual cells analyzed in three different experiments.
Our time-lapse data suggested that mitotic spindle assembly was not greatly perturbed in Cep135-deficient cells. Indeed, fixed-cell analysis indicated that mitotic spindles in the majority of Cep135-deficient cells were bipolar, with γ-tubulin localized exclusively at the spindle poles and the chromosomes aligned at the metaphase plate, as in wild-type cells (Figure 4A). We observed, however, a small increase in the number of Cep135-deficient cells with monopolar spindles compared with wild-type cells (Figure 4B). Bipolar spindle lengths were 7.66 ± 0.8 μm in Cep135-deficient cells and 7.41 ± 1.0 μm in wild-type controls, showing no significant difference (Supplemental Figure S2). To allow accurate scoring of centriole numbers, we arrested wild-type and Cep135-deficient cells in mitosis by monastrol, a small-molecule inhibitor of Eg5, the kinesin responsible for centrosome separation. Monastrol treatment therefore causes formation of monopoles that contain both centrosomes, each with two centrioles (Figure 4C). Most monopoles in wild-type cells contained the normal centriole complement of four, with only 4% of the poles displaying one or two centrioles. Some of these may correspond to centriole pairs that could not be resolved by confocal microscopy. In contrast, 26% of monopoles in Cep135-deficient cells contained only one or two centrioles (Figure 4D). Collectively these data demonstrate that although Cep135 is not essential for centriole biogenesis, its loss dysregulates control of centrosome number in a small proportion of cycling cells. Despite the increase in monopolar spindles and reduction in centriole numbers in Cep135-deficient cells, we observed only minor changes in proliferation rate and mitotic index. It is conceivable that our assays are not sensitive enough to detect subtle changes. Indeed, DT40 cells lacking another centrosomal protein, CEP63, exhibit >30% monopolarity and a dramatic reduction in centriole numbers, yet we observe only a 15% increase in doubling time (Sir et al. 2011).
FIGURE 4:
Analysis of mitosis and MT nucleation in Cep135-deficient cells. (A) Microscopy analysis of mitotic phases in wild-type and Cep135 KO cells. Immunofluorescence images of mitotic cells of the indicated genotype stained with antibodies against α-tubulin (red) and γ-tubulin (green); DNA is labeled with Hoechst 33258 (blue). Bar, 10 μm. (B) Bipolar, multipolar, and monopolar spindles in wild-type and Cep135 KO B1 and B2 cells were quantified in four independent experiments. Histogram shows mean + SEM of data from 150 mitotic cells per experiment. *p ≤ 0.05 in comparison with wild-type cells. (C) DT40 WT and Cep135 KO B1 were treated with 125 μM monastrol for 16 h. Cells were stained for antibodies against TACC3 (red) and centrin3 (green), and DNA is labeled with Hoechst 33258 (blue). Scale bar, 5 μm. (D) The number of centrioles within monopoles derived from monastrol-treated wild-type and KO B2 cells was counted in two independent experiments. Histogram shows mean + SEM of data from 50 mitotic cells per experiment. *p ≤ 0.05 in comparison with wild-type cells. (E) Immunofluorescence microscopy analysis of microtubule nucleation in wild-type DT40 cells before and after release from 3-h arrest in 1 μg/ml nocodazole at 4°C. Cells were stained with antibodies to α-tubulin (green) and centrin2 (red). DNA (blue) was visualized with 4′,6-diamidino-2-phenylindole (DAPI) staining before imaging. Scale bar, 10 μm. (F) Quantitation of percentage of cells of the indicated genotype with aster nucleation after microtubule regrowth for 1 min at 39.5°C. Histogram shows mean ± SD of three separate experiments in which at least 200 cells per experiment were counted. (G) Transmission electron micrographs of wild-type and Cep135−/− cells showing centriolar appendages and microtubules associated with the centrosome. Scale bar, 100 nm.
We next examined the ability of Cep135-deficient centrosomes to nucleate microtubules after depolymerization of microtubules (MTs) after cold and nocodazole treatment. We found that in the absence of Cep135, cells were capable of nucleating MTs as well as wild-type cells, implying that Cep135 is not essential for efficient microtubule nucleation in DT40 cells (Figure 4, E and F). Additional evidence for normal microtubule nucleation in Cep135 KO cells came from transmission electron microscopy (TEM) analysis. As shown in Figure 4G, Cep135-deficient centrosomes associate with microtubules. Together with the data showing normal mitoses in Cep135 KO cells, these results support the conclusion that there is no major effect of Cep135 disruption on the microtubule-organizing activity of the centrosome in DT40 cells.
