Wiskott-Aldrich syndrome protein and SCAR homologue (WASH) is an important regulator of vesicle trafficking. The authors show that, in Dictyostelium, WASH is required for lysosomal hydrolase recycling and function. WASH is therefore crucial for both autophagic and phagocytic degradation, and cells lacking WASH are unable to either survive starvation or grow on certain bacteria.
Abstract
Wiskott-Aldrich syndrome protein and SCAR homologue (WASH) is an important regulator of vesicle trafficking. By generating actin on the surface of intracellular vesicles, WASH is able to directly regulate endosomal sorting and maturation. We report that, in Dictyostelium, WASH is also required for the lysosomal digestion of both phagocytic and autophagic cargo. Consequently, Dictyostelium cells lacking WASH are unable to grow on many bacteria or to digest their own cytoplasm to survive starvation. WASH is required for efficient phagosomal proteolysis, and proteomic analysis demonstrates that this is due to reduced delivery of lysosomal hydrolases. Both protease and lipase delivery are disrupted, and lipid catabolism is also perturbed. Starvation-induced autophagy therefore leads to phospholipid accumulation within WASH-null lysosomes. This causes the formation of multilamellar bodies typical of many lysosomal storage diseases. Mechanistically, we show that, in cells lacking WASH, cathepsin D becomes trapped in a late endosomal compartment, unable to be recycled to nascent phagosomes and autophagosomes. WASH is therefore required for the maturation of lysosomes to a stage at which hydrolases can be retrieved and reused.
INTRODUCTION
The Wiskott-Aldrich syndrome protein and SCAR homologue (WASH) is an evolutionarily conserved regulator of the Arp2/3 complex. Like other members of the Wiskott-Aldrich syndrome protein (WASP) family, the primary role of WASH is to activate Arp2/3 and regulate the spatial and temporal formation of actin networks (Linardopoulou et al., 2007). Specifically, WASH generates actin subdomains on endocytic vesicles and is important for several sorting and maturation steps. These include retrograde transport from endosomes to the trans-Golgi via direct regulation of the retromer (Derivery et al., 2009; Gomez and Billadeau, 2009; Duleh and Welch, 2010; Harbour et al., 2010) and the sorting and trafficking of other signaling molecules, such as integrins, the epidermal growth factor (EGF), and transferrin receptors (Derivery et al., 2009, Zech et al., 2011).
A comparable role has also been described in Dictyostelium, whereby WASH-generated actin sequesters the vacuolar (v)-ATPase to late endosomal subdomains, leading to v-ATPase removal via recycling vesicles (Carnell et al., 2011). The v-ATPase is the proton pump responsible for establishing and maintaining an acidic pH in the lumen of lysosomes. Therefore its WASH-mediated removal leads to the neutralization and maturation of lysosomes into postlysosomes prior to exocytosis (Neuhaus et al., 2002). In Dictyostelium, the loss of WASH therefore arrests lysosome maturation before v-ATPase removal, blocking both neutralization and exocytosis (Carnell et al., 2011).
Like other WASP-family proteins, WASH acts as part of a complex (Jia et al., 2010; Veltman and Insall, 2010). Our recent work showed that, while disruption of the Strumpellin, ccdc53, or SWIP subunits leads to total loss of WASH activity, the FAM21 subunit has a unique role, driving the recycling of WASH (Park et al., 2013). Therefore loss of FAM21 gives hyperactive WASH, and, although cells retain the ability to recycle v-ATPase, endocytic maturation is blocked at a later stage, at which WASH is itself recycled. Importantly, these contrasting phenotypes of WASH and FAM21 mutants now allow us to dissect the pathway in more detail and discriminate direct and indirect effectors of WASH.
In addition to its specific trafficking roles, WASH appears to be more generally important in maintaining endosomal and lysosomal integrity. In mammalian cells, small interfering RNA (siRNA) leads to altered endosome morphology (Derivery et al., 2009; Duleh and Welch, 2010), while full deletion causes lysosomal collapse (Gomez et al., 2012; Piotrowski et al., 2012). We therefore hypothesized that WASH may also be important for normal lysosomal function.
Lysosomes are responsible for the degradation of endosomal contents. They are therefore essential for breaking down endocytic cargo, such as membrane receptors, as well as the contents of autophagosomes and bacteria captured within phagosomes. Autophagy is the name given to a family of pathways leading to the capture of cytoplasmic constituents and their delivery to lysosomes. This includes both microautophagy, wherein selected proteins are directly transported across the lysosomal membrane, and macroautophagy, wherein double-membrane vesicles are formed de novo, capturing parts of the cytoplasm and fusing with lysosomes (Xie and Klionsky, 2007). In this study, we have specifically studied macroautophagy, which we will simply refer to as autophagy for clarity.
Cells use autophagy for a number of purposes, and defective autophagy is associated with a number of medical conditions, including aging, cancer, infection, and neurodegeneration (Mizushima et al., 2008). The best-understood function of autophagy is during starvation, when degradation of the cytosol provides nutrients and energy to the cell, enabling survival. In professional phagocytes such as Dictyostelium, this is similar to the phagocytic pathway, in which the cell uses the same digestive pathway to liberate nutrients from captured bacteria (Deretic, 2008). Autophagy and phagocytosis therefore both require lysosomal delivery and digestion in order to feed the cell.
The WASH complex has been assigned a number of roles in vesicular trafficking. In this study, we assess the importance of WASH in lysosomal function within the context of both the autophagic and phagocytic pathways. We show that WASH is required for both processes in Dictyostelium and describe a new role for WASH in maintaining lysosomal flux.
RESULTS
WASH is not required for autophagosome formation
Previous work has shown that the WASH complex is involved in multiple stages of the endocytic pathway. Autophagy intersects with the endocytic pathway and relies on many of the same processes, such as v-ATPase and lysosomal hydrolase trafficking. We therefore directly assessed the role of WASH in autophagy, using the model system Dictyostelium discoideum, for which there are well-defined assays for autophagosome induction and formation (Calvo-Garrido et al., 2010, King et al., 2011).
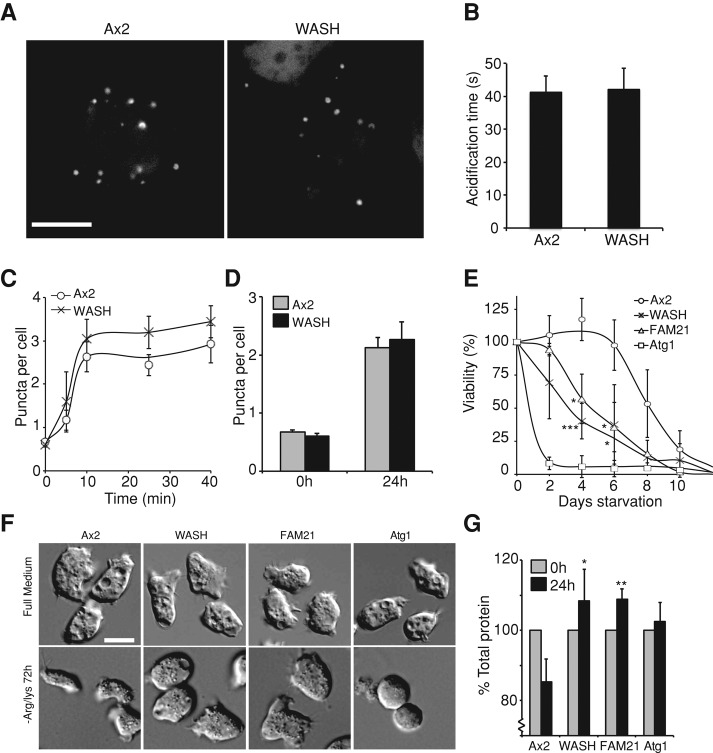
To directly observe autophagosome formation, we used a green fluorescent protein (GFP)-Atg8 fusion probe. During autophagosome formation, GFP-Atg8 is lipidated and incorporated into the expanding phagophore membrane (Ichimura et al., 2000) and can therefore be used to label autophagosomes until they are fully formed. When the autophagosomes are completed, the cytosolically exposed GFP-atg8 is cleaved by Atg4, the pH-sensitive GFP fluorescence on the interior is quenched by acidification, and the GFP signal is lost (Kirisako et al., 2000). Using cellular compression to both induce autophagy and improve imaging (King et al., 2011), we observed no gross defects in autophagosome formation or morphology in WASH-null cells compared with wild-type (Ax2) controls (Figure 1A and Supplemental Movies S1 and S2).
FIGURE 1:
Autophagy in WASH mutants. (A) High-resolution imaging of autophagosome formation in GFP-atg8–expressing cells compressed under agar. Stills are taken from Movies S1 and S2. (B) Quantification of the time taken for the GFP signal of individual autophagosomes in cells treated as in (A) to fade after autophagosome formation was complete. (n > 50 autophagosomes in total for each strain). (C) Autophagy was induced by placing cells expressing GFP-atg8 in SIH-Arg/Lys. The number of autophagosomes was counted at each time point. (D) The steady-state levels of autophagosomes after 24 h of starvation. (E) Survival of mutants under Arg/Lys starvation. At each time point, viability was measured by the ability to reform colonies on bacterial plates. (F) Morphology of cells under starvation. DIC images of cells after 3 d in full medium or SIH-Arg/Lys. (G) Total protein of cells after 24 h of Arg/Lys starvation. All values plotted are the means ± SD of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.005 (Student's t-test). Scale bars: 5 μm.
Previous work in Dictyostelium has demonstrated an important role for the WASH complex in v-ATPase trafficking and, consequently, the regulation of vesicular pH (Carnell et al., 2011). During maturation, autophagosomes are also acidified, due to delivery of the v-ATPase from the lysosomal compartment. The rate of acidification and maturation is represented by the loss of GFP-Atg8 fluorescence after phagophore completion, allowing quantification (see Movies S1 and S2). When we compared the time taken for the fluorescent signal to be removed in Ax2 and WASH mutants (n > 50, across three independent experiments), no difference was observed, which indicated there were no defects in acidification (Figure 1B).
In the above assays, autophagy was induced by mechanical compression, via pathways that are poorly understood (King, 2012). We therefore also tested the role of WASH under amino acid starvation. When WASH-null cells were placed in the defined SIH medium lacking arginine and lysine (SIH-Arg/Lys, which induces autophagy, but not the development of Dictyostelium; King et al., 2011) we found no differences in either the rate of induction or steady-state levels of autophagosomes after 24 h (Figure 1, C and D). We therefore conclude that WASH is not required for the induction, formation, or maturation of autophagosomes.
