A genome-wide approach to screen for haploinsufficient mutants that confer genome instability identified γ-TuSC genes. γ-TuSC is an important regulator in establishing the correct number and type of microtubules so that a spindle with the proper composition is assembled to maintain genome stability.
Abstract
How subunit dosage contributes to the assembly and function of multimeric complexes is an important question with implications in understanding biochemical, evolutionary, and disease mechanisms. Toward identifying pathways that are susceptible to decreased gene dosage, we performed a genome-wide screen for haploinsufficient (HI) genes that guard against genome instability in Saccharomyces cerevisiae. This led to the identification of all three genes (SPC97, SPC98, and TUB4) encoding the evolutionarily conserved γ-tubulin small complex (γ-TuSC), which nucleates microtubule assembly. We found that hemizygous γ-TuSC mutants exhibit higher rates of chromosome loss and increases in anaphase spindle length and elongation velocities. Fluorescence microscopy, fluorescence recovery after photobleaching, electron tomography, and model convolution simulation of spc98/+ mutants revealed improper regulation of interpolar (iMT) and kinetochore (kMT) microtubules in anaphase. The underlying cause is likely due to reduced levels of Tub4, as overexpression of TUB4 suppressed the spindle and chromosome segregation defects in spc98/+ mutants. We propose that γ-TuSC is crucial for balanced assembly between iMTs and kMTs for spindle organization and accurate chromosome segregation. Taken together, the results show how gene dosage studies provide critical insights into the assembly and function of multisubunit complexes that may not be revealed by using traditional studies with haploid gene deletion or conditional alleles.
INTRODUCTION
Multisubunit protein complexes regulate a variety of processes central to genome stability, including assembling mitotic spindles by the centrosome, attaching chromosomes to microtubules by the kinetochore, mediating cohesion between sister chromatids by cohesion complexes, and protecting the genome from accumulating errors by the DNA damage and spindle checkpoint signaling complexes (Hirano, 2000; Musacchio and Hardwick, 2002; Jaspersen and Winey, 2004; Lisby and Rothstein, 2004; Cheeseman and Desai, 2008). Although there has been considerable work on understanding how these complexes act and are regulated, much less is known with regard to how they are assembled with the correct stoichiometries and how altered subunit dosage affects their assembly and activities. This is of particular significance when considered within the context of diseases that are caused in part by altered gene dosage due to either increases or decreases in gene copy number, chromosome number, or deregulated transcription. Of note, such changes in gene dosage are highly prevalent in a large number of cancers. More recently, decreases in gene dosage as a result of whole or regional chromosome loss have been proposed to underlie the development of some cancers (Berger et al., 2011; Greenman, 2012). This is believed to arise as a result of haploinsufficiency, a condition in which a single functional copy of a gene is not enough to sustain normal activity in diploid organisms. The mechanism(s) of haploinsufficiency, however, remain enigmatic in many cases. The ability to perform genome-wide screens for haploinsufficient (HI) genes is at present not technically feasible in mammalian systems. It is possible to carry out such a comprehensive screen, however, in the budding yeast, Saccharomyces cerevisiae.
Considering the evolutionary conservation of genome stability pathways (Paulovich et al., 1997; Meraldi et al., 2006), identifying HI genes in yeast is likely to provide insights into orthologous HI genes in higher eukaryotes, including humans. Prior genome-scale studies of haploinsufficiency in budding yeast largely focused on fitness effects, and a limited number of reports investigated actin and DNA damage repair haploinsufficiencies (Deutschbauer et al., 2005; Haarer et al., 2007, 2011; Delneri et al., 2008; Strome et al., 2008; de Clare and Oliver, 2013). Finding a diverse group of biological processes that display genome instability (GI) as a result of haploinsufficient mutations provides new insight into the role of gene dosage and its specific effect on genome stability, an area that remains largely unexplored. We sought to identify HI genes that lead to GI, using a genome-wide approach that covers nearly all essential and nonessential yeast genes. We identified a variety of genes, including those encoding the γ-tubulin small complex (γ-TuSC). Although γ-TuSC is an evolutionarily conserved complex known to nucleate microtubule assembly, it is not fully understood how γ-TuSCs contribute to spindle organization (Kollman et al., 2011). We found that hemizygous deletions of γ-TuSC genes resulted in elevated rates of chromosome loss and a variety of spindle defects not previously reported in haploid studies of γ-TuSC mutants (Geissler et al., 1996; Knop et al., 1997; Vogel and Snyder, 2000; Vogel et al., 2001; Keck et al., 2011; Lin et al., 2011). We discovered that when γ-TuSC sites are limiting, rather than an overall decrease in microtubules, we observe more rapid kinetochore microtubule (kMT) shortening, increased density of interpolar microtubules (iMTs) during anaphase, and elevated chromosome loss rates. To our knowledge, this is the first report of a systematic genome-wide screen for HI genes that result in GI. Moreover, our studies of hemizygous γ-TuSC mutants suggest that iMT and kMT assembly is in part regulated by γ-TuSC such that a spindle of the correct composition is formed to mediate accurate chromosome segregation. Furthermore, this work highlights how studying haploinsufficiency mechanisms can yield novel insights into fundamental biological processes that may not be uncovered in studies with conditional alleles or whole-gene deletions.
RESULTS
Genome-wide identification of HI genes that guard against genome instability
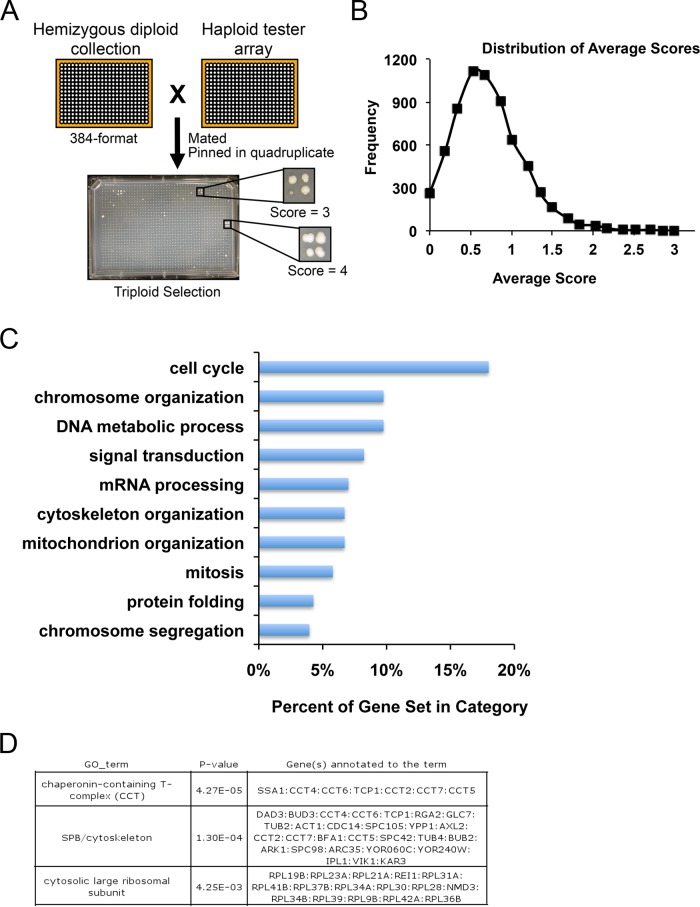
To develop a comprehensive screen that includes virtually all nonessential and essential genes and identify genes that are HI for genome stability, we used a hemizygous yeast deletion library in which one copy of each of the ∼6500 open reading frames was replaced with the KanMX gene (Winzeler et al., 1999; Giaever et al., 2002). Diploid yeast are typically nonmaters; however, loss of one copy of chromosome III or homozygosis at the MAT locus results in diploid maters (Herskowitz, 1988). We reasoned that mutants with elevated rates of chromosome loss or recombination should also have higher rates of mating. Therefore we screened for hemizygous mutants that mated with relatively high frequency to form triploids. By using Gene Ontology (GO) Slim Mapper for biological process terms, the screen identified genes required in cell cycle, chromosome structure and function, and other processes (Figure 1 and Supplemental Table S1). GO Term Finder analysis of the top 333 mutants, defined as those scoring ≥2 SDs above the mean, revealed significant enrichment for genes that encode for subunits of macromolecular complexes, the chaperonin-containing T complex (4.27 × 10−5), the cytoskeleton/spindle pole body (SPB; 1.3 × 10−4), and the large ribosomal subunit (4.25 × 10−3; Figure 1C, bottom). Applying GO Slim Mapper for macromolecular complexes terms revealed that ∼40% of the genes identified in the screen encode for subunits of the ribosome, spindle, kinetochore, DNA replication preinitiation, histone deacetylase, and ubiquitin ligase complexes and others (Supplemental Table S2). In addition, we identified genes that were not associated with a macromolecular complex term, such as the protein kinase A inhibitor BCY1, which we and others previously reported having a role in chromosome segregation (Li et al., 2005; Magtanong et al., 2011; Ma et al., 2012), as well as the small GTPase, IRA1, and a methionine-R-sulfoxide reductase, YKL069W. Although the results of the screen point to diverse processes encoded by haploinsufficient genes, it is not clear whether the effects on genome stability are direct or indirect. Nonetheless, the list of genes is a valuable resource in exploring a range of multisubunit complexes and functions, some of which are likely to directly affect genome stability.