To investigate the effects of Cep135 deficiency on centrosome composition, we next used immunofluorescence microscopy with antibodies specific to proteins localizing to different components of the centrosome. Analysis of components of the centrioles (centrin2, Cep76, ninein, glutamylated tubulin), cartwheel (Sas-6), PCM (γ-tubulin, Aurora A), and pericentriolar satellites (PCM1) revealed no difference between wild-type and Cep135 KO cells (Figure 5, A and B), indicating that Cep135 deficiency did not have any detectable effect on centrosome composition or maturation. We also examined the centrosome ultrastructure in the absence of Cep135 by TEM. As shown in Figure 6A, Cep135-deficient cells, like wild-type controls, contained orthogonally arranged centrioles of nine triplet microtubules, surrounded by electron-dense PCM, with the procentrioles elongating orthogonally from the mother centriole. Inner centriole diameters were 183 ± 23 nm in single sections of wild-type cells and 173 ± 18 nm in Cep135-null cells. Longitudinal sections of wild-type centrioles showed the mean centriole length to be 370 ± 28 nm (N = 20), whereas the Cep135-null centrioles averaged 350 ± 30 nm (N = 29). Together these data showed no major effect of Cep135 deficiency on centriole size. Of interest, we observed an unusual, electron-dense structure in longitudinal sections of Cep135-knockout centrioles that was not present in sections of wild-type centrioles (Figure 6B). We detected this structure in 34 of 40 Cep135-deficient centrosomes analyzed but not in any of 47 wild-type centrosomes. This structure was observed roughly halfway along the length of the centriole (mean 0.42 ± 0.09 centriole length from the proximal end; N = 20). Because such structures were observed in 34 of 40 Cep135-deficient centrioles, they are unlikely to be restricted to mother or daughter centrioles. Observation of the structure in three independently generated Cep135-null cell lines confirms that it arose due to the loss of Cep135. Based on the known effects of Cep135 or Bld10 deficiency in other model systems (Matsuura et al., 2004; Hiraki et al., 2007; Kleylein-Sohn et al., 2007), our prediction had been that Cep135 loss would strongly impair centriole organization. The absence of such effect beyond the appearance of the electron-dense structure was surprising.
FIGURE 5:

Centrosome composition is normal in Cep135-deficient cells. (A) Immunofluorescence microscopy analysis of wild-type and Cep135 KO cells stained with the following antibodies to centrosomal or centrosome-associated proteins: γ-tubulin (red), centrin2, Cep76, Sas-6, PCM1, ninein, glutamylated tubulin, and Aurora A (green). Cells were then counterstained with DAPI (blue). Scale bar, 5 μm. (B) Immunoblot of Sas-6 levels in cells of the indicated genotype. α-Tubulin was used as a loading control, and cell extract from human HCT116 cells served as a positive control for the Sas-6 antibody.
FIGURE 6:
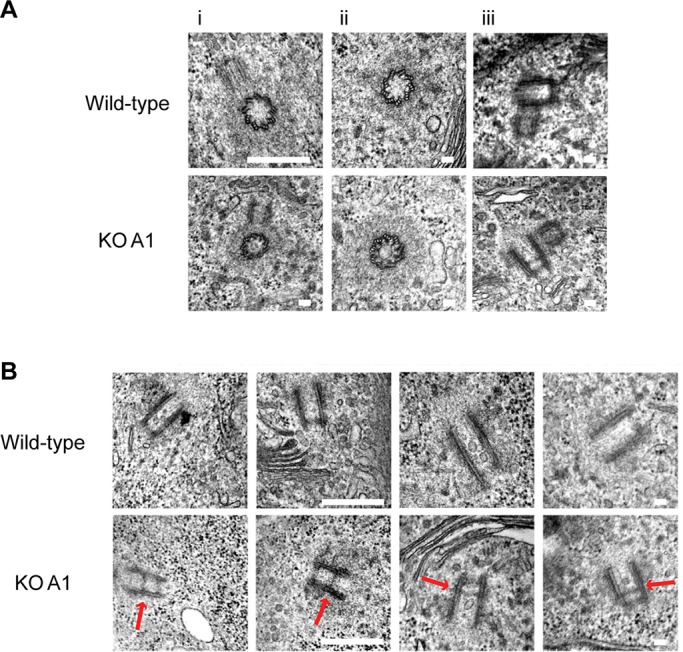
An atypical centriolar structure in longitudinal sections of Cep135-deficient centrioles (A) Transmission electron micrographs of wild-type and Cep135-deficient (KO A1) cells showing i) centriole duplication, ii) composition of nine triplet microtubules, iii) PCM integrity on centrosomes. (B) Transmission electron micrographs of wild-type and Cep135 KO centrioles. Note that the unusual electron-dense structure (indicated by the red arrow) is only observed in Cep135 KO cells. Short scale bar, 100 nm; long scale bar, 500 nm.