WASH is required for cytoplasmic degradation and surviving starvation
Although not required for autophagosome formation, we hypothesized that WASH may be necessary for subsequent processing of autophagosomes. The canonical role for autophagy is during starvation, when the degradation and recycling of cytoplasmic components is essential for cells to maintain nutrient levels and survive. To test whether the autophagic pathway was fully functional in WASH-null cells, we determined their ability to survive Arg/Lys starvation. While Ax2 cells were able to survive for almost 7 d before their viability decreased, WASH mutants died significantly faster, losing almost 50% viability after only 4 d (Figure 1E). Their survival was, however, better than that of Atg1 mutants, which are completely deficient in autophagy and started dying after just 24 h. WASH is therefore required for effective survival of Arg/Lys starvation.
WASH exists in a complex with four other proteins. We recently determined that one of the complex subunits, FAM21, is required to recycle WASH from endosomes. Loss of FAM21 therefore leads to persistent WASH activity, blocking the endocytic pathway at a later stage, after v-ATPase removal (Park et al., 2013). FAM21 mutants were also unable to survive starvation, to the same degree as WASH mutants (Figure 1E), indicating that the defect in survival is due to a trafficking event downstream of WASH and v-ATPase recycling.
The function of autophagy under starvation is to degrade the cytosol, which results in reduction of both cell size and mass (Otto et al., 2003). Total protein levels were decreased by 15% in Ax2 cells upon 24 h Arg/Lys starvation, but remained constant in Atg1-null cells (Figure 1G). Consistent with their inability to survive starvation, both WASH and FAM21 mutants also had no reduction in total cellular protein and retained their size over several days (Figure 1, F and G). Therefore, while autophagosomes form and acidify normally in WASH mutants, total protein is not reduced, and the mutant cells are unable to survive starvation. This indicates a defect in autophagic degradation and nutrient recycling.
Phagocytic proteolysis is reduced in WASH mutants
Autophagy is not the only pathway that breaks down vesicular cargo. In soil, Dictyostelium is a professional phagocyte, relying on the capture and digestion of bacteria within phagosomes to provide nutrients. After formation, phagosomes and autophagosomes are processed in a similar manner, with the same requirement for acidification and hydrolytic enzymes in order to release nutrients (Deretic, 2008). We therefore tested whether WASH is also required for phagocytic digestion.
Like autophagosome formation, loss of either WASH or FAM21 did not affect the rate of phagocytosis (Supplemental Figure S1A). We were therefore able to directly measure phagocytic proteolysis using beads coated with self-quenching fluorescently labeled DQ-BSA (bovine serum albumin), which becomes dequenched upon proteolysis (Gopaldass et al., 2012). With this assay, the rate of proteolysis was reduced by ∼50% in both WASH and FAM21 mutants compared with Ax2, and rescued by reexpression of GFP-WASH in WASH-null cells (Figure 2A). WASH is therefore required for both phagocytic and autophagic digestion, indicating a general role for WASH in vesicular degradation. Importantly, this cannot be due to a defect in acidification, as this is unaffected in both WASH and FAM21 mutants (Figure 4E; Carnell et al., 2011; Park et al., 2013).
FIGURE 2:
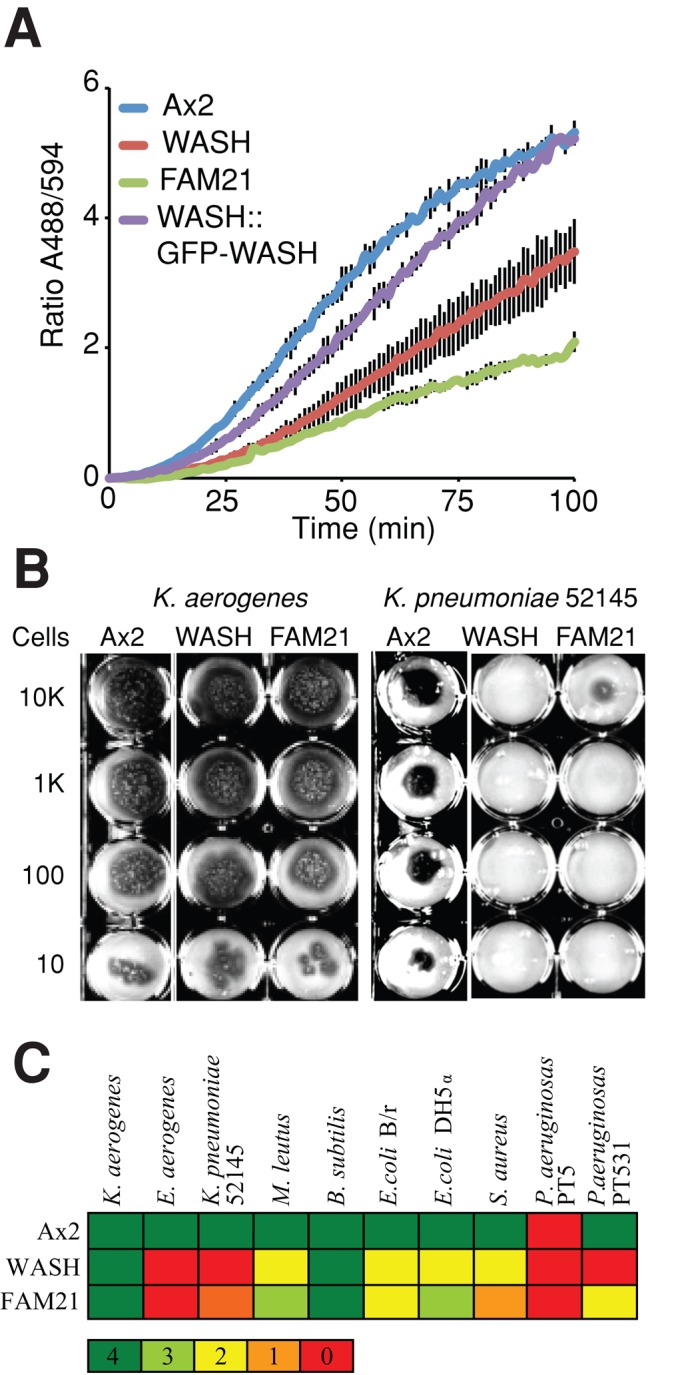
Phagocytic defects in WASH mutant cells (A) Proteolytic activity is decreased in WASH mutants, as measured by feeding cells DQ-BSA–labeled beads and measuring unquenching of the fluorophore over time. (B) WASH- and FAM21-null cells are unable to grow on certain bacteria. The indicated number of cells were seeded on the bacterial lawn, and Dictyostelium growth is indicated by the formation of dark plaques where the bacteria have been consumed. (C) Summary of mutant growth on various bacteria. The color code refers to the number of wells in (B) in which Dictyostelium were able to grow, indicating the severity of the growth defect.
FIGURE 4:
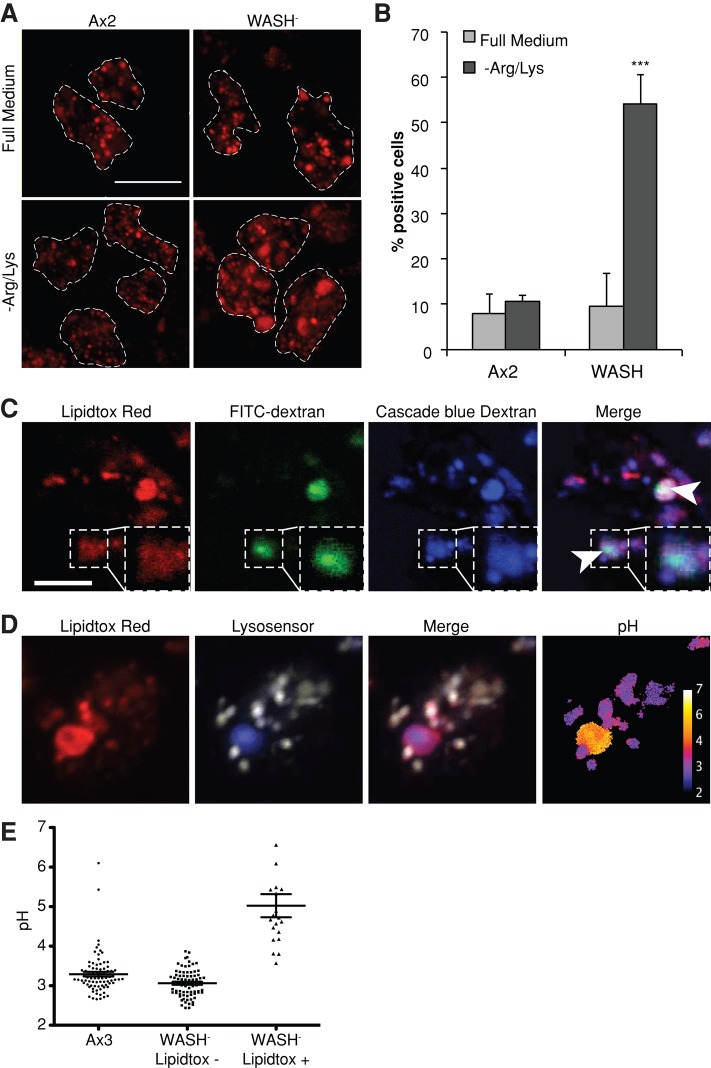
Phospholipid accumulation in WASH-null cells. (A) Cells grown for 24 h in either full SIH or SIH-Arg/Lys in the presence of LipidTOX Red. Arrows indicate large phospholipid accumulations. Images are confocal maximum intensity projections. Scale bar: 10 μm. The proportion of cells containing these large accumulations (>1.5-μm diameter) is quantified in (B). More than 50 cells were scored for each sample, and the values plotted are the mean ± SD of three independent experiments. ***, p < 0.005 (Student's t-test). (C) Colocalization of LipidTOX Red accumulations with both FITC (pH-sensitive) and Cascade Blue (pH-insensitive) dextran. WASH-null cells were grown overnight in SIH-Arg/Lys supplemented with LipidTOX Red and both dextrans. Arrows indicate subdomains of brighter FITC-dextran fluorescence. Image shown is a single confocal plane. Scale bar: 5 μm. (D) Endocytic pH of cells starved overnight in medium containing LysoSensor yellow/blue dextran and LipidTOX Red. The LysoSensor emission at 450 nm and 510 nm is colored blue and yellow, respectively, in the LysoSensor panel and was used to calculate the pH in the false-colored final image. The distribution of pH in individual vesicles is shown in (E). All the LipidTOX Red–negative vesicles in 10 cells of each strain were measured and are compared with the pH within the LipidTOX Red accumulations in >20 WASH-null cells.