FIGURE 1:
Genome-wide screen identifies genes that are HI for genome stability. (A) A hemizygous deletion library (∼6500 genes) was arrayed onto ∼16 plates (384 format; left). Each colony was pinned onto plates containing haploids and incubated overnight (right). Quadruplicate samples from each mating were pinned onto minimal media plates selecting for triploids (bottom). Scores from 0 to 4, depending on the growth of triploids, were assigned from each sample. (B) The mean score was 0.7, and the highest-scoring 333 mutants represent ≥2 SDs above the mean. (C) Bar graph showing a subset of results from GO Slim Mapper for biological process terms used to categorize the highest-scoring genes identified in the screen (≥2 SDs above the mean). (D) GO Term Finder analysis of the 333 mutants revealed enrichment for genes that encode subunits of the chaperonin-containing T complex, SPB/cytoskeleton, and large ribosomal subunit.
γ-TuSC and SPB haploinsufficiency leads to genome instability
As a test case for the value of the HI screen for dissecting the function of multisubunit complexes, we investigated the γ-TuSC and SPB genes. In a first step, we sought to validate the screen and ensure that the mutants are not specific to any defects related to the mating pathway. We constructed hemizygous deletions in a diploid reporter strain to quantitatively measure chromosome loss and recombination at chromosome V (Supplemental Figure S1A). We focused on the SPB, as this serves as a model to elucidate mechanisms underlying haploinsufficiencies often observed in multisubunit complexes. The SPB contains 18 subunits, all but two of which are essential (Jaspersen and Winey, 2004). Included within the SPB is the γ-TuSC, composed of three subunits, Spc97, Spc98, and Tub4 in a ratio of 1:1:2 (Kollman et al., 2010). The γ-TuSC nucleates the assembly of all microtubules (Kollman et al., 2011), yet we do not fully understand how γ-TuSC contributes to spindle organization and genome stability.
Our screen identified the γ-TuSC subunits and one of the inner SPB components encoded by SPC42 (Supplemental Table S1). We found higher chromosome loss rates in γ-TuSC hemizygous mutants spc97/+, spc98/+, and tub4/+ and the inner SPB subunit mutant spc42/+ compared with wild-type cells (Figure 2A), confirming that the SPB mutants displayed GI. Except for spc42/+ mutants, all strains displayed nearly wild-type levels of recombination (Figure 2B). We conclude that haploinsufficiency in the γ-TuSC subunits primarily affects chromosome segregation.
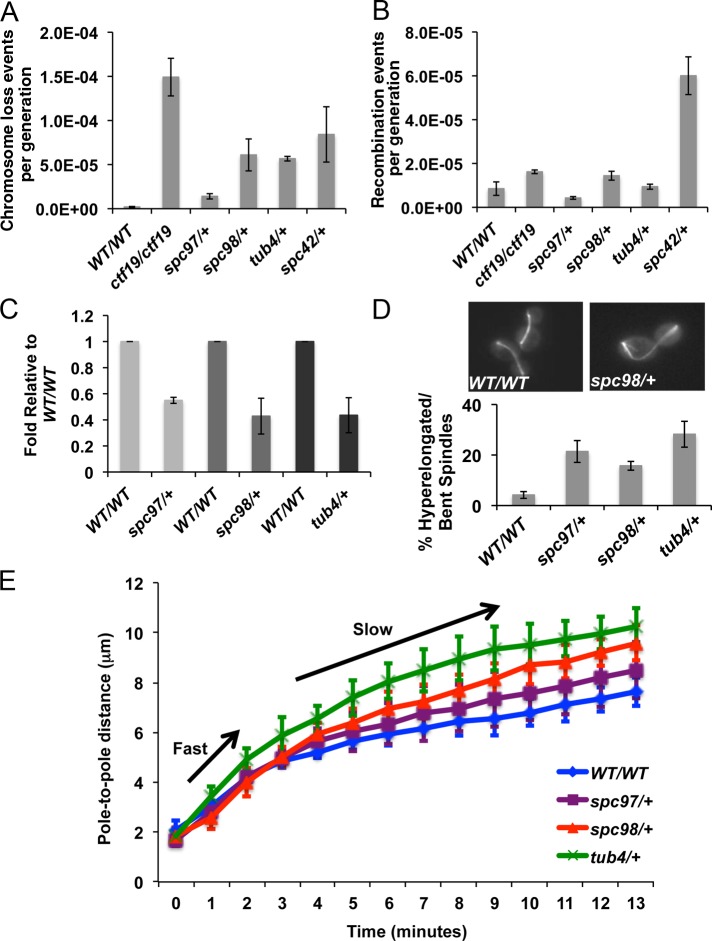
FIGURE 2:
γ-TuSC haploinsufficiency leads to higher rates of chromosome loss and altered anaphase spindle length and elongation velocity. (A, B) Chromosome loss and recombination rates were determined by fluctuation analysis for indicated strains (Supplemental Figure S1). ctf19/ctf19 has high chromosome loss rates and was used as a positive control (Hyland et al., 1999). Mean rates from three biological replicates are plotted; error bars represent SE. (C) Quantification of mRNA levels for γ-TuSC genes in asynchronously growing cells. Mean mRNA levels, normalized to wild type, from three biological replicates are plotted; error bars represent SE. (D) Asynchronously growing wild-type or mutant cells that express Tub1-GFP were examined. Mean number of anaphase cells with spindles that are abnormally long, bent, and/or have a fish-hook appearance, from three independent experiments; error bars represent SE. (E) Wild-type and spc98/+, spc97/+, and tub4/+ mutant cells expressing Tub1-GFP to label microtubules and Spc42-GFP to label spindle poles were imaged by live-cell confocal microscopy. Plotted are mean spindle lengths (pole-to-pole distance) as a function of time as cells transition from late metaphase through anaphase. Arrows indicate phases of fast and slow spindle elongation for wild-type (n = 8) and spc98/+ (n = 8), spc97/+(n = 6), and tub4/+ (n = 8) mutant cells.