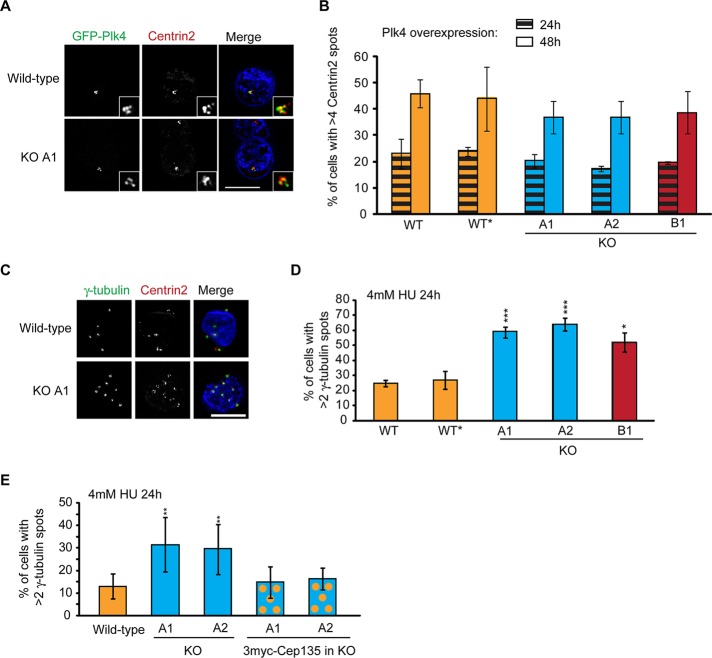
Cep135 is implicated in the Plk4-controlled cascade that regulates centriole duplication. Downstream of Plk4, Sas-6, CENPJ (CPAP), and CP110 are recruited to the nascent procentrioles at different stages during centriole formation (Leidel et al., 2005; Kleylein-Sohn et al., 2007; Peel et al., 2007). Centriole duplication induced by PLK4 overexpression was reduced in human U2OS cells after RNA interference (RNAi) depletion of Cep135 (Kleylein-Sohn et al., 2007). We therefore tested whether the ability of Plk4 to induce centriole formation is affected in the absence of Cep135 in DT40 cells. We observed supernumerary centriole formation in Cep135 KO cells upon overexpression of chicken Plk4 (Steere et al., 2011; Figure 7, A and B), suggesting that in DT40 cells Cep135 is probably dispensable for Plk4-induced centriole overduplication. Immunofluorescence microscopy analysis of the localization of Sas-6 and CP110 in Cep135-deficient cells showed that Cep135 deficiency had no detectable effect on the expression and localization of these proteins (Figure 5, A and B), further supporting the conclusion that Cep135 deficiency has no major effect on centriole assembly in DT40 cells.
FIGURE 7:
HU-induced centrosome amplification is increased in Cep135−/– cells. (A) Wild-type, wild-type*, and Cep135 KO A1, A2, and B1 cell lines were transiently transfected with GFP-Plk4 and fixed 24 and 48 h after treatment. Micrographs showing the expression and effect of GFP-Plk4 (green) transfection after 48 h. Cells were stained with an antibody against centrin2 (red) and counterstained with DAPI. Insets show supernumerary centrosomes. Wild-type*, parental DT40s for the B1 and B2 clones, included as an additional control. Scale bar, 5 μm. (B) Levels of centrosome amplification were quantified by immunofluorescence microscopy using an anti-centrin2 antibody in cells positive for GFP-Plk4. In wild-type, A1, and A2 cells, <9% of cells contained more than two centrosomes when not expressing GFP-Plk4. Data represent mean ± SD of three separate experiments where at least 200 cells were counted for each time point. (C) Micrograph showing HU-treated, wild-type and Cep135 KO A1 with multiple centrosomes. Cells were stained with antibodies against centrin2 (red) and γ-tubulin (green) and counterstained with DAPI. Scale bar, 5 μm. (D) Quantitation of HU-induced centrosome amplification using γ-tubulin as a marker. Data show mean ± SD of three separate experiments in which at least 200 cells were scored. *p ≤ 0.05, ***p ≤ 0.001 in comparison with wild type, as calculated by a two-tailed unpaired Student's t test. (E) Quantitative analysis of wild-type, Cep135 KO A1 and A2 DT40, and rescued Cep135 KO A1 and A2 cell lines (Cep135−/−::3myc-Cep135) 24 h after treatment with 4 mM HU. Cells with more than two γ-tubulin spots were counted. Data show mean ± SD of three separate experiments in which at least 200 cells were scored. **p ≤ 0.01 in comparison with wild type, as calculated by a two-tailed unpaired Student's t test.
Like many other cell types, DT40 cells undergo centrosome overduplication during prolonged cell cycle arrests that occur in response to genotoxic stresses, such as DNA double-strand break induction or replication fork stalling (Balczon et al., 1995; Dodson et al., 2004; Bourke et al., 2007). To test whether this response depends on Cep135, we examined the centrosome reduplication capability of Cep135-deficient cells after hydroxyurea (HU) treatment. As shown in Figure 7, C and D, HU treatment induced significantly higher levels of centrosome amplification in Cep135 KO cell lines than in wild-type populations. Of importance, transgenic expression of 3myc-Cep135 in the Cep135-deficient cells restored HU-induced centrosome amplification levels to wild-type levels, confirming that this overamplification effect was dependent on Cep135 (Figure 7E). Searching for a mechanism for how HU affects Cep135- deficient cells differently from wild-type cells, we tested whether Cdk2 level or Cdk2 phosphorylation at Thr-160 was changed in the absence of Cep135, as these are required for centrosome overduplication after HU treatment (Hinchcliffe et al., 1999; Matsumoto et al., 1999) or ionizing radiation (Bourke et al., 2010). Immunoblot analysis revealed, however, no effect of Cep135 deficiency on Cdk2 levels or Cdk2 phosphorylation at Thr-160 after HU treatment (data not shown). Taken together, these results suggest that Cep135 plays a role in limiting HU-induced centrosome reduplication but has no clear effect on Plk4-induced centrosome amplification in DT40 cells.