Efficient phagocytic digestion is essential for Dictyostelium to survive on bacteria as a food source. We therefore tested the ability of WASH-null Dictyostelium to grow on several bacterial strains. While WASH- and FAM21-null cells grew normally on our laboratory strains of Klebsiella aerogenes and Bacillus subtilis, both mutants had dramatically reduced growth on the majority of the other strains tested (Figure 2, B and C), demonstrating a physiological requirement for WASH in growth on bacteria.
Lysosomal components are reduced in the phagocytic compartment
Phagosome maturation is a highly regulated and organized process during which lysosomal components are sequentially delivered and retrieved (Clarke et al., 2002; Gotthardt et al., 2002, 2006a). To establish the mechanism underlying the defects in degradation, we used quantitative comparative proteomics to identify differentially represented proteins in the isolated phagosomes of Ax2, WASH, and FAM21 mutants (Dieckmann et al., 2012; Gotthardt et al., 2006a). To confirm this approach, we first looked at the levels of v-ATPase. In Dictyostelium, WASH is required for v-ATPase recycling, and WASH mutants retain v-ATPase on their endosomes (Carnell et al., 2011). Consistent with this, all nine subunits of the v-ATPase complex were elevated in the WASH-null phagocytic compartment compared with wild-type (Figure 3B). In contrast, FAM21 mutants retain WASH activity (Park et al., 2013), and several of the v-ATPase subunits were identified as decreased in FAM21-null phagosomes relative to Ax2. This validates the data set and confirms that we are able to identify differentially trafficked proteins with this method.
FIGURE 3:
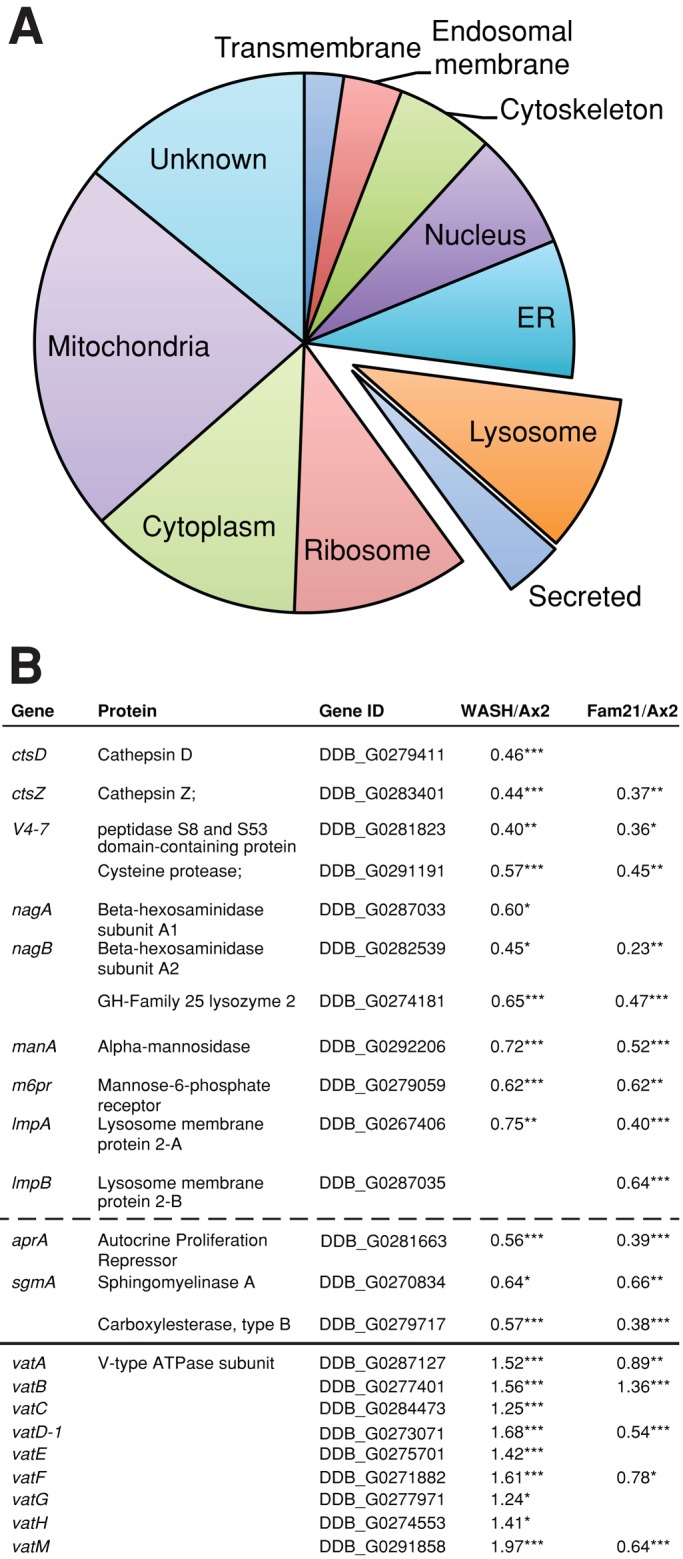
Proteomic analysis of mutant phagosomes. (A) The relative proportion of proteins from different compartments is reduced by at least 30% in both WASH and FAM21 mutant phagosomes. Eighty-five proteins in all were identified by this criteria. (B) Identity and relative abundance of lysosomal and secreted (below dashed line) proteins identified in this screen. Lysosomal proteins only identified in one mutant are also listed. The relative abundance of v-ATPase subunits is included in the lower section. *, p < 0.05; **, p < 0.005; ***, p < 0.001 (Student's t-test).
Although the loss of WASH or FAM21 blocks the endocytic pathway at different points, both mutants have comparable defects in phagocytosis and autophagy. We therefore looked for differences in the phagocytic pathway common to both mutants. Of 85 proteins that were decreased by at least 30% in both mutants, 11 (13%) were lysosomal proteins (Figure 3A and Supplemental Table S1). Several additional lysosomal proteins were also decreased in either WASH or FAM21 mutants, indicating a general loss of lysosomal enzymes from the phagocytic compartment (Figure 3B). These include several proteases, such as cathepsins D and Z, as well as a cysteine protease, explaining the measured reduction in proteolytic activity. A form of lysozyme (DDB_G0274181) was also reduced in both mutant phagosomes. As lysozymes are necessary to break down bacterial cell walls, this reduction would also contribute to the defective growth of the mutants on bacteria.
In addition to known endo/lysosomal proteins, our proteomic analysis also identified differences in a number of mitochondrial and ribosomal components that would not normally be expected to be present in phagosomes. As the data show relative changes, these components are unlikely to be contaminants, and their absolute abundance is not known. Instead, we speculate that these proteins indicate the convergence of the autophagic and phagocytic pathways and represent either changes in autophagosome trafficking or global transcriptional changes.
Interestingly, the reduction in hydrolytic enzymes was accompanied by a decreased mannose-6-phosphate receptor (M6PR) in both mutants. It should be noted, however, that this M6PR is most similar to the cation-dependent (CD)-M6PR family. Unlike the cation-independent (CI)-M6PRs, CD-M6PRs do not interact with the retromer (Arighi et al., 2004; Seaman, 2004) and are therefore unlikely to be directly mediated by WASH, as recently described (Gomez and Billadeau, 2009).
WASH is also required for autophagic lipid catabolism
In addition to proteolytic enzymes, several enzymes involved in lipid catabolism were also reduced in WASH mutant phagosomes. These include both subunits of β-hexosaminidase, as well as sphingomyelinase A. In humans, loss of these enzymes causes the accumulation of large quantities of lipid within lysosomes, leading to Tay-Sachs and Sandhoff syndromes (β-hexoaminidase deficiency) and Niemann-Pick disease types A and B (sphingomyelinase A deficiency). Inhibition of sphingomyelinase has also been proposed to be a major cause of drug-induced phospholipidosis (Yoshida et al., 1985). Therefore, to test whether lipid catabolism was disrupted in WASH-null cells, we used a fluorescently labeled phospholipid (LipidTOX Red) to label phospholipid accumulations in live cells.
In full medium, both Ax2 and WASH mutant cells were indistinguishable (Figure 4A). However, after 16 h of Arg/Lys starvation, WASH-null cells contained large accumulations of phospholipid, with 53% of cells containing a structure >1.5 μm in diameter (Figure 4, A and B). To determine whether these structures were endocytic in origin, we also incubated the cells with fluorescently labeled dextrans. These colocalized with the large phospholipid structures, indicating the lipid was accumulating within the endocytic system (Figure 4C).
In WASH-null cells, the largest phospholipid-rich structures colocalized with pH-sensitive fluorescein isothiocyanate (FITC)-dextran, indicating they were neutralized. To confirm this, we directly measured the pH of these vesicles using LysoSensor yellow/blue dextran, which has both pH-sensitive and pH-insensitive emission peaks (Diwu et al., 1999). This again accumulated in the phospholipid-rich structures and indicated they were pH 5, in contrast to pH 3.1 found in normal endocytic vesicles (Figure 4, D and E). This was surprising, because WASH is essential for v-ATPase recycling in Dictyostelium, and neutral compartments are normally completely absent in WASH-null cells (Carnell et al., 2011). Elevated lysosomal pH has, however, been reported in macrophages forced to accumulate cholesterol and in several lipid storage disorders (Bach et al., 1999; Holopainen et al., 2001; Cox et al., 2007). It is therefore likely that the excessive accumulation of lipids somehow disrupts acidification. These compartments also frequently contained a subdomain of brighter FITC-fluorescence (Figure 4C, arrows), indicating they must contain some intravesicular structures.