Next, to confirm that hemizygous γ-TuSC mutants exhibited haploinsufficiency, we tested whether the γ-TuSC genes were expressed at 50% of wild-type levels. This control is important, since changes in gene copy number can lead to compensation in transcription to maintain gene dosage (Prestel et al., 2010). Such regulation is rare in yeast, however, as suggested in a study of ∼700 genes (Springer et al., 2010). We measured mRNA levels for each γ-TuSC subunit and observed no changes in expression of the wild-type copy, as the heterozygous spc97/+, spc98/+, and tub4/+ mutants expressed ∼50% of wild-type levels for SPC97, SPC98, and TUB4, respectively (Figure 2C). We conclude that the chromosome segregation defects in γ-TuSC mutants are a result of haploinsufficiency.
Hemizygous γ-TuSC mutants have altered spindle morphology during anaphase
Previous studies of haploid γ-TuSC mutants reported failure to develop normal spindles at nonpermissive temperatures (Geissler et al., 1996; Knop et al., 1997; Vogel and Snyder, 2000; Vogel et al., 2001; Keck et al., 2011; Lin et al., 2011). Thus we investigated whether γ-TuSC hemizygous mutants might also have spindle defects. We observed that a higher proportion (∼16–25%) of spc97/+, spc98/+, and tub4/+ mutants displayed hyperelongated spindles, some with a “fish-hook” appearance, compared with wild-type cells (∼5%; Figure 2D). These results prompted us to determine whether γ-TuSC mutants have altered spindle elongation kinetics. We constructed strains in which the pole-to-pole distance as a function of time could be measured using live-cell fluorescence microscopy. In each of the γ-TuSC mutants, the elongation velocity was nearly identical to wild type during the early fast phase of anaphase, but rates of elongation during the slow phase were increased compared with wild-type cells. In early anaphase, the rates for spc98/+, tub4/+, and spc97/+ were similar to those of the wild-type strain (1.23 ± 0.083 [n = 8], 1.299 ± 0.115 [n = 8], 1.24 ± 0.108 [n = 6], and 1.17 ± 0.075 μm/min [n = 8], respectively; all mutants compared with wild type had p ≈ 0.6). During the slower phase of anaphase, however, spc98/+, tub4/+, and spc97/+ mutants elongated their spindles at a markedly greater rate than wild-type cells (0.362 ± 0.018 [n = 8] vs. wild type, p < 0.0001; 0.383 ± 0.0401 [n = 8] vs. wild type, p = 0.009; 0.311 ± 0.0292 [n = 6] vs. wild type, p = 0.0078; and 0.222 ± 0.011 μm/min [n = 8], respectively). These findings suggest that the hyperelongated spindles in γ-TuSC mutants are associated with increased spindle elongation rates.
Interpolar MTs, which emanate from opposite poles, overlap, and interdigitate at the midzone, facilitate spindle elongation during anaphase (Winey et al., 1995; Goshima and Scholey, 2010). Changes in the length distribution of iMTs affect the length of spindles and, in some cases, the rate of spindle elongation (Goshima and Scholey, 2010; Avunie-Masala et al., 2011). As a first approximation of the length distribution of iMTs, we measured the levels of Tub1–green fluorescent protein (GFP) in metaphase and anaphase spindles in wild-type and spc98/+ mutants. During metaphase, spc98/+ mutants had a distribution of Tub1-GFP that was similar to that of wild-type cells (Supplemental Figure S1B). In contrast, anaphase spindles in spc98/+ mutants contained less Tub1-GFP near the spindle poles and more at the midzone than wild-type cells, suggesting that the length distribution of spindle microtubules during anaphase is disrupted in spc98/+ mutants (Figure 3A). To determine whether the increase in Tub1-GFP at the midzone might be a result of greater polymerization of microtubules in spc98/+ mutants compared with wild-type cells, we performed fluorescence recovery after photobleaching (FRAP) experiments. This allowed us to determine the exchange of Tub1-GFP and assembly at the region where photobleaching occurs. Because we were most interested in iMT assembly at the midzone, our experimental design was similar to FRAP experiments reported by Maddox et al. (2000), in which we photobleached mid-anaphase spindles at the mother–daughter bud neck region. We analyzed only anaphase spindles in spc98/+ mutants and wild-type cells that started with a length of 5.3–6.2 μm and ended with a length of 7.9–8.2 μm (Figure 4E). By doing so, we could ensure that the measurements were being made on comparable anaphase spindles. We observed that fluorescence was nearly fully recovered by ∼210 s in spc98/+ mutants (Figure 4E). Near-complete recovery in wild-type cells, however, was not reached until ∼270 s (Figure 4E). Taken together, these results indicate that spc98/+ mutants have greater iMT assembly compared with wild-type cells.
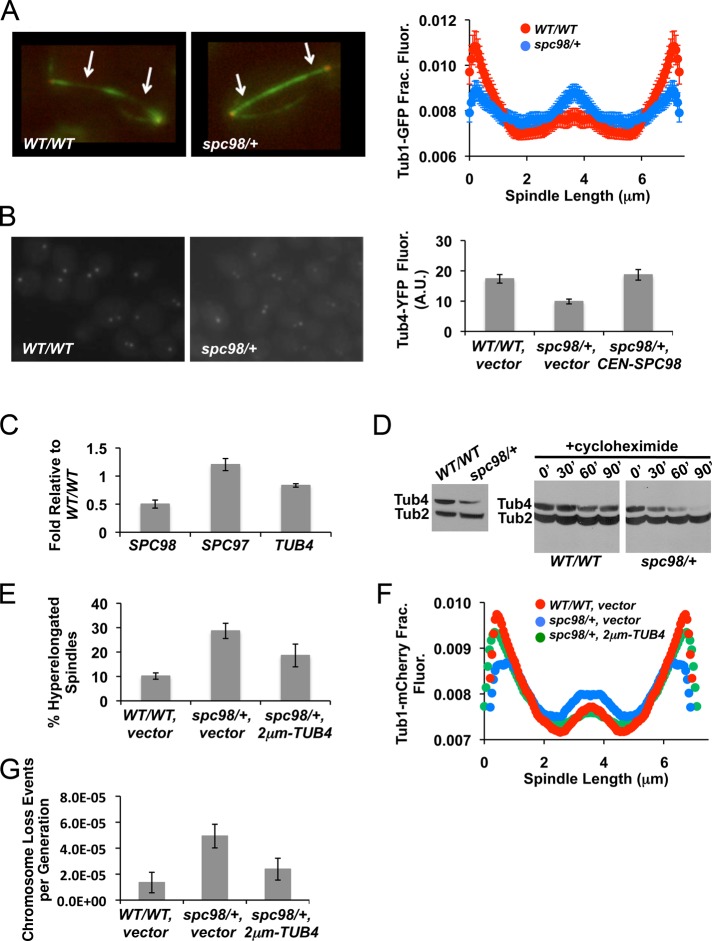
FIGURE 3:
γ-Tubulin levels are crucial for anaphase spindle organization and accurate chromosome segregation. (A) Wild-type and spc98/+ mutant cells expressing Tub1-GFP were imaged by TIRF microscopy. Levels of Tub1-GFP were measured across the length of anaphase spindles. Mean intensity values normalized to the total fluorescence from each strain; error bars represent SE. The differences in Tub1-GFP distribution between wild type and spc98/+ are significant, as determined by the Kolmogorov–Smirnov test; p = 0.0011. Arrows point to regions flanking the midzone. (B) Metaphase wild type, spc98/+ mutant cells, and spc98/+ mutant cells carrying a low-copy (CEN) plasmid of SPC98 expressing Tub4-YFP were imaged, and total YFP fluorescence was quantified using MetaMorph. Mean fluorescence from ∼30 cells; error bars represent SE. (C) The levels of mRNA were measured using reverse transcription-PCR. The average amount of each transcript in spc98/+ mutants relative to wild-type cells in three independent replicates; error bars represent SE. (D) Western blot analysis for total Tub4-HA levels (left) and rate of turnover (right) in asynchronously growing cells or during cycloheximide treatment at various time points (in minutes) in wild type and spc98/+ mutants, respectively. Tub2 was used as a loading control. Two independent replicates were performed with similar results. (E) Asynchronously growing spc98/+ mutant cells that expressed Tub1-GFP with vector (2 μm) or a high-copy plasmid containing TUB4 (2 μm-TUB4) were imaged by fluorescence microscopy. Mean number of anaphase cells with spindles that are bent or have a fish-hook appearance from three independent experiments; error bars represent SE. (F) Levels of Tub1-GFP were measured across anaphase spindles from wild type with vector, spc98/+ with vector, or spc98/+ with 2 μm-TUB4. Mean intensity values normalized to the total fluorescence for each strain; error bars represent SE. Based on the Kolmogorov–Smirnov test, Tub1-GFP distributions in spc98/+ carrying 2 μm-TUB4 are the same as those of wild type with vector (p = 0.025), but spc98/+ carrying vector is significantly different from wild type with vector (p = 0.00014). (G) spc98/+ cells carrying 2 μm-TUB4 or vector control were assayed for chromosome loss as described in Figure 2. Mean chromosome loss rate from three independent replicates; error bars represent SE.