In summary, we generated multiple independently derived clones of DT40 cells in which the Cep135 locus was disrupted by gene targeting. Cep135-deficient cells proliferate normally and possess functional centrosomes that contain centrioles with normal ninefold symmetry. Because knockdown experiments indicate that Cep135 is required for normal centriole structures and Plk4-induced centriole overduplication in human cells (Kleylein-Sohn et al., 2007; Lin et al., 2013), we did not expect to find such apparently normal centrioles in Cep135-deficient cells. The Cep135 orthologue, Bld10, is needed for centriole formation in Chlamydomonas (Matsuura et al., 2004) and Paramecium (Jerka-Dziadosz et al., 2010), with the overexpression of mutant forms of the protein giving rise to centrioles that have lost their ninefold symmetry (Hiraki et al., 2007). Similarly, RNAi experiments in Drosophila described a moderate effect of Cep135 knockdown on centrosome duplication (Dobbelaere et al., 2008). Whereas Drosophila bld10-mutant spermatocytes displayed mild perturbations of centriole structure and aberrant central-pair microtubules in the axoneme, mutant somatic cells showed no impairment in centrosome duplication (Mottier-Pavie and Megraw, 2009; Carvalho-Santos et al., 2012; Roque et al., 2012). It is therefore possible that, whereas the role of Cep135 in stabilizing centriole structures is universal, rapidly cycling, transformed cells such as DT40s can assemble centrioles of normal appearance in the absence of Cep135. The small reduction in overall centriole numbers in Cep135-deficient cells indicates that in these cells, centriole biogenesis is mildly impaired.
Current understanding of cartwheel formation involves the association of Sas-6 homodimers through their N-terminal domains into a circular hublike structure from which the coiled-coil Sas-6 C-termini extend to form the spokes (Kitagawa et al., 2011; van Breugel et al., 2011). Localization of Cep135 to these spokes in a number of organisms suggests that it might play a role in the cartwheel, at least in Paramecium, Tetrahymena, and Chlamydomonas (Matsuura et al., 2004; Hiraki et al., 2007; Jerka-Dziadosz et al., 2010; Bayless et al., 2012). In Drosophila somatic cells, Bld10 localizes as a punctate area on both old and nascent centrioles, whereas in human cells Cep135 appears as a tight ring around the wall of the proximal end of the mother centrioles, with a much weaker signal overlapping with the cartwheel of the procentriole (Fu and Glover, 2012; Sonnen et al., 2012). Similarly, in DT40 cells, we observed a strong CEP135 localization on the proximal ends of mother centrioles throughout the cell cycle, with a prominent signal on procentrioles by early mitosis (Figure 2E).
Consistent with an evolutionarily conserved functional link between Sas-6 and Cep135, Drosophila bld10 mutants display unstable cartwheels and impaired connection between the cartwheel and centriolar microtubules (Roque et al., 2012). Cep135 seems to be required for the stabilization of Sas-6 rather than for the actual assembly of the cartwheel (Nakazawa et al., 2007; Jerka-Dziadosz et al., 2010), in keeping with the direct interaction between Cep135 and Sas-6 observed in experiments using immunoprecipitation and recombinant protein pull downs (Lin et al., 2013). In addition to cartwheel assembly, Drosophila Sas-6 also promotes centriole cohesion (Stevens et al., 2010). A role for Bld10/CEP135 in stabilizing basal body organization has been hypothesized from the analysis of ciliary function in Drosophila and Tetrahymena mutants of Bld10/CEP135 (Bayless et al., 2012; Carvalho-Santos et al., 2012). Loss of appropriate cartwheel-centriole attachment is therefore a possible explanation for both the atypical centriolar lumen structure that we see and the hyperamplification of centrioles in cells treated with HU. Indeed, the electron-dense nature and uniform appearance of this luminal structure could be consistent with a misplaced cartwheel. It is also feasible that the structure represents an ectopic cartwheel, which forms during or after centriole elongation. In mammalian cells, cartwheels are detectable in procentrioles but not in mature centrioles (Alvey, 1986; Strnad and Gönczy, 2008; Guichard et al., 2010). This is likely to hold true in chicken cells, since cartwheels are rarely seen in our electron micrographs. Intriguingly, we observed the lumen structure in 34 of 40 Cep135-deficient centrioles, implying that unlike a normal cartwheel, this structure remains visible in mature centrioles. Further experiments will be necessary to establish the identity and biological meaning of this structure.
We propose that Cep135 promotes the maintenance of the cartwheel and thus contributes to cohesion between parental and nascent centrioles. Progressive loss/displacement of cartwheel structures was observed in Drosophila bld10 mutants (Roque et al., 2012). Because DT40 cells exhibit a rapid 8-h cell cycle, premature loss of the cartwheel and centriole cohesion may have little effect on centriole formation or amplification during normal cell cycles. An unstable cartwheel structure could nonetheless lead to abortive procentriole assembly, consistent with the small increase in monopolar spindles and observed reduction in centriole numbers in these cells.