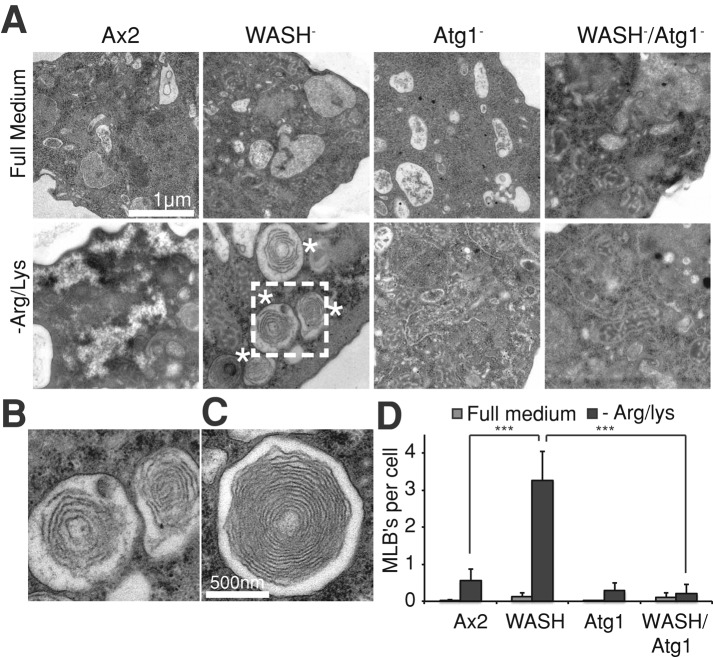
To look at these structures more closely, we analyzed mutant cells by transmission electron microscopy (TEM). Confirming our live-cell experiments, starved WASH-null cells contained many large multilamellar structures that were almost completely absent in both wild-type and full medium controls (Figure 5). Lysosomal multilamellar bodies (MLBs) are the classical hallmarks of many lysosomal storage disorders, including Tay-Sachs and Niemann-Pick disease, consistent with a reduction in β-hexosaminidase and sphingomyelinase activity in WASH mutant cells (Schmitz and Muller, 1991). WASH is therefore required for lysosomal lipid catabolism during starvation. Note also that, while the cytosol of Ax2 cells showed signs of digestion, becoming patchy upon starvation, WASH mutants retained a dense cytoplasm (Figure 5A). This is comparable with previous observations of autophagy mutants, confirming that WASH-null cells do not lose mass upon starvation (Otto et al., 2003).
FIGURE 5:
Ultrastructural analysis of starved WASH mutants. (A) Cells were placed in either full or Arg/Lys-deficient medium for 36 h before processing for TEM. Micrographs show the formation of large MLBs in starved WASH mutant cells, as indicated by asterisks. (B) and (C) Enlarged images of MLBs, (B) is the boxed region in (A). (D) Quantification of MLB frequency. At cells were grown as indicated, and the number of MLBs was scored per section, per cell. At least 20 cells were scored for each sample, and values are the mean ± SD of three experiments. ***, p < 0.005 (Student's t-test).
MLBs are induced in WASH-null cells by starvation. As autophagy is highly active under these conditions, we tested whether autophagy directly contributed to this phenotype. Examination of WASH/Atg1 double mutants by TEM indicated the accumulation of MLBs was blocked (Figure 5, A and D). The extensive phospholipid accumulation in WASH mutant cells is therefore dependent on autophagy. We interpret this to mean WASH is required for the membranes and contents of autophagosomes to be efficiently degraded, causing collapse of the perturbed lysosomal system in WASH-null cells.
Aberrant cathepsin D and β-hexosaminidase trafficking in WASH mutants
To establish why the degradative capacity was reduced in WASH mutants, we examined the activity and localization of lysosomal enzymes. Despite a 50% reduction in the phagosomal compartment, when we measured the total protein level of cathepsin D (catD) by Western blot, there was no difference between WASH-null and Ax2 cells, nor any accumulation of partly processed enzyme (Figure 6, A and B). We then measured the total enzymatic activity of lysosomal enzymes, using fluorogenic substrates for cathepsin D/E and β-hexosaminidase. Again, despite reduced protein levels in the phagocytic compartment, WASH-null cells contained 50% more cathepsin D/E and more than threefold more β-hexosaminidase activity than wild-type (Figure 6, C–E).
FIGURE 6:
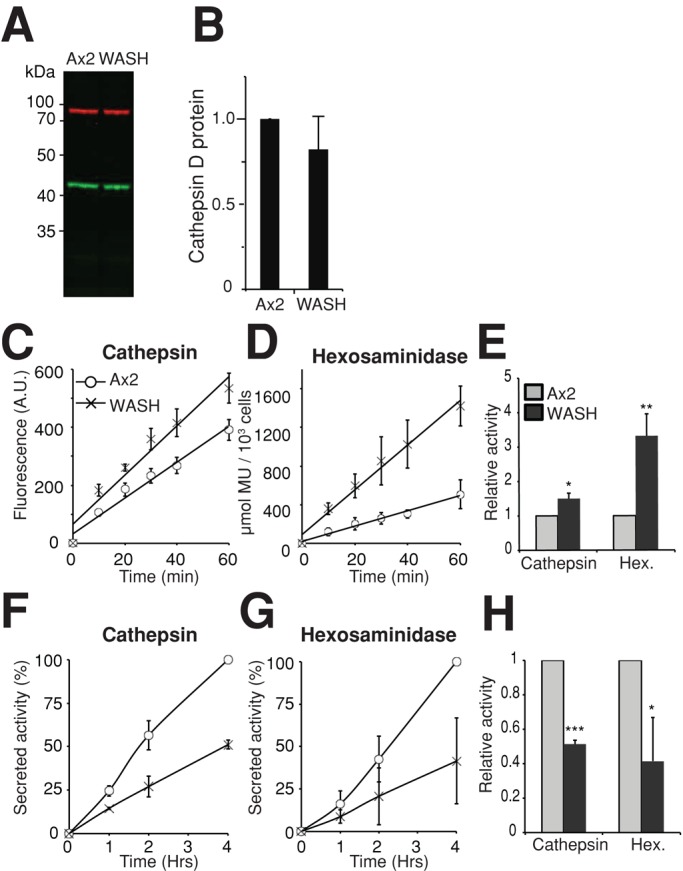
Hydrolytic activities in WASH mutants. (A) Western blot of catD (green) with 3-methylcrotonyl-CoA carboxylase α (MCCC1) as loading control in red. Quantitation of three independent blots shown in (B). (C–E) hydrolytic activity in Ax2 (circles) and WASH-null (crosses) whole-cell lysates using fluorogenic substrates for (C) cathepsin D/E and (D) β-hexosaminidase. Activity is indicated by the increase in fluorescence over time. (E) The relative activity of WASH-null cells (dark gray) compared with Ax2 (light gray). (F–H) Secreted activity of (F) catD and (G) β-hexosaminidase. Cells were placed in fresh media for the times indicated, and the activity in the media was measured as above. (H) Relative rates of secretion are plotted in (H). All values are the means ± SD of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.005 (Student's t-test).
As Dictyostelium cells constitutively secrete lysosomal components, and WASH is essential for the secretion of insoluble material such as dextran (Dimond et al., 1981; Carnell et al., 2011), we asked whether lysosomal enzyme secretion was also blocked. In WASH-null cells, the secretion of both cathepsin D/E and β-hexosaminidase activity was reduced by 50% (Figure 6, F–H). This explains the increase in total activities in the mutant but also demonstrates that, unlike dextran, lysosomal enzymes are partly secreted by a WASH-independent pathway.
These experiments indicate that, despite the presence of elevated total levels of lysosomal hydrolases, their delivery to both phagosomes and autophagosomes is blocked. WASH is therefore needed for the correct trafficking, rather than the synthesis or processing, of the digestive enzymes.
WASH is required for lysosomal enzyme recycling
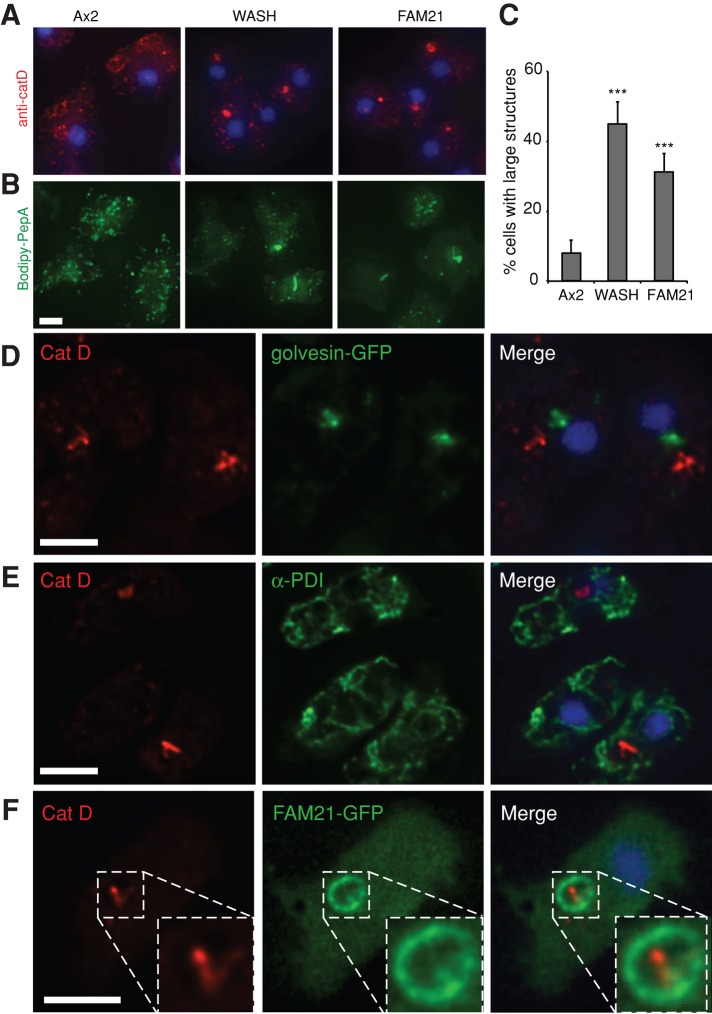
To determine where lysosomal trafficking was disrupted, we examined the localization of catD in mutant cells by immunofluorescence. As previously reported, in Ax2 cells fixed with ultracold methanol, the catD antibody labeled spots on several large ring structures (endocytic vesicles) and numerous smaller puncta (Figure 7A; Neuhaus et al., 2002). In both WASH- and FAM21-null cells however, catD frequently accumulated in a single large structure, with a significant reduction in other small puncta (Figure 7, A and C). This was confirmed using an alternative probe, fluorescently labeled pepstatin A, which specifically labels active catD and gave identical results (Figure 7B; Chen et al., 2000).