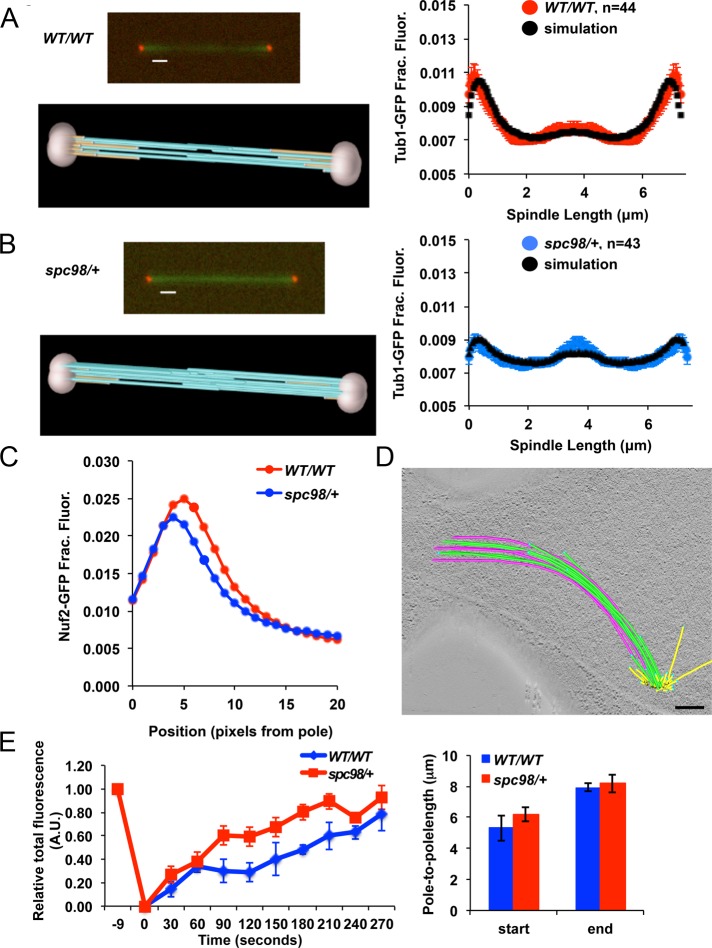
FIGURE 4:
Model convolution simulation, fluorescence microscopy, electron tomography, and FRAP reveal increased altered iMT and kMT assembly in spc98/+ mutants. (A, B) Model convolution simulation of Tub1-GFP distributions for wild-type and spc98/+ mutant cells. The simulations predict fewer kMTs (gold) and more iMTs (light blue) in spc98/+ mutants than in wild-type cells. (C) The distance between KT clusters and SPBs was measured in anaphase wild-type (red curve) and spc98/+ mutant cells expressing Spc110-mCherry to mark the spindle poles, Tub1-GFP to label microtubules, and Nuf2-GFP to mark kinetochores (blue curve). The plot indicates that spc98/+ mutants contain shorter kMTs than do wild-type cells; n = 42 for spc98/+ mutants, and n = 57 for wild-type cells. Average spindle length is 7.4 ± 1.5 μm in wild-type cells (mean ± SD) and 7.8 ± 2.3 μm in spc98/+ mutants. (D) Electron tomography revealed that spc98/+ spindles contained abnormal SPB positioning and spindle curvature (Figures 2D and 3A). More iMTs (green and pink) are present and have a much longer region of interdigitation than is typically found in wild-type cells. See also Supplemental Figure S2 and Supplemental Videos S1–S3. Bar, 200 nm. (E) FRAP (left) was performed on mid-anaphase spindles in spc98/+ mutant (n = 7) and wild-type cells (n = 4). A rectangular region (1.2 × 0.6 μm) over the mother–bud neck was photobleached, and the rate at which fluorescence was recovered normalized to total spindle fluorescence is plotted. In both strains the starting and ending spindle lengths are comparable (right).
Hemizygous γ-TuSC mutants have reduced level of Tub4 at the SPB
Structural studies show that the γ-TuSC forms a 1:1:2 stoichiometric complex of Spc98-Spc97-Tub4, respectively (Kollman et al., 2010). Erlemann et al. (2012) reported a slightly different stoichiometry of γ-TuSC subunits in vivo, suggesting that additional molecules of Tub4 and Spc98 are present. Considering that the stoichiometry between γ-TuSC subunits might be altered in spc98/+mutants, we hypothesized that altered length distributions of anaphase spindle microtubules may be due to defective γ-TuSC structures assembling at the SPB. We quantified the level of Tub4–yellow fluorescent protein (YFP) associated with the SPB and observed a reduction of ∼40% in spc98/+ mutants compared with wild-type cells (Figure 3B). Transformation of spc98/+ mutants with a centromeric plasmid expressing SPC98 restored Tub4-YFP at SPB to wild-type levels. This shows that the lower level of Tub4-YFP is due to decreased gene dosage of SPC98 (Figure 3B). The levels of TUB4 mRNA were nearly identical between wild-type and spc98/+ mutant cells, demonstrating that TUB4 transcription is unchanged in spc98/+ mutants (Figure 3C). Western blot analysis showed lower levels of Tub4–hemagglutinin (HA) in spc98/+ mutants than in wild-type cells (Figure 3D, left). It is possible that excess Tub4 not bound to Spc98 in spc98/+ mutants is unstable, as previously reported for other proteins (Chen and Archer, 2005). Hence, we compared the rate at which Tub4-HA is turned over in spc98/+ mutants to that in wild-type cells. In spc98/+ mutants we observed a marked decrease in Tub4-HA levels after 30 min of cycloheximide treatment, and it was virtually undetected by 90 min (Figure 3D, right). In contrast, Tub4-HA levels remained relatively unchanged in wild-type cells (Figure 3D, right). Therefore we conclude that Tub4-HA is more rapidly turned over in spc98/+ mutants than in wild-type cells and this contributes to reduced levels of Tub4 at the SPB.
