Abstract
Micelles were prepared from polymer-peptide block copolymer amphiphiles containing substrates for protein kinase A, protein phosphatase-1 and matrix metalloproteinases 2 and 9. We examine reversible switching of the morphology of these micelles through a phosphorylation-dephosphorylation cycle and study peptide-sequence directed changes in morphology in response to proteolysis. Furthermore, the exceptional uniformity of these polymer-peptide particles makes them amenable to cryo-TEM reconstruction techniques lending insight into their internal structure.
In biology, stimuli-responsive multisubunit assemblies are ubiquitous, and mimicking these systems via synthetic approaches is of increasing interest. Interfacing such synthetic materials with biological systems is particularly promising for a range of biomedical applications including targeted drug delivery and molecular diagnostics.1 Within this class of materials are particles capable of changing morphology in response to stimuli. Enzymes are attractive and unique stimuli with great potential in this regard, as they propagate an amplified response via catalytic reactions,2 can be highly substrate specific, and have expression patterns sometimes associated with disease states.3 Nanoscale assemblies of block copolymer amphiphiles are well-suited for the development of functional, stimuli-responsive systems because changes in the chemical or physical nature of the amphiphile4 can lead to formation, destruction, or morphological transformations.5 However, while there are elegant examples of enzyme-responsive formation and destruction of such materials,6 there are no examples of enzymatic switches of micellar morphology.7 This is despite the tremendous interest in enzymes as stimuli for responsive materials in general,8 and the power of tunable amphiphilicity for switching the shape and size of nanoscale particles, as demonstrated for a range of other stimuli.7
To develop nanoparticles capable of enzyme-directed morphological transformations, we hypothesized that peptides, as enzyme substrates, could be utilized as hydrophilic head groups in polymeric amphiphiles (Figure 1).9, 10 When properly designed, these polymer-peptide amphiphiles would aggregate to generate enzymatically responsive micelles. To validate this hypothesis, we explored enzymatic modulation of particle morphology via common post-translational modification processes utilized to manipulate biomolecular assemblies in natural systems. Furthermore, cryoelectron microscopy (cryo-TEM) and three-dimensional (3D) image reconstruction were used to confirm the spherical micellar morphology and uniformity of the particles and to determine their radial density profile.11
Figure 1.
Peptide-substrate polymeric amphiphiles assemble into spherical micelles. The peptide substrates within the micelle corona interact with enzymes to generate a variety of morphologies of polymeric amphiphile aggregates depending on the design of the peptide substrate and enzymes added.
Amphiphilic polymer-peptides were designed, containing substrates for four different cancer-associated enzymes: protein kinase A (PKA),12 protein phosphatase-1 (PP1),13 and matrix-metalloproteinases MMP-2 and MMP-9.3b-d By incorporating these enzyme substrates into the polar head groups of the copolymers, the micelle morphology and aggregation behaviour of the materials can be modified using the following mechanisms: 1) phosphorylation by PKA at serine residues, 2) dephosphorylation by PP1 at serine residues, 3) peptide cleavage by MMPs at Gly-Leu peptide bonds. We reasoned that enzymatic reactions occurring within the shell of the particles would facilitate changes in the steric bulk, and electrostatic properties of the amphiphiles, and would result in changes to the overall architecture via the establishment of new equilibria for surfactant aggregation.4 Furthermore, we expected enzyme-directed responses to be influenced by the design of the peptide substrate. To test this hypothesis, spherical micelles (M1, M2) were prepared from amphiphilic peptide-brush copolymers that differed only in the relative ordering of the peptide substrates (Table 1). Ring-opening metathesis polymerization (ROMP)1n was utilized to synthesize a block copolymer of hydrophobic sidechains (phenyl groups) and N-hydroxysuccinimide sidechains14 for subsequent conjugation with peptides. To prepare micelles M1 and M2, the block copolymer amphiphiles were dissolved in DMSO/DMF (1:1) and dialyzed against buffered water over 24 h (Table 1, Supporting Information, Figure 1S).
Table 1.
The peptide-shell polymeric amphiphiles and resulting parameters for micelles M1 and M2.
| Peptide substratea | mb | nb | Polymer Mn, g/molc | Mw Mnc | Dhd | PDId | |
|---|---|---|---|---|---|---|---|
| M1 |
|
34 | 6 | 19470 | 1.01 | 24 | 0.27 |
| M2 |
|
34 | 5 | 19430 | 1.17 | 33 | 0.19 |
PKA/PP1 and MMP substrates are shown in red and blue, respectively, with phosphorylation or cleavage sites bolded and underlined. Peptides are conjugated to the polymer through the amino termini.