FIGURE 7:
Localization of catD in WASH mutant cells. (A) Ultracold methanol fixed cells stained with anti-catD antibody. (B) Paraformaldehyde-fixed cells stained with Bodipy-pepstatin A. (C) Quantification of large aggregate frequency in cells stained as in (A); ***, p < 0.005 (Student's t-test). For further characterization of these structures, colocalization of catD with (D) golvesin-GFP (Golgi), (E) anti-PDI antibody staining, and (F) FAM21-GFP was examined in fixed, WASH-null cells. Deconvolved wide-field images shown in all panels are all maximum intensity projections, except (E) and (F), which are single planes. All scale bars: 5 μm.
These observations indicate that catD is retained within a specific cellular compartment in WASH mutants. These concentrated catD structures, however, neither colocalized with the Golgi marker golvesin-GFP (Schneider et al., 2000) nor with the endoplasmic reticulum marker protein disulfide isomerase (PDI; Figure 7, D and E). Recently we showed that FAM21-GFP is able to localize to maturing endosomes independently of the other WASH complex members (Park et al., 2013). When we expressed FAM21-GFP in WASH-null cells, it clearly localized to a vesicular membrane surrounding the catD structure, indicating that the catD accumulates where WASH would normally be active.
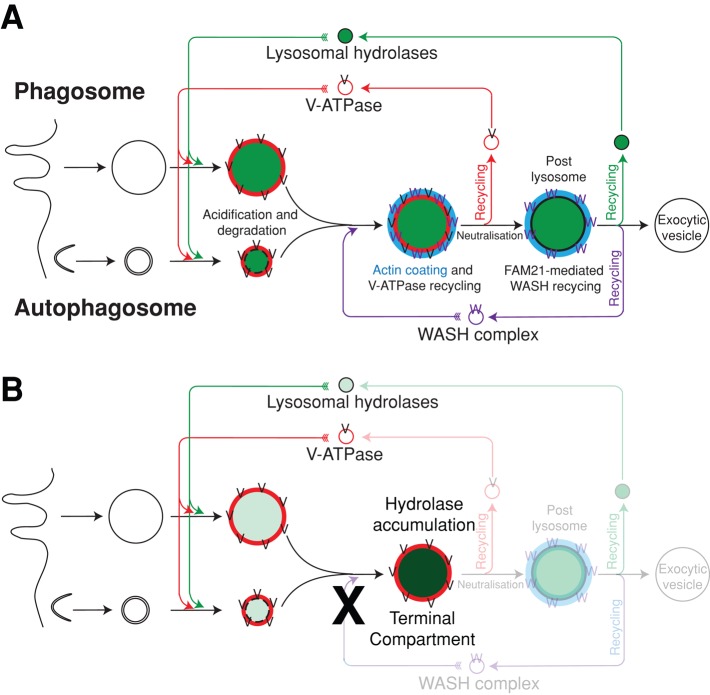
Previous analysis of Dictyostelium phagosome maturation has shown that the lysosomal hydrolases, including catD, are recovered from the phagosome at a late stage, just prior to exocytosis of the indigestible material (Gotthardt et al., 2002). Loss of WASH must therefore block the pathway before this recycling can take place. Lysosomal proteins therefore accumulate in a terminal compartment, unable to fuse with or be delivered to nascent phagosomes and autophagosomes. Furthermore, the identical results obtained in FAM21-null cells (in which the pathway proceeds past v-ATPase recycling and neutralization before stalling) indicate that hydrolase recycling occurs at the same time as or after FAM21-mediated removal of WASH and is independent of v-ATPase retrieval (Figure 8).
FIGURE 8:
(A) Schematic model of WASH and the endocytic pathway in Dictyostelium. After the formation of phagosomes and autophagosomes, v-ATPase and lysosomal hydrolases are delivered. These digest the cargo until WASH is recruited, sorting the v-ATPase into small recycling vesicles and neutralizing the compartment. Subsequently the FAM21-mediated recycling of WASH removes the actin coat, prior to exocytosis. At the same late stage, lysosomal hydrolases are also recovered for delivery to new (auto)phagosomes. (B) In the absence of WASH, v-ATPase recycling is blocked, and lysosomes remain acidic. The pathway can proceed no further, which traps the hydrolases in this terminal compartment and blocks their recycling and delivery to nascent autophagosomes and phagosomes.
DISCUSSION
The WASH complex, and the actin it polymerizes on vesicles, has been associated with an increasing number of trafficking roles. In this paper, we demonstrate that disruption of WASH causes the accumulation of lysosomal hydrolases in a late endocytic compartment, where they get trapped and are thus unavailable for recycling to nascent degradative compartments.
The reduced delivery of lysosomal hydrolases in WASH mutants has several physiological implications. The primary defect in these mutants is a decreased capacity to degrade lipids and proteins. This means that both phagocytosis and autophagy are compromised, and the cell is unable to use either bacteria or its own cytoplasm to provide nutrients. Therefore, while WASH mutants grow normally on our standard laboratory strain of K. aerogenes, they have much more difficulty growing on other bacteria. It should be noted that both the Dictyostelium and laboratory Klebsiella have been strongly selected for optimum growth over decades of coculture in the laboratory, and, although K. aerogenes, Klebsiella pneumoniae, and Enterobacter aerogenes are now classified as the same bacteria (Brisse et al., 2006), WASH mutants could grow only on our standard laboratory strain. In the wild, amoebae need to consume a wide range of bacteria, and so WASH function will be crucial for survival. While there is no obvious pattern in the types of bacteria that WASH mutants cannot use, it is likely that they have particularly high levels of fat or lipid or are simply more difficult to digest. In addition to enzymes required for protein and lipid catabolism, we also identified lysozyme as reduced in WASH mutant phagosomes. As lysozyme is required to break down the bacterial cell wall, this reduction will also disrupt the digestive process and explain this phenotype.
Consistent with a general lysosomal defect, WASH is also critical for autophagic degradation. While there are no overt defects in autophagosome formation or acidification, WASH-null cells are unable to digest their cytoplasm and survive starvation. In addition to known endocytic proteins, our proteomics assay also identified a number of components, such as mitochondrial and ribosomal proteins, that would not normally be expected to be present in phagosomes. As these data are ratiometric, it is unlikely that these are contaminants, and they instead may be the products of autophagy, indicative of altered autophagic trafficking and fusion with the endo/phagocytic pathway.
Interestingly, we also show that induction of autophagy leads to the accumulation of large quantities of phospholipid within lysosomal multilamellar bodies. This is characteristic of many lysosomal storage disorders and implies that autophagy is a major source of lipid delivery to the lysosome. Autophagy is also required for the biogenesis of physiological MLBs, such as those secreted by lung alveolar cells as a surfactant, indicating a general role for autophagy in the transport of lipids (Hariri et al., 2000). Autophagy is therefore a conserved source of lysosomal phospholipid and may therefore be a viable therapeutic target for the treatment of lysosomal storage disorders.
Previous work has shown that WASH is absolutely required for v-ATPase recycling and vesicle neutralization in Dictyostelium (Carnell et al., 2011), so we were surprised to find the largest accumulations of lipid within neutral vesicles (Figure 4C). However, lysosomal pH is elevated in several lipid storage diseases (Bach et al., 1999; Holopainen et al., 2001), and the autophagy-mediated accumulation of lysosomal cholesterol in Niemann-Pick type C disease also inhibits proteolytic activity (Elrick et al., 2012). It is therefore likely that the excessive lipid accumulation in WASH-null lysosomes disrupts lysosomal function in multiple ways, exacerbating the underlying defect in enzyme delivery.
In a number of respects, our observations contrast with the work of others on mammalian epithelial cells, indicating a novel mechanism by which WASH influences lysosomal trafficking. In HeLa cells, WASH was shown to directly regulate retromer-mediated retrograde transport of the CI-M6PR from endosomes to the Golgi. WASH knockdown therefore leads to CI-M6PR accumulation on endosomes (Gomez and Billadeau, 2009). In contrast, our proteomic data show a putative CD-M6PR decreased on both WASH- and FAM21-null phagosomes. Loss of FAM21 causes the constitutive activation of WASH in both Dictyostelium and mammalian cells (Park et al., 2013), and the FAM21-free complex remains able to mediate v-ATPase recycling from late endosomes. In this respect, WASH and FAM21 mutants have opposing phenotypes. Despite the contrasting defects in v-ATPase trafficking, the putative CD-M6PR is reduced in both mutant phagosomes. This implies either indirect regulation by WASH or that the hydrolases are recycled with the WASH complex independently of its actin polymerization activity.
Our data indicate that lysosomal hydrolases are retrieved after WASH-mediated neutralization. This is in good agreement with previous analysis of Dictyostelium phagosome maturation, which showed that different hydrolases are delivered to and recovered from phagosomes at distinct stages in their maturation (Gotthardt et al., 2002). It is not known how the soluble enzymes are selectively retrieved, or how different lysosomal populations are maintained, but WASH is clearly important to define distinct phases of phagosome maturation. Binding of M6PRs to lysosomal hydrolases is pH dependent, resulting in their release when they enter acidified lysosomes (Seaman, 2004). It is therefore plausible that a similar mechanism occurs during hydrolase recycling, requiring WASH-mediated neutralization before enzyme retrieval.
While the general mechanisms are conserved, there are clearly substantial differences in the trafficking pathways of Dictyostelium and mammalian epithelial and fibroblast cell lines. As a professional phagocyte, Dictyostelium is more representative of other phagocytic cells, such as leukocytes, macrophages, and dendritic cells, which have similar requirements for the delivery of lysosomal enzymes to phagosomes (Neuhaus and Soldati, 1999). A role of WASH in preparing phagosomes for exocytosis has also been shown in macrophages expelling the pathogenic yeast Cryptococcus neoformans, indicating conserved functionality (Carnell et al., 2011). Macrophages seem particularly susceptible to drug-induced phospholipidosis (Schmitz and Grandl, 2009), so a similar WASH-dependent pathway may also be important to protect cells against drug toxicity.
The secretion of lysosomal hydrolases is not unique to Dictyostelium. In specialized mammalian cells, lysosome-related organelles also undergo exocytosis (Raposo et al., 2002), and calcium can stimulate the secretion of lysosomes in fibroblasts (Andrews, 2000; Laulagnier et al., 2011). Whether the release of lysosomal enzymes is the primary function of such secretory events is doubtful, considering that most will be inactive at the extracellular pH. It is therefore possible that mechanisms exist to recover the hydrolases prior to release that might require WASH in a similar manner.