MATERIALS AND METHODS
Cell culture
Chicken DT40 cells were cultured in RPMI 1640 medium with 10% fetal calf serum (FCS; Lonza, Basel, Switzerland) and 1% chicken serum (Sigma-Aldrich, St. Louis, MO) as previously described (Takata et al., 1998). HCT116 cells were a gift from B. Vogelstein (Johns Hopkins University, Baltimore, MD) and were cultured in DMEM with 10% FCS. In centriole amplification experiments, 5 × 106 cells were resuspended in 100 μl of Kit T solution (Lonza) and mixed with 5 μg of endotoxin-free pEGFP-C1-Plk4 in 100 μl of the same solution (Steere et al., 2011). After nucleofection with program B-23 (Lonza), cells were resuspended in 10 ml of warmed medium. A similar approach was used for other transient transfections. For stable transfections, 10 × 106 cells were electroporated with 30 μg of linearized constructs in a Gene Pulser Apparatus (Bio-Rad, Hercules, CA) with a voltage of 550 V at capacitance of 25 μF and then selected using antibiotics as previously described (Takata et al., 1998). Irradiation of cells was performed using a 137Cs source (Mainance Engineering, Waterlooville, United Kingdom). HU was from Sigma-Aldrich, and a 1 M stock solution was made up in dimethyl sulfoxide (DMSO). Centrosome overduplication was induced by treating cells with 4 mM HU. Monastrol was provided by Tocris Bioscience (Ellisville, MO) and used in a final concentration of 125 μM for 16 h.
Cloning
Gene targeting constructs were generated by PCR on DT40 genomic DNA using the primers detailed in Table 1. Resistance cassettes were inserted into BamHI sites in the final construct. A TAP (Burckstummer et al., 2006) for insertion in-frame with Cep135 was a gift from K. J. Patel (Medical Research Council Laboratory of Molecular Biology, Cambridge, United Kingdom). For cloning of the 3myc-Cep135 construct, total RNA from DT40 cells was reverse transcribed using the SuperScript First-Strand synthesis kit (Invitrogen, Carlsbad, CA), and Cep135 was amplified using KOD Hot Start Polymerase (EMD, San Diego, CA). The Cep135 cDNA was subcloned into pGEM T-Easy, verified by sequencing, and subsequently cloned into pCMV-3TAG-2C (Stratagene, Santa Clara, CA). Similarly, chicken CP110 was cloned by RT-PCR from DT40 cells, sequenced, and then inserted into pCMV-3TAG (Agilent, Santa Clara, CA).
TABLE 1:
Primers used in constructing Cep135 targeting and expression constructs.
| Purpose | Primer sequence (5′–3′) |
|---|---|
| KO Cep135 construct A | |
| 5′ arm forward | GCGGCCGCTGTGGGTTGACCTGTCTCT |
| 5′ arm reverse | GGATCCTAAAATATCGTTTGTTACAGC |
| 3′ arm forward | GGATCCCTATACAGGATTTACTCTTG |
| 3′ arm reverse | GGTACCTTGTTTTGACTAATCAACAGC |
| KO Cep135 construct B | |
| 5′ arm forward | GAATTCTGCTACTATGCAACCAAATAACC |
| 5′ arm reverse | GGATCCTTATGATATCTCTAGCTTTTCTTGTAA |
| 3′ arm forward | GGATCCTAACTGCTGAACCAGCTTGAACAAGAA |
| 3′ arm reverse | GCGGCCGCTGCCAGTGAAGCATCTCCATCTTC |
| TAP Cep135 construct | |
| 5′ arm forward | aagcttGCTAACTGAAGTTGCTGTCT |
| 5′ arm reverse | gaattcCTCCTTGAAGCTCACGTTTC |
| 3′ arm forward | ggatccCCAGTACTTGAGCAATCAAA |
| 3′ arm reverse | actagtAGTTGATCGTATCCCTGACT |
| 3myc-Cep135 construct | |
| Chicken Cep135 forward | CCGCGGATGACGACAACAGCGGAGC |
| Chicken Cep135 reverse | GGATCCTTACTCCTTGAAGCTCACGTTT |
| KO Tsga10 construct | |
| 5′ arm forward | ATCGATTTGCAAGAGTGTGTGGAAGAA |
| 5′ arm reverse | GGATCCTTAGTTTGAATCCTGGGCTTTC |
| 3′ arm forward | GGATCCTGAACATCATCCCCTGAACG |
| 3′ arm reverse | ACTAGTTTGGTTCATCCACACTTCCA |
Diagnostic and quantitative PCR
Genomic DNA of clones was extracted using the Puregene Kit (Qiagen, Valencia, CA) and screened via PCR using the primers in Table 2. Primer locations on the Cep135 locus are diagrammed in Figures 1A and 2A and those for the TSGA10 locus in Supplemental Figure S1. For quantitative PCR (qPCR), total RNA from wild-type and Cep135 KO DT40 cells was isolated and reverse transcribed using the Superscript III First Strand Synthesis kit (Invitrogen). qPCR was performed using the Fast Sybr Green reaction mix (Applied Biosystems, Foster City, CA). Amplification and readout were carried out using the 7900HT Fast Real Time PCR System (Applied Biosystems) with the primers presented in Table 3. Data were analyzed applying the comparative CT method (Schmittgen and Livak, 2008).