Increased TUB4 gene dosage suppresses spindle defects and chromosome loss in spc98/+ mutants
Given that γ-tubulin nucleates microtubule assembly (Oakley et al., 1990; Felix et al., 1994; Stearns and Kirschner, 1994; Zheng et al., 1995) and that we observed reduced Tub4 levels at the SPB in spc98/+ mutants, we sought to determine whether high-copy TUB4 could suppress the spindle defects observed in spc98/+ mutants. Compared with spc98/+, the percentage of spc98/+ cells with high-copy TUB4 displaying hyperelongated spindles was lower (Figure 3E). We also observed suppression of the altered Tub1-GFP distribution in spc98/+ mutants when TUB4 was overexpressed (Figure 3F). We next examined whether high-copy TUB4 suppressed the chromosome loss phenotype of spc98/+ strains and found that, consistent with a role for Tub4 in spc98/+ haploinsufficiency, TUB4 overexpression returned the chromosome loss rate of spc98/+ mutants to near wild-type levels (Figure 3G). Conversely, deleting one copy of TUB4 in spc98/+ enhanced chromosome loss rates compared with either spc98/+ or tub4/+ single mutants (Supplemental Figure S1D).
Model convolution simulation and electron tomography reveal increases in iMTs and rapid shortening of kMTs in spc98/+ mutants
We used model convolution simulation to identify spindle defects that account for the altered Tub1-GFP distribution in spc98/+ mutants. This strategy has been used to evaluate iMT length distributions in yeast anaphase spindles (Gardner et al., 2008). In budding yeast, a “core bundle” of polar microtubules begins to assemble in preanaphase spindles (Winey et al., 1995). The core bundle forms the central spindle and is believed to aid in SPB separation and form the future iMT bundle (Winey et al., 1995). In anaphase, short microtubules near the SPB are typically associated with kMTs, whereas iMTs are present at the midzone and cross-linked by microtubule-binding proteins to push the spindle poles apart (Goshima and Scholey, 2010). Model convolution simulation of anaphase cells with similar spindle lengths predicted that spc98/+ mutants have an average decrease in kMT length and density, with a corresponding increase in average length and density of longer iMTs, compared with wild-type cells (Figure 4, A and B, and Supplemental Figure S1C).
The marked decrease in kMT-associated fluorescence near the spindle poles for spc98/+ mutants suggests that kMTs disassemble faster in spc98/+ mutants than in wild-type cells. We determined the position of the kinetochores with respect to the spindle poles during anaphase in spc98/+ and wild-type cells with spindle lengths measuring ∼7 μm, the stage when model convolution simulation revealed a difference in the amounts of kMTs and iMTs in spc98/+ and wild-type cells (Figure 4, A and B). The positions of kinetochores were marked by Nuf2-GFP, and SPBs were labeled with Spc110-mCherry, a stable spindle pole subunit (Jaspersen and Winey, 2004). Nuf2-GFP was found to be ∼25% farther from the spindle pole (∼85 nm) in wild-type spindles than in the spc98/+ mutants (Figure 4C). This suggests that kMTs shorten more rapidly in spc98/+ anaphase cells than in wild-type cells, since measurements were taken from spindles of similar length, which is consistent with the observed decrease in Tub1-GFP fluorescence near the poles in spc98/+ mutants (Figure 3, A and F).
We next used electron tomography to image the three-dimensional (3D) arrangement of preanaphase and anaphase spindles. Tomograms of spc98/+ spindles (Figure 4D, Supplemental Figure S3, and Supplemental Videos S1–S3) revealed altered spindle organization consistent with that observed by fluorescence microscopy of Tub1-GFP (Figures 2D and 3A). Diploid wild-type anaphase spindles contain three or four iMTs from each pole that interdigitate in the midzone and ∼32 short kinetochore microtubules at each pole (Supplemental Figure S2). In contrast, spc98/+ mutants in anaphase displayed an increase in number of iMTs and length of iMT interdigitation (Figure 4D and Supplemental Video S1). Analysis of preanaphase spc98/+ spindles showed that most (Supplemental Figure S3, spc98/+ cells 1–3 and 5) contained an altered core bundle, and three displayed hyperelongated off-axis spindle MTs (Supplemental Figure S3, spc98/+ cells 1, 2, and 5). In addition, abnormal SPB positioning was observed, and these short spindles often contained SPBs in nuclear envelope invaginations (Supplemental Figure S3, spc98/+ cells 2, 3, and 5, and Supplemental Video S3), indicating a possible defect in anaphase completion.
DISCUSSION
In this study, we sought to identify genes that encode subunits of multimeric complexes to better understand how altered gene dosage affects biological activities with consequences on genomic stability. To this end, we screened a library of ∼6500 hemizygous mutants for haploinsufficiency and observed enrichment for genes encoding subunits of macromolecular complexes, including the spindle pole body. We chose to further investigate the genes encoding the γ-TuSC, one of the spindle pole body subcomplexes that are evolutionarily conserved. The γ-tubulin complexes were first reported to be microtubule-nucleating structures in the 1990s (Zheng et al., 1995; Martin et al., 1998). We do not fully understand, however, how γ-TuSCs contribute to regulating the length and number of iMTs and kMTs and the rate of spindle elongation (i.e., pole-to-pole separation) so that a robust and stereotyped spindle structure assembles in every cell division. Our studies of hemizygous γ-TuSC mutants suggest that this is achieved in part by an antagonism between assembly of iMTs and kMTs. Imbalances such as those conferred by changes in γ-TuSC nucleation activity can lead to greater iMT assembly and result in kMT instability. This is an unexpected finding and provides the first evidence to show how γ-TuSC plays a crucial role in balancing the assembly dynamics between kMTs and iMTs so that a mitotic spindle of the correct size and composition is formed to maintain genome stability. To our knowledge, these observations are the first to show that haploinsufficiency of γ-TuSC alters assembly of iMTs and kMTs and causes chromosome missegregation. Furthermore, previous studies of conditional loss-of-function alleles of γ-TuSC did not reveal phenotypes of faster spindle elongation, increased iMT assembly, and rapid degradation of Tub4 in spc98/+ mutants. This underscores how studying haploinsufficiency may yield novel insights into fundamental biological processes that may not be uncovered in studies with haploid alleles.
Genes identified in our screen suggest that a variety of processes can affect genome stability either directly or indirectly. Using GO Slim Mapper, we found that the top 333 genes represented many GO Process categories and, further, that ∼40% encode for proteins that are part of known macromolecular complexes. The vast majority of genes within our list can be considered preliminary, since they still require validation, such as by secondary genome stability assays. The results with the γ-TuSC genes, however, indicate that our screen can provide a valuable beginning in exploring HI-based mechanisms of GI. Further validation of the genes will be important in developing our understanding of general HI mechanisms, a topic with relevance to basic and disease mechanisms. Indeed, there have been reports of different biological defects associated with haploinsufficiency in single or multiple genes, yet the underlying mechanism in virtually all cases is not well understood. Our work in dissecting the mechanism(s) that cause the spc98/+ haploinsufficiency provides new insight into this question. Notably, our observations suggest that one of the underlying causes for the haploinsufficiency in spc98/+ mutants is decreased levels of Tub4, the yeast γ-tubulin, at the spindle pole bodies. Tub4 is the γ-TuSC subunit that physically interacts with α/β tubulin subunits to mediate microtubule polymerization (Oakley and Oakley, 1989; Oakley et al., 1990). Using fluorescence microscopy, we observed diminished levels of Tub4 at the spindle pole body and by Western analysis an overall reduction in Tub4 in spc98/+ mutants. TUB4 mRNA levels were virtually identical between spc98/+ mutant and wild-type cells, ruling out an effect of transcription. Cycloheximide experiments to measure the turnover rate of Tub4, however, revealed that Tub4 is more rapidly degraded in spc98/+ mutants than in wild-type cells. We demonstrated that the decrease in levels of Tub4 may be the underlying cause for several of the defects observed in spc98/+ mutants by showing that high-copy TUB4 suppressed the spindle and chromosome segregation defects. These observations indicate that a threshold of Tub4 is crucial for proper regulation of spindle length and accurate chromosome segregation when only one copy of SPC98 is present. We propose that a single copy of SPC98 leads to higher rates of Tub4 degradation, which is analogous to other protein complexes, where loss in one subunit leads to unbound partner subunits that are more rapidly degraded (Chen and Archer, 2005). Taken together, our results provide new insights into mechanisms of haploinsufficiency that may apply to other complexes and systems.