Several groups have also reported the disruption of endosomal and lysosomal networks when WASH is ablated in mammalian cells (Derivery et al., 2009; Gomez et al., 2012; Piotrowski et al., 2012). While there is good evidence that WASH specifically regulates the trafficking of several transmembrane proteins, such as the v-ATPase, retromer, growth factor receptors, and integrins (Derivery et al., 2009; Gomez and Billadeau, 2009; Carnell et al., 2011; Zech et al., 2011), such severe disruption of the lysosomal system will have global physiological significance. In this study, we have shown that, in Dictyostelium at least, WASH has a general role in lysosomal maintenance and the efficient digestion of phagocytic and autophagic cargo.
MATERIALS AND METHODS
Cell strains and culture
For all experiments, except the proteomics analysis, the Dictyostelium Ax2 strain was the wild-type control and was the parent of all mutants. Ax3 parent and isogenic knockout strains were used for the phagosome proteomics. All WASH and FAM21 knockout strains were described recently (Park et al., 2013). Cells were routinely cultured in HL-5 axenic medium, except in starvation experiments, when they were grown in defined SIH medium (Han et al., 2004) and then washed three times in SIH-Arg/Lys. All media were supplied by Formedium (Hunstanton, UK). Cells were transformed by electroporation using standard methods.
The procedure to test growth on bacteria has been extensively described previously (Froquet et al., 2009). Briefly, 10–104 Dictyostelium cells are deposited on a bacterial lawn (50 μl of an overnight culture) and allowed to grow until the colonies become visible. The bacterial strains used for the growth tests were kindly provided by Pierre Cosson (Centre Médical Universitaire, Geneva) and were K. pneumoniae laboratory strain and 52145 isogenic mutant (Benghezal et al., 2006), the isogenic Pseudomonas aeruginosa strains PT5 and PT531 (rhlR-lasR avirulent mutant; Cosson et al., 2002), Escherichia coli DH5α (Life Technologies, Paisley, UK), E. coli B/r (Gerisch, 1959), nonsporulating B. subtilis 36.1 (Ratner and Newell, 1978), and Micrococcus luteus (Wilczynska and Fisher, 1994). E. aerogenes was obtained from the American Type Culture Collection (strain no. 51697; Manassas, VA).
Dictyostelium viability and total protein assays
Cells were washed three times in SIH–Arg/Lys before being resuspending at 5 × 106 cells/ml in shaking flasks. Every 2 d, samples were removed and plated onto SM agar plates (Formedium, Hunstanton, UK) in conjunction with a lawn of K. aerogenes, and viability was assessed by the proportion of cells able to form new colonies, as previously described (Otto et al., 2003). For determination of total protein upon starvation, cells were washed as above, and 1 × 106 cells were seeded in wells of a 24-well plate. At each time point, the contents of triplicate wells were resuspended, pelleted by centrifugation, and frozen. Pellets were then lysed in 100 μl 150 mM NaCl, 10 mM Tris, 1 mM ethylene glycol tetraacetic acid (EGTA), 1 mM EDTA, 1% NP40 (pH 7.5), and total protein measures were taken using Precision Red reagent (Cytoskeleton, Denver, CO). Protein levels were then normalized to the unstarved protein measurements for each strain.
Cloning and gene disruption
For generating an Atg1 knockout construct, the 5′ and 3′ fragments of the gene were amplified using the primers AAACAAATGAACCCTTTGCC/aagcttTTGGATCCCATAAGGAAGTGAAGAGGCG and GGATCCaaaagcttTATTCATCACCAACCGAGGC/TGAGTTCTCACTTCAAATGC. These were then combined by PCR, and the floxed blasticidin cassette from pLPBLP (Faix et al., 2004) was inserted as a BamHI/HindIII fragment to generate the final Atg1 knockout construct pJSK471. The Bsr cassette from WASH-null cells was removed using Cre recombinase (Faix et al., 2004), and pJSK471 was used to make both a WASH/Atg1 double and an Ax2 Atg1 single knockout. The FAM21-GFP construct used was described by Park et al. (2013), and the golvesin-GFP plasmid was generated by amplifying the full-length golvesin gene from cDNA, adding 5′BamHI/3′XbaI sites, and subcloning into the GFP-fusion vector pDM450 (Veltman et al., 2009).
Live-cell imaging and pH determination
Autophagosomes were observed using the GFP-Atg8 plasmid pDM430 (King et al., 2011). High-resolution images were obtained by overlaying cells with a layer of 1% agarose and compressing them by capillary action, as previously described (Yumura et al., 1984; King et al., 2011). Images were obtained on a Nikon (Tokyo, Japan) Eclipse TE 2000-U microscope equipped with a 100×/1.45 numerical aperture (NA) Nikon total internal reflection fluorescence oil-immersion objective, 473-nm diode laser illumination (Omicron, Rodgau, Germany), and a Cascade 512F EMCCD camera (Photometrics, Tucson, AZ).
For determination of autophagosome induction upon starvation, Z-series of images (0.25-μm step size) of uncompressed cells were taken at each time point, using an Olympus IX81 inverted wide-field microscope equipped with a 60×/1.42 NA Plan-ApoN objective. The number of visible puncta in maximum intensity projections was scored for >100 cells at each point. Differential interference contrast (DIC) images were captured on an inverted microscope fitted with a 100×/1.40 NA oil-immersion objective (Nikon).
For phospholipid labeling, cells were incubated with HCS LipidTOX Red (Molecular Probes) diluted 2000× in SIH or SIH–Arg/Lys for 24 h. The endocytic pathway was labeled by also including 0.2 mg/ml Cascade Blue and 0.4 mg/ml FITC-dextran (Sigma-Aldrich, St. Louis, MO). Live-cell images were captured on an Olympus FV1000 confocal microscope, using a Plan-ApoN 60×/1.4 NA objective. Ax2 control cells were used to calibrate the microscope laser intensities such that the pH-sensitive FITC fluoresced only in a subset of vesicles, and identical settings were used with all strains and conditions. For direct determination of vesicular pH, cells were incubated overnight with 0.2 mg/ml LysoSensor yellow/blue dextran (Life Technologies; Diwu et al., 1999). Confocal images were then collected, using 365-nm excitation and detecting the emission at both 450 and 510 nm. pH was determined by comparing the ratio of 450/510-nm emission to a standard curve generated by imaging LysoSensor in SIH medium at different pHs (Figure S1B). The false-colored vesicular pH images were then generated using ImageJ (http://rsbweb.nih.gov/ij) by thresholding the 450-nm channel to generate a mask and converting the 450/510-nm emission ratios to pH values. The emission ratio of individual vesicles was determined by manually outlining each vesicle and measuring the mean intensity of each channel.
Immunohistochemistry
Two methods of fixation were used. All antibody staining was done using cells fixed in ultracold methanol, as described by Hagedorn et al. (2006). Briefly, cells were seeded onto thin (no. 0) acid-washed coverslips and left to adhere. They were then plunged into a beaker of −80°C methanol on dry ice and left to fix for 30 min. After several washes in phosphate-buffered saline (PBS), coverslips were blocked with 3% bovine serum albumin (BSA) and stained. The rabbit polyclonal anti-catD antibody was a kind gift from Agnes Journet (Journet et al., 1999), and PDI was stained using a cocktail of five monoclonal antibodies. For Bodipy-pepstatin A staining, cells were fixed with 4% paraformaldehyde, 0.1% Brij35 in citrate buffer (50 mM sodium citrate, pH 4.1) for 20 min before being washed and then stained with 1 μM Bodipy-pepstatin A (Life Technologies; Chen et al., 2000). Coverslips were kept in citrate buffer at all stages to maintain cathepsin in the active conformation. All coverslips were mounted in ProLong Gold with 4′,6-diamidino-2-phenylindole (Life Technologies). Images were captured on an Olympus IX81 wide-field fluorescence microscope with a 100×/1.4 NA objective and an additional 1.6× optovar magnification. For deconvolution, a Z-stack of images (0.25-μm step size) was captured with a CoolSNAPHQ camera (Photometrics). Images were then deconvolved by Volocity software (Perkin Elmer-Cetus, Waltham, MA) using calculated point-spread functions.
Phagocytosis and in vivo proteolysis assay
Bead uptake was measured as previously described (Sattler et al., 2013). Fluorescent beads of 1-μm diameter (fluorescent YG-carboxylated beads; Polysciences, Warrington, PA) were added to cells at a 200:1 dilution; at each time point, cells were washed free of unbound beads, and the number of ingested beads was determined by flow cytometry. Proteolysis experiments were adapted from Yates et al. (2005) and carried out as described previously (Gopaldass et al., 2012). Briefly, cells were fed beads coupled to Alexa Fluor 594 succinimidyl ester (Life Technologies) and BSA labeled with DQ Green at a self-quenching concentration (Molecular Probes). On BSA proteolysis, DQ Green fluorescence increases due to dequenching of the fluorophore. Results were normalized to bead uptake by plotting the ratio of DQ Green to Alexa Fluor 594 fluorescence as a function of time.
TEM
For investigating subcellular structures using TEM, adherent Dictyostelium cells were fixed in HL5c medium with 2% glutaraldehyde and 1% paraformaldehyde for 1 h. Subsequently the cells were scraped off the dish, washed with buffer, and incubated for 1 h with 0.5% OsO4. The samples were washed several times; this was followed by an incubation step with 1% tannin for 20 min at room temperature. The cells were washed again, dehydrated, embedded in Epon resin, and processed for conventional electron microscopy. Grids were examined with a Tecnai Spirit transmission electron microscope (FEI, Eindhoven, Netherlands). Images were quantified by counting the number of MLBs in >20 cells for each strain/condition.
Cathepsin and hexosaminidase assays
For enzymatic assays, cells growing in HL5 were resuspended, washed twice in KK2 buffer (0.1 M potassium phosphate, pH 6.1), and lysed in 50 mM sodium citrate buffer (pH 4.2) with 1% Triton X100 at 5 × 104 cells/ml. For cathepsin activity, lysates were incubated at 37°C with 50 μM cathepsin fluorogenic substrate (BML-P145-001; Enzo Life Sciences, Exeter, UK) in 50 mM citrate buffer (pH 4.2), and samples were removed and quenched by being diluted at 1:10 in 10% trichloroacetic acid. Fluorescence at 330-nm excitation/410-nm emission was then detected in a fluorometer. For β-hexoaminidase activity, 2 mM 4-methylumbelliferyl-β-d-N-acetylglucosamine (Calbiochem, San Diego, CA) was used as substrate, and reactions were quenched by dilution at 1:10 in 0.2 M glycine, 0.2 M Na2CO3. Fluorescence was measured at 365-nm excitation/445-nm emission. For calculation of β-hexoaminidase activity, 4-methylumbelliferone was used to generate a standard curve.