TABLE 2:
Primers used in diagnostic PCR for gene targeting of Cep135.
| Primer | Primer sequence (5′–3′) |
|---|---|
| A | GACCCGTACATTGCAGACCT |
| B | ACAAGAAACTACCACCTAAAAAGCA |
| C | AGCCAAGCTGGGCGTAATCATTACGAGGAGCGCTTTTGTTTTGTATTG |
| D | GCATTCCAATGTTGCTGCTA |
| E | GAGGGGACAGCACTAGGACA |
| F for puromycin | GTCACCGAGCTGCAAGAACT |
| F for neomycin | CGTTGGCTACCCGTGATATT |
| F for blasticidin | CCCCCTGAACCTGAAACATA |
| G for puromycin cassettes | GTCACCGAGCTGCAAGAACT |
| G for neomycin cassettes | GACTTTCCTTTTCTGCCATCA |
| G for blasticidin cassettes | CCCCCTGAACCTGAAACATA |
| H | TTGTGATAATCGCCAAGGTG |
| I | CGGTGGTCTTCTCGTCCATA |
| J | TCTGAAAACACACTTGCTTCG |
| K | TCGAGATTTCACTCAGCTCAA |
| L | ACGATTCTGGCATCGAAGTT |
Letters indicate primers shown in Figures 1A and 2A and Supplemental Figure S1.
TABLE 3:
Primers for Cep135 qPCR.
| Primer | Primer sequence (5′–3′) |
|---|---|
| GAPDH forward | GCTCATCTGAAGGGTGGT |
| GAPDH reverse | CATAAGACCCTCCACAAT |
| Cep135 exon 4 forward | CCTCCATCAGCTGTTAGTGCT |
| Cep135 exon 4 reverse | GTCTGCAATGTACGGGTCCT |
| Cep135 exon 6 forward | AAGAAATAGAACGCCTCATGCT |
| Cep135 exon 6 reverse | CTCATTGCTTTTACTTCTTGATTCC |
| Cep135 exon 13 forward | CAGACTGAGAAGAGAAGCAAAACAT |
| Cep135 exon 13 reverse | CTGCCAGTGAAGCATCTCC |
Microtubule regrowth assay
For microtubule depolymerization, DT40 cultures were treated with 1 μg/ml nocodazole for 3 h. Cells were washed three times in ice-cold 1× phosphate-buffered saline (PBS; supplemented with 0.1% volume of DMSO) and adhered to poly-l-lysine slides for 10 min at 4°C on ice. Slides were submerged in growth media at 40°C for 1 min before methanol fixation and immunostaining.
Immunofluorescence microscopy
Cells were washed in PBS and adhered to poly-l-lysine cover slips. Fixation was performed in ice-cold methanol for 5 min at −20°C, followed by a brief permeabilization using 0.1% Tween 20 in PBS. Visualization of ninein, Sas-6, and Aurora A required an additional permeabilization step using 1% Triton X-100/0.5% NP-40 in PBS for 5 min. Cells were then blocked in 5% bovine serum albumin in PBS for 5 min and incubated with primary antibodies for 1h at 37°C. The primary antibodies used in this study and their dilutions were as follows: mouse polyclonal against Cep135 (Ohta et al., 2002) at 1:2000; mouse monoclonals against Sas-6 (BO1P; Abnova) at 1:50, α-tubulin (B512; Sigma-Aldrich) at 1:2000, γ-tubulin (GTU88; Sigma-Aldrich) at 1:150, γ-tubulin (C-20; Santa Cruz Biotechnology, Santa Cruz, CA) at 1:250, Aurora A (35C1; Abcam, Cambridge, MA) at 1:500, and glutamylated tubulin (GT335; a gift from C. Janke, Institut Curie, Paris, France; Wolff et al., 1992) at 1:300; and rabbit polyclonals against centrin2 (poly6288; BioLegend, San Diego, CA) at 1:250, Cep76 (a gift from W. Tsang and B. Dynlacht, New York University School of Medicine, New York, NY; Tsang et al., 2009) at 1:200, PCM1 (817; a gift from A. Merdes, University of Toulouse, Toulouse, France) at 1:5000, ninein (ab4447; Abcam) at 1:100, protein G (Abcam) at 1:1000, centrin3 (Abnova, San Jose, CA) at 1:1000, and TACC3 and CDK5RAP2 (Barr et al., 2010) at 1:1000.