It has long been suggested that microtubule motors and microtubule-associated proteins that cross-link microtubules regulate spindle length (Goshima and Scholey, 2010). In this article we show how the gene dosage of γ-TuSC is important in determining spindle length and organization. We propose that a critical threshold of γ-TuSC nucleation activity is required to balance the assembly of iMTs to kMTs so the correct number and length of each type of MT is formed. Using a variety of molecular biology, fluorescence microscopy, and electron tomography experiments, we found several defects in the anaphase spindle that are consistent with increased iMT assembly when the gene dosage of γ-TuSC subunits is reduced. By total internal reflection fluorescence (TIRF) microscopy and model convolution simulation, we observed an increase in midzone fluorescence of Tub1-GFP in spc98/+ anaphase spindles compared with wild-type cells, suggesting that there are more iMTs or there is a denser region of interdigitated iMTs. Electron tomography of several spindles in preanaphase (∼1.5–2 μm) is consistent with greater overlap, since several spc98/+ mutant spindles contain MTs that are much longer than the length of the spindle. We envision that these MTs may potentially form the iMTs, and thus, as the spindle elongates, there will inevitably be a larger region of iMT overlap. A longer region of overlap can facilitate longer pole-to-pole separation (Saunders et al., 1995, 1997) leading to the observed increase in spindle elongation velocity during the slow phase of anaphase in γ-TuSC mutants and the more frequent appearance of hyperelongated “fish-hook” spindles. Furthermore, FRAP experiments revealed that spc98/+ mutant anaphase spindles have faster Tub1-GFP exchange, consistent with faster iMT assembly, compared with wild-type cells. Moreover, measurements of kMT lengths (i.e., Nuf2-to-SPB distance) and density by TIRF microscopy (i.e., Tub1-GFP near spindle poles) and model-convolution simulations are consistent with more rapid anaphase shortening of kMTs in spc98/+ mutants compared with wild-type cells. These results are consistent with antagonism between iMT and kMT assembly. We also observed a reduction in Tub4-YFP levels at the spindle pole bodies in spc98/+ mutants and suppression of spc98/+ mutant phenotypes by overexpression of TUB4. Previously, Tub4 was shown to interact with α/β tubulin subunits in mediating MT polymerization (Oakley and Oakley, 1989; Oakley et al., 1990). We suggest that the changes in Tub4 levels might result in less stable γ-TuSC sites that affect nucleation activity. A recent report showed that the in vivo stoichiometry of γ-TuSCs at the spindle pole body appears to differ from that observed in in vitro studies by having additional molecules of Tub4 and Spc98 (Erlemann et al., 2012). The authors proposed that the excess Tub4 and Spc98 may act to stabilize the γ-TuSC ring or help to position the MT seam. We propose that the spindle defects in spc98/+ mutants might reflect a reduction in the additional Tub4 molecules that provide stability to the γ-TuSC and affect the assembly of MTs. This is consistent with the suppression of the spindle and chromosome segregation defects in spc98/+ mutants by high-copy TUB4. Thus in the γ-TuSC mutants the limiting γ-TuSC nucleation activity leads to greater iMT assembly, which then compromises the stability of kMTs, possibly due to limited pools of free tubulin subunits. In turn, more rapid depolymerization of kMTs during anaphase could result in premature kinetochore detachment of one sister before they are fully separated (i.e., before cohesin is fully dissociated and sisters are free), which could result in chromosome missegregation. This would be consistent with the observed elevated chromosome loss rate in spc98/+ mutants. Taken together, our results provide a previously undefined role for γ-TuSC in determining the stability of iMTs relative to kMTs during anaphase to maintain genome stability.
Although the role of γ-TuSC haploinsufficiency in the development of human disease remains to be determined, the conservation between yeast and humans suggests that orthologous genes in humans may be HI for genome stability. Indeed, a search of the Sanger Human Cancer Genome Project Copy Number Analysis in Cancer database revealed that each of the human orthologues of the γ-TuSC genes undergoes loss of heterozygosity (LOH). Most strikingly, in ∼50% of breast, lung, and esophageal cancer cell lines, there was LOH for TUBGCP3, the human orthologue of SPC98. Moreover, a predictive algorithm developed by Huang et al. (2010) also finds TUBGCP3 and TUBG2, a human γ-tubulin orthologue, to be probable HI genes. Future studies with genes identified in our screen will provide insights into orthologous pathways that contribute to GI in human cells and potentially uncover novel mechanisms that are vulnerable to gene dosage defects.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast media and techniques were performed as described (Guthrie and Fink, 1991). Yeast strains were derived from BY4741 or W303 as indicated (Supplemental Table S2). Hemizygous deletions and epitope tag fusions were constructed by directly transforming diploids (YMB6541) with a kanMX or natMX cassette as described (Longtine et al., 1998). Transformants were confirmed by colony PCR using an upstream primer from the 5′ untranslated region of the gene of interest and a primer within the G418MX or NATMX cassettes. Plasmids pRS316-SPC98 and 2 μm-TUB4 were gifts from the Trisha Davis (University of Washington, Seattle, WA) and Charlie Boone (University of Toronto, Toronto, Canada) laboratories, respectively.
HI screen of hemizygous deletion library
The S. cerevisiae hemizygous deletion library (∼6500 strains) was purchased from Open Biosystems (Huntsville, AL; YSC1055). The strains were arrayed onto solid media (yeast extract/peptone/dextrose [YPD]) with a VersArray Colony Arrayer (Bio-Rad, Hercules, CA) robotic arm and allowed to grow for 2–3 d at 30°C. To qualitatively measure mating proficiency, we used a high-throughput approach in which each of the ∼6500 hemizygous diploids was mated to MATa or MATα haploids. Four samples from each mating reaction were pinned onto plates that would support only the growth of triploid cells. Depending on the number of replicates that showed growth of triploids, which was tested in quadruplicate, we assigned a score from 0 (no growth in any of the four replicates) to 4 (growth observed in all four replicates; Figure 1A). The screen was performed in triplicate and the average distribution of scores plotted.
Chromosome loss and recombination assays
A diploid strain (YMB6541) was constructed in which one homologue of chromosome V carries a deletion in CAN1 and a replacement of HOM3 with LEU2, allowing us to select for diploids with both homologues before measurement of chromosome loss. A similar assay was previously described (Hartwell and Smith, 1985) in which one copy of chromosome V carries mutations in CAN1 and HOM3. Loss of one homologue confers canavanine resistance and threonine auxotrophy. On the other hand, recombination or mutation at the CAN1 locus results in diploids that are canavanine resistant but prototrophic for threonine. Diploids were grown in medium lacking threonine and leucine (SC-Thr-Leu) to select for cells carrying both copies of chromosome V, and three colonies were isolated and streaked out on YPD plates to measure chromosome loss rates. Typically, nine colonies from each original streak were assayed for canavanine resistance and threonine prototrophy. Each colony was resuspended in 500 μl to 1 ml of sterile water, and 25 μl was spread onto SC-R+Can (60 μg/ml). Another 25 μl of a 1:5000 dilution was spread onto YPD plates to measure how many cells were assayed. Colonies grown on SC+Can were replica plated onto SC-Thr medium. The number of colonies on each of the three plates was then used to measure chromosome loss rates using the method of Lea and Coulson (Lea and Coulson, 1949; Zheng et al., 2011). For chromosome loss rates of diploids with plasmids we used medium selective for the plasmid rather than YPD.