For measuring secreted enzyme activity, cells were washed three times in fresh media and seeded at 2 × 106 cells per well of a 24-well plate in 500 μl of fresh medium. At each time point, the medium was carefully removed and spun for 30 s at 3000 rpm in a benchtop microfuge to pellet any cells, and 400 μl of the supernatant was taken. Activity was determined by incubating samples of the media with the appropriate substrates as above for 1 h at 37°C.
Mass spectrometry and analysis
Phagosomes were isolated by density centrifugation after cells were fed latex beads, as described previously, using a pulse–chase of 15 min/2 h 45 min (Gotthardt et al., 2006b). The reduction, alkylation, digestion, and tandem mass tag (TMT) labeling was mainly performed as described by Dayon et al. (2008). Briefly, phagosome pellets were resuspended at 100 μg proteins per 33 μl of triethylammonium hydrogen carbonate buffer (TEAB: 0.1 M, pH 8.5, 6 M urea). After addition of 50 mM Tris-(2-carboxyethyl) phosphine hydrochloride (TCEP), samples were reduced for 1 h at 37°C. Alkylation was then performed at room temperature in the dark for 30 min after addition of 1 μl of 400 mM iodoacetamide. Then 67 μl of TEAB and 2 μg trypsin were added, and digestion was performed overnight at 37°C. Each sample was then labeled with a TMT reagent according to the manufacturer's instructions (Proteome Sciences, Frankfurt, Germany) and evaporated under a speed-vacuum.
Off-gel electrophoresis was performed according to the manufacturer's instructions (Agilent, Santa Clara, CA). After being desalted, pooled samples were reconstituted in OFFGEL solution. Isoelectric focusing was performed using a 12-well frame on an Immobiline DryStrip (pH 3–10, 13 cm) and run at 8000 V, 50 μA, 200 mW, until 20 kVh was reached. The 12 fractions were then recovered and desalted using C18 MicroSpin columns.
ESI LTQ-OT (electrospray ionization linear trap quadropole orbitrap) mass spectrometry (MS) was performed on a LTQ Orbitrap Velos (Thermo Electron, Waltham, MA) equipped with a NanoAcquity system (Waters). Peptides were trapped on a 5-μm, 200-Å Magic C18 AQ (Michrom) 0.1-mm × 20-mm precolumn and separated on a 5-μm, 100-Å Magic C18 AQ (Michrom, Auburn, CA) 0.75-mm × 150-mm column with a gravity-pulled emitter. Analytical separation was run for 65 min using a gradient of H2O/FA 99.9%/0.1% (solvent A) and CH3CN/FA 99.9%/0.1% (solvent B). The gradient was run as follows: 95% A and 5% B at 0–1 min, then to 65% A and 35% B at 55 min, and 20% A and 80% B at 65 min at a flow rate of 220 nl/min. For MS survey scans, the OT resolution was set to 60,000 and the ion population was set to 5 × 105, with an m/z window of 400–2000. A maximum of three precursors were selected for both collision-induced dissociation (CID) in the LTQ and high-energy C-trap dissociation (HCD) with analysis in the OT. For tandem MS (MS/MS) in the LTQ, the ion population was set to 7000 (isolation width of 2 m/z), while for MS/MS detection in the OT, it was set to 2 × 105 (isolation width of 2.5 m/z), with resolution of 7500, first mass at m/z = 100, and maximum injection time of 750 ms. The normalized collision energies were set to 35% for CID and 60% for HCD.
Protein identification was performed with the EasyProt platform (Gluck et al., 2012). After peak list generation, the CID and HCD spectra were merged for simultaneous identification and quantification (Dayon et al., 2010). The parent ion tolerance was set to 10 ppm. TMT-sixplex amino terminus and TMT-sixplex lysine (229.1629 Da), carbamidomethylation of cysteines were set as fixed modifications. All data sets were searched once in the forward and once in the reverse Dictyostelium Uniprot_sprot database. For identification, only proteins matching two different peptide sequences were kept.
Isobaric quantification was performed using the IsoQuant module of EasyProt. TMT sixplex was selected as the reporter, and a mass tolerance of 0.05 m/z was used to calculate WASH/Ax2 and FAM21/Ax2 protein ratios. Statistics were performed using EasyProt's Mascat and Libra statistical methods to generate a list of proteins that significantly differed between samples.
Western blotting
Cells growing in HL5 were resuspended, washed in KK2 buffer, and lysed in 150 mM NaCl, 10 mM Tris, 1 mM EGTA, 1 mM EDTA, 1% NP40 (pH 7.5). Samples were then normalized to total protein content and boiled in SDS buffer (Life Technologies) before being loaded on a 10% Bis-Tris acrylamide Nu-PAGE gel (Life Technologies). Gels were then transferred onto nitrocellulose membrane before being probed with anti-catD antibody diluted 1:5000 in PBS (Journet et al., 1999) and a DyLight 800 anti-rabbit fluorescent secondary antibody (Pierce, Rockford, IL). As a loading control, we used Alexa Fluor 680–labeled streptavidin (Life Technologies), which, in Dictyostelium, recognizes a single protein at ∼77 kDa corresponding to the mitochondrial methylcrotonyl-CoA carboxylase (Davidson et al., 2013). Blots were analyzed on a fluorescent gel imager, and images were quantified using ImageJ.
Supplementary Material
Acknowledgments
We thank Agnes Journet for kindly providing us with catD antibody; Pierre Cosson for sharing the bacterial strains; David Strachan for help with image processing; and Alex Scherl, Carla Pasquarello, Patrizia Arboit, and Alexandre Hainard from the Proteomic Core Facility (CMU, Geneva) for their proteomics expertise and help. We are also very grateful to Pete Thomason, Tobias Zech, and Laura Machesky for many helpful discussions and critiques of the manuscript and to Pete Watson for informed suggestions. This work was supported by Cancer Research–UK core funding to R.H.I. and a grant from the Swiss National Science Foundation to the T.S. laboratory.
Abbreviations used:
- BSA
bovine serum albumin
- catD
cathepsin D
- CD
cation dependent
- CI
cation independent
- CID
collision-induced dissociation
- EGF
epidermal growth factor
- EGTA
ethylene glycol tetraacetic acid
- FITC
fluorescein isothiocyanate
- GFP
green fluorescent protein
- HCD
high-energy C-trap dissociation
- M6PR
mannose-6-phosphate receptor
- MLB
multilamellar body
- MS
mass spectrometry
- NA
numerical aperture
- PBS
phosphate-buffered saline
- PDI
protein disulfide isomerase
- siRNA
small interfering RNA
- TCEP
Tris-(2-carboxyethyl) phosphine hydrochloride
- TEAB
triethylammonium hydrogen carbonate buffer
- TEM
transmission electron microscopy
- TMT
tandem mass tag
- v-ATPase
vacuolar ATPase
- WASH
Wiskott-Aldrich syndrome protein and scar homologue
- WASP
Wiskott-Aldrich syndrome protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-02-0092) on July 24, 2013.
Present addresses: *Department of Biomedical Sciences, Firth Court, Sheffield University, Sheffield, S10 2TN, United Kingdom;
†Département de Biochimie, Université de Lausanne, Chemin des boveresses 155, CH-1066 Epalinges, Switzerland.