For Cep135−/− cell lines A1 and A2, images were acquired using a BX51 microscope (Olympus, Tokyo, Japan) with 60× (numerical aperture [NA] 1.4) and 100× (NA 1.35) objectives and Openlab software (Improvision/PerkinElmer, Waltham, MA). Deconvolved (nearest-neighbor differential interference contrast) images were saved as Photoshop CS4 files (Adobe, San Jose, CA). For Cep135−/− cell lines B1 and B2, cells were analyzed with a confocal Nikon eclipse 90i microscope equipped with a 100× oil (NA 1.40) objective (Nikon, Melville, NY). We acquired 0.5-μm-step Z-stacks using an Eclipse C1Si camera driven by EZ-C1 software (Nikon). Z-stacks were projected in Volocity, version 5.0 (PerkinElmer), and exported as two-dimensional volume-rendered images into Photoshop CS4. Superresolution microscopy was performed using a three-dimensional structured illumination microscope by OMX DeltaVision (Applied Precision, Issaquah, WA). Cells were imaged with a 100× (NA 1.40) Olympus (Tokyo, Japan) objective. Data were reconstructed using SoftWorx software (Applied Precision).
Live cell imaging
For live cell imaging, cells were allowed to attach to poly-d-lysine–coated dishes (MatTek, Ashland, MA) for 2–3 h in medium supplemented with 12.5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5. Timing and light exposure were kept minimal by using reduced excitation light. Images were taken every 3 min for 3 h on an integrated microscope system (DeltaVision) using a PlanApo N 60× oil objective (NA 1.42) and a 39.5°C environmental chamber (WeatherStation; Precision Control, Sammamish, WA). Quick projections were made with SoftWorx software.
Transmission electron microscopy
DT40 cells were prepared for TEM as previously described (Dantas et al., 2011). Sections were cut on a microtome (Reichert-Jung Ultracut E; Leica, Wetzlar, Germany), stained with uranyl acetate and lead citrate, and then viewed on an electron microscope (H-7000; Hitachi, Tokyo, Japan). Images were taken with an ORCA-HRL camera (Hamamatsu Photonics, Hamamatsu, Japan) and processed using AMT, version 6 (Advanced Microscopy Techniques, Woburn, MA).
Flow cytometry
Cell cycle analysis by flow cytometry involved the treatment of cells with 5-bromo-2′-deoxyuridine (BrdU; Sigma-Aldrich) and visualization with antibodies to BrdU or phospho-histone H3 as previously described (Wang et al., 2013).
Western blot
To obtain whole-cell extracts for Western blotting, cells were washed in PBS and lysed in 250 mM Tris, pH 7.6, 0.1% Triton X-100, 2.5 mM NaF, 1 mM Na3VO4, 2.5 mM ethylene glycol tetraacetic acid, 12.5 mM sodium pyrophosphate, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM aprotinin, and 1 mM pepstatin. The primary antibodies used in this study were polyclonal rabbit anti-Cep135 (Ohta et al., 2002), anti-Scc1 (Stephan et al., 2011), anti–protein G (ab 7250; Abcam), Sas-6, anti–α-tubulin, and anti–β-actin (ab 8227; Abcam).
Supplementary Material
Acknowledgments
We are indebted to a PRTLI4 grant to fund the National Biophotonics and Imaging Platform Ireland (www.nbipireland.ie), which supported the TEM. Research at F.G.’s laboratory is funded by Cancer Research UK and a Royal Society University Research Fellowship. Work in C.M.’s lab was supported by Science Foundation Ireland Principal Investigator Awards 08/IN.1/B1029 and 10/IN.1/B2972.
Abbreviations used:
- BrdU
5-bromo-2’-deoxyuridine
- DMSO
dimethyl sulfoxide
- FCS
fetal calf serum
- HU
hydroxyurea
- KO
knockout
- MT
microtubule
- NA
numerical aperture
- PCM
pericentriolar material
- RFP
red fluorescent protein
- RNAi
RNA interference
- TAP
tandem affinity purification
- TEM
transmission electron microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-03-0149) on July 17, 2013.
*These authors contributed equally to this article and are listed alphabetically.
REFERENCES
- Alvey PL. Do adult centrioles contain cartwheels and lie at right angles to each other. Cell Biol Int Rep. 1986;10:589–98. doi: 10.1016/0309-1651(86)90136-0. [DOI] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer WE, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr AR, Kilmartin JV, Gergely F. CDK5RAP2 functions in centrosome to spindle pole attachment and DNA damage response. J Cell Biol. 2010;189:23–39. doi: 10.1083/jcb.200912163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayless BA, Giddings TH, Jr, Winey M, Pearson CG. Bld10/Cep135 stabilizes basal bodies to resist cilia-generated forces. Mol Biol Cell. 2012;23:4820–4832. doi: 10.1091/mbc.E12-08-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- Bourke E, Brown JA, Takeda S, Hochegger H, Morrison CG. DNA damage induces Chk1-dependent threonine-160 phosphorylation and activation of Cdk2. Oncogene. 2010;29:616–624. doi: 10.1038/onc.2009.340. [DOI] [PubMed] [Google Scholar]