Spindle morphology and elongation rates
Wild type or hemizygous mutants containing Tub1-GFP were grown asynchronously for 4–5 h at 30°C, fixed with 3.7% formaldehyde for 10 min, washed once with 80% ethanol, and resuspended in 1× phosphate-buffered saline. Cells were examined using an upright Zeiss Axioskop microscope (Carl Zeiss, Jena, Germany) with GFP filter sets. Only large-budded cells that had entered anaphase with long spindles were counted. For measuring spindle elongation rates, diploids carrying both Tub1-GFP and Spc42-GFP were used. Approximately 2 μl of cell suspension was placed into a slide chamber, and an agarose slab containing 1% agarose and synthetic media was placed over the cells. A Zeiss LSM510 confocal microscope (Carl Zeiss, Jena, Germany) with a 100× objective/numerical aperture (NA) ∼1.4 was used to obtain 12-bit Z-stacks comprising 5 × 0.4–μm slices every minute for 1 h. Projections from each time frame were constructed and combined into individual movies using ImageJ software (National Institutes of Health, Bethesda, MD). The length of the spindle (distance from each Spc42-GFP focus) was measured for each time interval and plotted using Excel (Microsoft, Redmond, WA).
Quantification of Tub4-YFP at the spindle pole body
Diploids carrying one copy of Tub4-YFP were grown at 30°C for 3 h before imaging at room temperature. The same slide chambers and cell volume were used as those described for the spindle elongation experiments. To ensure collection of the total fluorescence from every cell, the Z-stack for each field of cells was imaged such that the entire depth of the cells would be included. A DeltaVision microscope workstation (Applied Precision, Issaquah, WA) was used with 100× objective/NA ∼1.4. MetaMorph software (Molecular Devices, Sunnyvale, CA) was used to construct projections from each set of Z-stacks, and the maximum fluorescence for each YFP signal was quantified. YFP signals were collected only from cells with short spindles, indicated by duplicated YFP signals that were ∼2 μm apart.
RNA and protein analysis
Total RNA was extracted from asynchronously growing cells using hot phenol (Basrai et al., 1999). Approximately 1 μg of total RNA was treated with DNase I in 20-μl reaction volume. Reverse transcription-PCR was performed using the Access RT-PCR System (Promega, Madison, WI). Reactions were then separated on 1.4% agarose gel electrophoresis, stained with ethidium bromide, and imaged. Images of DNA products were quantified using GeneSnap software (Syngene, Frederick, MD). Primer sequences are available upon request. Western blot analysis to determine the turnover of Tub4-HA after cycloheximide treatment was performed as described previously (Au et al., 2008). Total protein was extracted from asynchronously growing cells to measure total Tub4-HA using the trichloroacetic acid method (Mishra et al., 2011). Protein samples were separated by 4–12% PAGE and transferred to nitrocellulose membranes. Blots were probed with monoclonal anti-HA (clone 12CA5; Roche, Indianapolis, IN) or polyclonal anti-Tub2 (custom made; Covance, Berkeley, CA), followed by standard chemiluminescence.
TIRF imaging and model convolution simulation
Live-cell imaging of fluorescently labeled strains was performed using a Nikon Ti-E inverted microscope with perfect focus (Nikon, Melville, NY). The microscope is equipped with a 100× TIRF objective combined with a 2.5× projection lens and an iXon EMCCD camera (final pixel size, 64 × 64 nm; Andor, Belfast, United Kingdom). Automation of the microscope and digital image acquisition was performed using Nikon Elements software. Cells were prepared for imaging by resuspending them from a liquid culture into water and 4% glucose and then flowing them into a flow chamber preincubated with concanavalin A to allow cells to adhere to the coverslip. After incubation for ∼15 min, nonadhered cells were removed from the flow cells via vacuum, and fresh water/glucose was introduced into the flow chamber for imaging. Imaging was performed using 488- and 561-nm TIRF lasers, with laser angles adjusted to a “pseudo-TIRF” position to allow for imaging depths of 500–1000 nm. Images were taken in a single focus plane, and spindles in which both poles were clearly in focus were selected for analysis. Image analysis was performed using ImageJ and custom-made Matlab scripts (MathWorks, Natick, MA). Model convolution simulation was performed as previously described (Gardner et al., 2008).
FRAP
FRAP experiments were carried out on a Zeiss 510 confocal microscope with a 100×/numerical aperture 1.3 oil immersion objective. Cells were imaged with a 488-nm laser line from a 40-mW argon laser at low laser intensity (0.6%) to reduce bleaching due to imaging. A rectangular region of 1.2 × 0.6 μm was photobleached in the middle of the spindles of defined length (5.3–6.2 to 7.9–8.2 μm) with a short (32 ms) laser pulse operating at 40% of the laser power. Fluorescence recovery was monitored at 30-s time intervals. We collected Z-stacks of three focal planes with 500 nm step size before and after the photobleach and measured the intensity from maximal intensity projections of the complete 3D stack onto a single plane. Image background was subtracted from those measurements. Intensity of the bleached area was divided by the intensity for the whole spindle in the same cell to correct for bleaching due to imaging. The resulting curve was normalized to the prebleach level of brightness. Normalized curves from the individual cells were averaged.
Preparation of cells for electron microscopy
Strains were prepared for electron microscopy as previously described (Giddings et al., 2001). Briefly, aliquots from logarithmic-phase cultures were prepared by high-pressure freezing using a Wohlwend Compact 02 HPF (Sennwald, Switzerland). The frozen samples were then freeze substituted in 0.25% glutaraldehyde and 0.1% uranyl acetate in acetone, followed by embedding in Lowicryl HM20. Serial sections (250 nm) were collected onto Formvar-coated copper slot grids and poststained using 2% aqueous uranyl acetate, followed by Reynold's lead citrate. Colloidal gold particles (15 nm) were affixed to both sides of the grids to serve as alignment markers.
Electron tomography
Tomography was performed using a Tecnai F20 or F30 electron microscope (FEI, Hillsboro, OR) operating at 200 or 300 kV as described (O'Toole et al., 1999, 2002). The SerialEM program (Mastronarde, 2005) was used to automatically acquire images every 1 deg over a ±60-deg range using a Gatan (Pleasanton, CA) 2K × 2K charge-coupled device camera at a pixel size of 1–1.5 nm. Tomograms from two to six serial sections were calculated using the IMOD software package (Mastronarde, 1997) and joined to produce a final volume containing mitotic spindles. We recorded three wild-type and eight spc98/+ mitotic spindles.
Modeling and analysis of tomographic data
Tomograms were displayed and analyzed using the IMOD program 3dmod (Kremer et al., 1996). Spindle microtubules originating from each pole were modeled as green or purple lines, respectively. In anaphase cells, the iMTs were modeled in green lines, and the kMTs were modeled as yellow lines. Blue spheres represented the positions of microtubule plus ends. The projections of the 3D models were displayed and rotated to study their 3D geometry.