REFERENCES
- Andrews NW. Regulated secretion of conventional lysosomes. Trends Cell Biol. 2000;10:316–321. doi: 10.1016/s0962-8924(00)01794-3. [DOI] [PubMed] [Google Scholar]
- Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol. 2004;165:123–133. doi: 10.1083/jcb.200312055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach G, Chen CS, Pagano RE. Elevated lysosomal pH in Mucolipidosis type IV cells. Clin Chim Acta. 1999;280:173–179. doi: 10.1016/s0009-8981(98)00183-1. [DOI] [PubMed] [Google Scholar]
- Benghezal M, et al. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 2006;8:139–148. doi: 10.1111/j.1462-5822.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- Brisse S, Grimont F, Grimont PAD. The genus Klebsiella. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes. New York: Springer; 2006. pp. 159–196. [Google Scholar]
- Calvo-Garrido J, Carilla-Latorre S, Kubohara Y, Santos-Rodrigo N, Mesquita A, Soldati T, Golstein P, Escalante R. Autophagy in Dictyostelium: genes and pathways, cell death and infection. Autophagy. 2010;6:686–701. doi: 10.4161/auto.6.6.12513. [DOI] [PubMed] [Google Scholar]
- Carnell M, Zech T, Calaminus SD, Ura S, Hagedorn M, Johnston SA, May RC, Soldati T, Machesky LM, Insall RH. Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J Cell Biol. 2011;193:831–839. doi: 10.1083/jcb.201009119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CS, Chen WN, Zhou M, Arttamangkul S, Haugland RP. Probing the cathepsin D using a BODIPY FL-pepstatin A: applications in fluorescence polarization and microscopy. J Biochem Biophys Methods. 2000;42:137–151. doi: 10.1016/s0165-022x(00)00048-8. [DOI] [PubMed] [Google Scholar]
- Clarke M, Köhler J, Arana Q, Liu T, Heuser J, Gerisch G. Dynamics of the vacuolar H+-ATPase in the contractile vacuole complex and the endosomal pathway of Dictyostelium cells. J Cell Sci. 2002;115:2893–2905. doi: 10.1242/jcs.115.14.2893. [DOI] [PubMed] [Google Scholar]
- Cosson P, Zulianello L, Join-Lambert O, Faurisson F, Gebbie L, Benghezal M, Van Delden C, Curty LK, Kohler T. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J Bacteriol. 2002;184:3027–3033. doi: 10.1128/JB.184.11.3027-3033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BE, Griffin EE, Ullery JC, Jerome WG. Effects of cellular cholesterol loading on macrophage foam cell lysosome acidification. J Lipid Res. 2007;48:1012–1021. doi: 10.1194/jlr.M600390-JLR200. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, King JS, Insall RH. The use of streptavidin conjugates as immunoblot loading controls and mitochondrial markers for use with Dictyostelium discoideum. Biotechniques. 2013;55:39–41. doi: 10.2144/000114054. [DOI] [PubMed] [Google Scholar]
- Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- Dayon L, Pasquarello C, Hoogland C, Sanchez JC, Scherl A. Combining low- and high-energy tandem mass spectra for optimized peptide quantification with isobaric tags. J Proteomics. 2010;73:769–777. doi: 10.1016/j.jprot.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Deretic V. Autophagosome and phagosome. Methods Mol Biol. 2008;445:1–10. doi: 10.1007/978-1-59745-157-4_1. [DOI] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Dieckmann R, Gueho A, Monroy R, Ruppert T, Bloomfield G, Soldati T. The balance in the delivery of ER components and the vacuolar proton pump to the phagosome depends on myosin IK in Dictyostelium. Mol Cell Proteomics. 2012;11:886–900. doi: 10.1074/mcp.M112.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimond RL, Burns RA, Jordan KB. Secretion of lysosomal enzymes in the cellular slime mold, Dictyostelium discoideum. J Biol Chem. 1981;256:6565–6572. [PubMed] [Google Scholar]
- Diwu Z, Chen CS, Zhang C, Klaubert DH, Haugland RP. A novel acidotropic pH indicator and its potential application in labeling acidic organelles of live cells. Chem Biol. 1999;6:411–418. doi: 10.1016/s1074-5521(99)80059-3. [DOI] [PubMed] [Google Scholar]
- Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken) 2010;67:193–206. doi: 10.1002/cm.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrick MJ, Yu T, Chung C, Lieberman AP. Impaired proteolysis underlies autophagic dysfunction in Niemann-Pick type C disease. Hum Mol Genet. 2012;21:4876–4887. doi: 10.1093/hmg/dds324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Kreppel L, Shaulsky G, Schleicher M, Kimmel AR. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 2004;32:e143. doi: 10.1093/nar/gnh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froquet R, Lelong E, Marchetti A, Cosson P. Dictyostelium discoideum: a model host to measure bacterial virulence. Nat Protoc. 2009;4:25–30. doi: 10.1038/nprot.2008.212. [DOI] [PubMed] [Google Scholar]
- Gerisch G. Ein submerskulturver fahren fur entwicklungsphysiologische untersuchungen an Dictyostelium discoideum. Naturwissenschaften. 1959;46:654–656. [Google Scholar]
- Gluck F, et al. EasyProt—an easy-to-use graphical platform for proteomics data analysis. J Proteomics. 2012;79:146–160. doi: 10.1016/j.jprot.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Gorman JA, de Narvajas AA, Koenig AO, Billadeau DD. Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol Biol Cell. 2012;23:3215–3228. doi: 10.1091/mbc.E12-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopaldass N, et al. Dynamin A, myosin IB and Abp1 couple phagosome maturation to F-actin binding. Traffic. 2012;13:120–130. doi: 10.1111/j.1600-0854.2011.01296.x. [DOI] [PubMed] [Google Scholar]
- Gotthardt D, Blancheteau V, Bosserhoff A, Ruppert T, Delorenzi M, Soldati T. Proteomics fingerprinting of phagosome maturation and evidence for the role of a Gα during uptake. Mol Cell Proteomics. 2006a;5:2228–2243. doi: 10.1074/mcp.M600113-MCP200. [DOI] [PubMed] [Google Scholar]
- Gotthardt D, Dieckmann R, Blancheteau V, Kistler C, Reichardt F, Soldati T. Preparation of intact, highly purified phagosomes from Dictyostelium. Methods Mol Biol. 2006b;346:439–448. doi: 10.1385/1-59745-144-4:439. [DOI] [PubMed] [Google Scholar]
- Gotthardt D, Warnatz HJ, Henschel O, Bruckert F, Schleicher M, Soldati T. High-resolution dissection of phagosome maturation reveals distinct membrane trafficking phases. Mol Biol Cell. 2002;13:3508–3520. doi: 10.1091/mbc.E02-04-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn M, Neuhaus EM, Soldati T. Optimized fixation and immunofluorescence staining methods for Dictyostelium cells. Methods Mol Biol. 2006;346:327–338. doi: 10.1385/1-59745-144-4:327. [DOI] [PubMed] [Google Scholar]
- Han SI, Friehs K, Flaschel E. Cultivation of Dictyostelium discoideum on an improved synthetic medium in a conventional bioreactor. Process Biochem. 2004;39:925–930. [Google Scholar]
- Harbour ME, Breusegem SY, Antrobus R, Freeman C, Reid E, Seaman MN. The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J Cell Sci. 2010;123:3703–3717. doi: 10.1242/jcs.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri M, Millane G, Guimond MP, Guay G, Dennis JW, Nabi IR. Biogenesis of multilamellar bodies via autophagy. Mol Biol Cell. 2000;11:255–268. doi: 10.1091/mbc.11.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holopainen JM, Saarikoski J, Kinnunen PK, Jarvela I. Elevated lysosomal pH in neuronal ceroid lipofuscinoses (NCLs) Eur J Biochem. 2001;268:5851–5856. doi: 10.1046/j.0014-2956.2001.02530.x. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, Billadeau DD. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc Natl Acad Sci USA. 2010;107:10442–10447. doi: 10.1073/pnas.0913293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet A, Chapel A, Jehan S, Adessi C, Freeze H, Klein G, Garin J. Characterization of Dictyostelium discoideum cathepsin D. J Cell Sci. 1999;112:3833–3843. doi: 10.1242/jcs.112.21.3833. [DOI] [PubMed] [Google Scholar]
- King JS. Mechanical stress meets autophagy: potential implications for physiology and pathology. Trends Mol Med. 2012;18:583–588. doi: 10.1016/j.molmed.2012.08.002. [DOI] [PubMed] [Google Scholar]
- King JS, Veltman DM, Insall RH. The induction of autophagy by mechanical stress. Autophagy. 2011;7:1490–1499. doi: 10.4161/auto.7.12.17924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulagnier K, Schieber NL, Maritzen T, Haucke V, Parton RG, Gruenberg J. Role of AP1 and Gadkin in the traffic of secretory endo-lysosomes. Mol Biol Cell. 2011;22:2068–2082. doi: 10.1091/mbc.E11-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 2007;3:e237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus EM, Almers W, Soldati T. Morphology and dynamics of the endocytic pathway in Dictyostelium discoideum. Mol Biol Cell. 2002;13:1390–1407. doi: 10.1091/mbc.01-08-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus EM, Soldati T. Molecular mechanisms of membrane trafficking. What do we learn from Dictyostelium discoideum. Protist. 1999;150:235–243. doi: 10.1016/S1434-4610(99)70026-X. [DOI] [PubMed] [Google Scholar]
- Otto GP, Wu MY, Kazgan N, Anderson OR, Kessin RH. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem. 2003;278:17636–17645. doi: 10.1074/jbc.M212467200. [DOI] [PubMed] [Google Scholar]
- Park L, Thomason PA, Zech T, King JS, Veltman DM, Carnell M, Ura S, Machesky LM, Insall RH. Cyclical action of the WASH complex: FAM21 and capping protein drive WASH recycling, not initial recruitment. Dev Cell. 2013;24:169–181. doi: 10.1016/j.devcel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Piotrowski JT, Gomez TS, Schoon RA, Mangalam AK, Billadeau DD. WASHout T cells demonstrate defective receptor trafficking, proliferation and effector function. Mol Cell Biol. 2012;33:958–973. doi: 10.1128/MCB.01288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Fevrier B, Stoorvogel W, Marks MS. Lysosome-related organelles: a view from immunity and pigmentation. Cell Struct Funct. 2002;27:443–456. doi: 10.1247/csf.27.443. [DOI] [PubMed] [Google Scholar]
- Ratner DI, Newell PC. Linkage analysis in Dictyostelium discoideum using multiply marked tester strains: establishment of linkage group VII and the reassessment of earlier linkage data. J Gen Microbiol. 1978;109:225–236. doi: 10.1099/00221287-109-2-225. [DOI] [PubMed] [Google Scholar]
- Sattler N, Monroy R, Soldati T. Quantitative analysis of phagocytosis and phagosome maturation. Methods Mol Biol. 2013;983:383–402. doi: 10.1007/978-1-62703-302-2_21. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Grandl M. Endolysosomal phospholipidosis and cytosolic lipid droplet storage and release in macrophages. Biochim Biophys Acta. 2009;1791:524–539. doi: 10.1016/j.bbalip.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Muller G. Structure and function of lamellar bodies, lipid-protein complexes involved in storage and secretion of cellular lipids. J Lipid Res. 1991;32:1539–1570. [PubMed] [Google Scholar]
- Schneider N, Schwartz JM, Kohler J, Becker M, Schwarz H, Gerisch G. Golvesin-GFP fusions as distinct markers for Golgi and post-Golgi vesicles in Dictyostelium cells. Biol Cell. 2000;92:495–511. doi: 10.1016/s0248-4900(00)01102-3. [DOI] [PubMed] [Google Scholar]
- Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman DM, Akar G, Bosgraaf L, Van Haastert PJ. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 2009;61:110–118. doi: 10.1016/j.plasmid.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Insall RH. WASP family proteins: their evolution and its physiological implications. Mol Biol Cell. 2010;21:2880–2893. doi: 10.1091/mbc.E10-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska Z, Fisher PR. Analysis of a complex plasmid insertion in a phototaxis-deficient transformant of Dictyostelium discoideum selected on a Micrococcus luteus lawn. Plasmid. 1994;32:182–194. doi: 10.1006/plas.1994.1054. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yates RM, Hermetter A, Russell DG. The kinetics of phagosome maturation as a function of phagosome/lysosome fusion and acquisition of hydrolytic activity. Traffic. 2005;6:413–420. doi: 10.1111/j.1600-0854.2005.00284.x. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Arimoto K, Sato M, Sakuragawa N, Arima M, Satoyoshi E. Reduction of acid sphingomyelinase activity in human fibroblasts induced by AY-9944 and other cationic amphiphilic drugs. J Biochem. 1985;98:1669–1679. doi: 10.1093/oxfordjournals.jbchem.a135438. [DOI] [PubMed] [Google Scholar]
- Yumura S, Mori H, Fukui Y. Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J Cell Biol. 1984;99:894–899. doi: 10.1083/jcb.99.3.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T, Calaminus SD, Caswell P, Spence HJ, Carnell M, Insall RH, Norman J, Machesky LM. The Arp2/3 activator WASH regulates α5β1-integrin-mediated invasive migration. J Cell Sci. 2011;124:3753–3759. doi: 10.1242/jcs.080986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.