- Bourke E, Dodson H, Merdes A, Cuffe L, Zachos G, Walker M, Gillespie D, Morrison CG. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 2007;8:603–609. doi: 10.1038/sj.embor.7400962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Bennett KL, Preradovic A, Schutze G, Hantschel O, Superti-Furga G, Bauch A. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods. 2006;3:1013–1019. doi: 10.1038/nmeth968. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, et al. BLD10/CEP135 is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev Cell. 2012;23:412–424. doi: 10.1016/j.devcel.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. Stepwise evolution of the centriole-assembly pathway. J Cell Sci. 2010;123:1414–1426. doi: 10.1242/jcs.064931. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardtii. J Cell Sci. 1974;16:529–556. doi: 10.1242/jcs.16.3.529. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Dantas TJ, Wang Y, Lalor P, Dockery P, Morrison CG. Defective nucleotide excision repair with normal centrosome structures and functions in the absence of all vertebrate centrins. J Cell Biol. 2011;193:307–318. doi: 10.1083/jcb.201012093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M, Gonczy P. The arithmetic of centrosome biogenesis. J Cell Sci. 2004;117:1619–1630. doi: 10.1242/jcs.01128. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Josué F, Suijkerbuijk S, Baum B, Tapon N, Raff J. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J. 2004;23:3864–3873. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson H, Wheatley SP, Morrison CG. Involvement of centrosome amplification in radiation-induced mitotic catastrophe. Cell Cycle. 2007;6:364–370. doi: 10.4161/cc.6.3.3834. [DOI] [PubMed] [Google Scholar]
- Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2:120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Chrétien D, Marco S, Tassin AM. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 2010;29:1565–1572. doi: 10.1038/emboj.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:851–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001;15:1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Hussain MS, et al. A truncating mutation of CEP135 causes primary microcephaly and disturbed centrosomal function. Am J Hum Genet. 2012;90:871–878. doi: 10.1016/j.ajhg.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerka-Dziadosz M, Gogendeau D, Klotz C, Cohen J, Beisson J, Koll F. Basal body duplication in Paramecium: the key role of Bld10 in assembly and stability of the cartwheel. Cytoskeleton (Hoboken) 2010;67:161–171. doi: 10.1002/cm.20433. [DOI] [PubMed] [Google Scholar]
- Kim K, Lee S, Chang J, Rhee K. A novel function of CEP135 as a platform protein of C-NAP1 for its centriolar localization. Exp Cell Res. 2008;314:3692–3700. doi: 10.1016/j.yexcr.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Lin YC, Chang CW, Hsu WB, Tang CJ, Lin YN, Chou EJ, Wu CT, Tang TK. Human microcephaly protein CEP135 binds to hSAS-6 and CPAP, and is required for centriole assembly. EMBO J. 2013;32:1141–1154. doi: 10.1038/emboj.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- Matsuura K, Lefebvre PA, Kamiya R, Hirono M. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J Cell Biol. 2004;165:663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottier-Pavie V, Megraw TL. Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol Biol Cell. 2009;20:2605–2614. doi: 10.1091/mbc.E08-11-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Ohta T, Essner R, Ryu JH, Palazzo RE, Uetake Y, Kuriyama R. Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J Cell Biol. 2002;156:87–99. doi: 10.1083/jcb.200108088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque H, Wainman A, Richens J, Kozyrska K, Franz A, Raff JW. Drosophila Cep135/Bld10 maintains proper centriole structure but is dispensable for cartwheel formation. J Cell Sci. 2012;125:5881–5886. doi: 10.1242/jcs.113506. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sir JH, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, Reichelt S, D'Santos C, Woods CG, Gergely F. A primary microcephaly protein complex forms a ring around parental centrioles. Nat Genet. 2011;43:1147–1153. doi: 10.1038/ng.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steere N, Wagner M, Beishir S, Smith E, Breslin L, Morrison CG, Hochegger H, Kuriyama R. Centrosome amplification in CHO and DT40 cells by inactivation of cyclin-dependent kinases. Cytoskeleton (Hoboken) 2011;68:446–458. doi: 10.1002/cm.20523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan AK, Kliszczak M, Dodson H, Cooley C, Morrison CG. Roles of vertebrate smc5 in sister chromatid cohesion and homologous recombinational repair. Mol Cell Biol. 2011;31:1369–1381. doi: 10.1128/MCB.00786-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Roque H, Raff JW. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev Cell. 2010;19:913–919. doi: 10.1016/j.devcel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Gönczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–96. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Tsang WY, Spektor A, Vijayakumar S, Bista BR, Li J, Sanchez I, Duensing S, Dynlacht BD. Cep76, a centrosomal protein that specifically restrains centriole reduplication. Dev Cell. 2009;16:649–660. doi: 10.1016/j.devcel.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A, Takeda S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Terada Y, Matuliene J, Kuriyama R. Interaction of Cep135 with a p50 dynactin subunit in mammalian centrosomes. Cell Motil Cytoskeleton. 2004;58:53–66. doi: 10.1002/cm.10175. [DOI] [PubMed] [Google Scholar]
- van Breugel M, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- Wang Y, Dantas TJ, Lalor P, Dockery P, Morrison CG. Promoter hijack reveals pericentrin functions in mitosis and the DNA damage response. Cell Cycle. 2013;12:635–646. doi: 10.4161/cc.23516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff A, de Nechaud B, Chillet D, Mazarguil H, Desbruyeres E, Audebert S, Edde B, Gros F, Denoulet P. Distribution of glutamylated alpha and beta-tubulin in mouse tissues using a specific monoclonal antibody, GT335. Eur J Cell Biol. 1992;59:425–432. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.