Supplementary Material
Acknowledgments
We thank Tom Giddings and Christina Clarissa for electron microscopy specimen preparation; Janet Meehl-Fox for yeast cultures; Charlie Boone, Duncan Clarke, Trisha Davis, and Manuel Mendoza for reagents; Jackie Vogel for critical reading of the manuscript and sharing unpublished results; Hannah Klein for advice on fluctuation analysis; and Basrai laboratory members for discussions. E.O.T. is supported by Grant 8P41GM103431 from the National Institutes of General Medical Sciences to Andreas Hoenger (Department of Molecular, Cellular and Developmental Biology, University of Colorado–Boulder). M.K.G. and B.M.S. are supported by a grant from the Pew Scholars Program in the Biomedical Sciences and National Institutes of Health Grant GM-100122. M.W. is supported by National Institutes of Health Grant GM51312. J.S.C. and M.A.B. are supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health.
Abbreviations used:
- GI
genome instability
- γ-TuSC
gamma-tubulin small complex
- HI
halploinsufficient
- iMT
interpolar microtubule
- kMT
kinetochore microtubule
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-12-0902) on July 3, 2013.
REFERENCES
- Au WC, Crisp MJ, DeLuca SZ, Rando OJ, Basrai MA. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics. 2008;179:263–275. doi: 10.1534/genetics.108.088518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avunie-Masala R, Movshovich N, Nissenkorn Y, Gerson-Gurwitz A, Fridman V, Koivomagi M, Loog M, Hoyt MA, Zaritsky A, Gheber L. Phospho-regulation of kinesin-5 during anaphase spindle elongation. J Cell Sci. 2011;124:873–878. doi: 10.1242/jcs.077396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai MA, Velculescu VE, Kinzler KW, Hieter P. NORF5/HUG1 is a component of the MEC1-mediated checkpoint response to DNA damage and replication arrest in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7041–7049. doi: 10.1128/mcb.19.10.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Chen J, Archer TK. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol Cell Biol. 2005;25:9016–9027. doi: 10.1128/MCB.25.20.9016-9027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Clare M, Oliver SG. Copy-number variation of cancer-gene orthologs is sufficient to induce cancer-like symptoms in Saccharomyces cerevisiae. BMC Biol. 2013;11:24. doi: 10.1186/1741-7007-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delneri D, et al. Identification and characterization of high-flux-control genes of yeast through competition analyses in continuous cultures. Nat Genet. 2008;40:113–117. doi: 10.1038/ng.2007.49. [DOI] [PubMed] [Google Scholar]
- Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlemann S, Neuner A, Gombos L, Gibeaux R, Antony C, Schiebel E. An extended gamma-tubulin ring functions as a stable platform in microtubule nucleation. J Cell Biol. 2012;197:59–74. doi: 10.1083/jcb.201111123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA, Antony C, Wright M, Maro B. Centrosome assembly in vitro: role of gamma-tubulin recruitment in Xenopus sperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, et al. The microtubule-based motor Kar3 and plus end-binding protein Bim1 provide structural support for the anaphase spindle. J Cell Biol. 2008;180:91–100. doi: 10.1083/jcb.200710164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Giddings TH, Jr, O'Toole ET, Morphew M, Mastronarde DN, McIntosh JR, Winey M. Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 2001;67:27–42. doi: 10.1016/s0091-679x(01)67003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. Control of mitotic spindle length. Annu Rev Cell Dev Biol. 2010;26:21–57. doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- Greenman CD. Cancer. Haploinsufficient gene selection in cancer. Science. 2012;337:47–48. doi: 10.1126/science.1224806. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:3–933. [PubMed] [Google Scholar]
- Haarer B, Aggeli D, Viggiano S, Burke DJ, Amberg DC. Novel interactions between actin and the proteasome revealed by complex haploinsufficiency. PLoS Genet. 2011;7:e1002288. doi: 10.1371/journal.pgen.1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B, Viggiano S, Hibbs MA, Troyanskaya OG, Amberg DC. Modeling complex genetic interactions in a simple eukaryotic genome: actin displays a rich spectrum of complex haploinsufficiencies. Genes Dev. 2007;21:148–159. doi: 10.1101/gad.1477507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985;110:381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Chromosome cohesion, condensation, and separation. Annu Rev Biochem. 2000;69:115–144. doi: 10.1146/annurev.biochem.69.1.115. [DOI] [PubMed] [Google Scholar]
- Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland KM, Kingsbury J, Koshland D, Hieter P. Ctf19p: A novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J Cell Biol. 1999;145:15–28. doi: 10.1083/jcb.145.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Keck JM, et al. A cell cycle phosphoproteome of the yeast centrosome. Science. 2011;332:1557–1561. doi: 10.1126/science.1205193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Merdes A, Mourey L, Agard DA. Microtubule nucleation by gamma-tubulin complexes. Nat Rev Mol Cell Biol. 2011;12:709–721. doi: 10.1038/nrm3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- Li JM, Li Y, Elledge SJ. Genetic analysis of the kinetochore DASH complex reveals an antagonistic relationship with ras/protein kinase A pathway and a novel subunit required for Ask1 association. Mol Cell Biol. 2005;25:767–778. doi: 10.1128/MCB.25.2.767-778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TC, Gombos L, Neuner A, Sebastian D, Olsen JV, Hrle A, Benda C, Schiebel E. Phosphorylation of the yeast gamma-tubulin Tub4 regulates microtubule function. PLoS One. 2011;6:e19700. doi: 10.1371/journal.pone.0019700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R. DNA damage checkpoint and repair centers. Curr Opin Cell Biol. 2004;16:328–334. doi: 10.1016/j.ceb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Ma L, Ho K, Piggott N, Luo Z, Measday V. Interactions between the kinetochore complex and protein kinase A pathway in Saccharomyces cerevisiae. G3 (Bethesda) 2012;2:831–841. doi: 10.1534/g3.112.002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magtanong L, et al. Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell. Nat Biotechnol. 2011;29:505–511. doi: 10.1038/nbt.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin OC, Gunawardane RN, Iwamatsu A, Zheng Y. Xgrip109: a gamma tubulin-associated protein with an essential role in gamma tubulin ring complex (gammaTuRC) assembly and centrosome function. J Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, Foltz DR, Basrai MA. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 2011;7:e1002303. doi: 10.1371/journal.pgen.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Hardwick KG. The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- O'Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole ET, Winey M, McIntosh JR, Mastronarde DN. Electron tomography of yeast cells. Meth Enzymol. 2002;351:81–95. doi: 10.1016/s0076-6879(02)51842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Oakley CE, Oakley BR. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- Paulovich AG, Toczyski DP, Hartwell LH. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- Prestel M, Feller C, Becker PB. Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol. 2010;11:216. doi: 10.1186/gb-2010-11-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M, Weissman JS, Kirschner MW. A general lack of compensation for gene dosage in yeast. Mol Syst Biol. 2010;6:368. doi: 10.1038/msb.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of gamma-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Strome ED, Wu X, Kimmel M, Plon SE. Heterozygous screen in Saccharomyces cerevisiae identifies dosage sensitive genes that affect chromosome stability. Genetics. 2008;178:1193–1207. doi: 10.1534/genetics.107.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Drapkin B, Oomen J, Beach D, Bloom K, Snyder M. Phosphorylation of gamma-tubulin regulates microtubule organization in budding yeast. Dev Cell. 2001;1:621–631. doi: 10.1016/s1534-5807(01)00073-9. [DOI] [PubMed] [Google Scholar]
- Vogel J, Snyder M. The carboxy terminus of Tub4p is required for gamma-tubulin function in budding yeast. J Cell Sci. 2000;113:3871–3882. doi: 10.1242/jcs.113.21.3871. [DOI] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O'Toole ET, Mastronarde DN, Giddings TH, Jr, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Zheng X, Epstein A, Klein HL. Methods to study mitotic homologous recombination and genome stability. Methods Mol Biol. 2011;745:3–13. doi: 10.1007/978-1-61779-129-1_1. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.