Block size of m (“phenyl block”) was determined by SEC-MALS (Mn = 8553 g/mol), and n was estimated via SEC-MALS and UV-Vis as described in supporting information.
Polymer Mn and Mw/Mn determined by SEC-MALS.
Hydrodynamic diameter and micelle PDI (polydispersity) were determined by DLS.
The diameters of the spherical particles, as confirmed by transmission electron microscopy (TEM), scanning electron microscopy (SEM), and dynamic light scattering (DLS: hydrodynamic diameter, Dh), were between 24 and 33 nm (Table 1). We further examined M1 using cryo-TEM followed by single-particle, 3D image reconstruction to characterize and define the particles in a native hydrated state (Figure 2).15 Intriguingly, the radial density profile for the spherically averaged reconstruction (Figure 2c) has a similar shape to that simulated and measured for other copolymer and surfactant-based micelles as determined by alternative techniques.16 In particular, the materials show low density at the central core and a region of higher density at the edge of the core and in the surrounding shell. This profile is consistent with a hydrophobic core radius of 5 to 7 nm. From individual particle images we assigned radii to the micelles, defined as the distance from the center at which the minimum density occurs (Supporting information, Figure 2S). The results indicate that the micelles have radii that vary continuously between approximately 10.5 and 13.5 nm. Taken together we conclude that the particles are spherical micellar architectures, 24 nm in average diameter with a maximum variation of 3 nm.
Figure 2.

TEM characterization of M1. (a) Micrograph of M1 sample, stained with 1% uranyl acetate. (b) Micrograph of unstained vitrified M1 sample. (c) Reconstructed radial density plot from cryo-TEM data (Supporting information, Figure 2S).
To establish that micelle morphology can be reversibly altered through enzymatic reactions, the micelles were subjected to phosphorylation by PKA and subsequent dephosphorylation by PP1 (Figure 3 for M2 data and Supporting Information Figure 3S for M1 data). When treated with PKA and ATP (2 mM) for 24 hrs at 30 °C, the initially spherical M2 (at 20 μM, with respect to polymer-peptide amphiphile), changed dramatically in morphology (Figure 3a-b). A 50-fold increase in hydrodynamic diameter was observed (Figure 3e) together with the appearance of amorphous structures in TEM images. The phase transition occurs as phosphate group introduction into the shell of the micellar aggregates produces a significant change in structure and charge of the polymer-peptide amphiphiles.17 However, rather than an increase in hydrophilicity causing an increase in surface curvature resulting in smaller micelles, we observe aggregation into larger structures. Therefore, it is plausible that aggregation is the result of salt bridge formation between phosphorylated particle shells, or that particles aggregate as a result of dipole-induced-dipole interparticle interactions.
Figure 3.
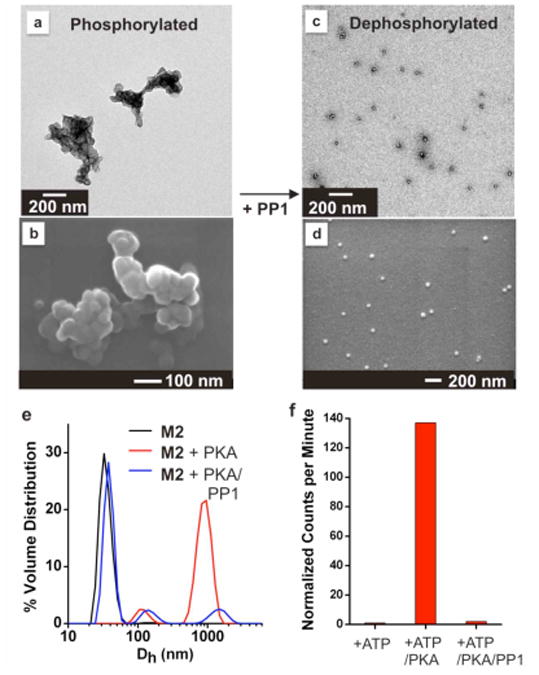
Response of M2 to sequential additions of PKA and PP1. (a) TEM and (b) SEM: M2 (20 μM with respect to polymer-peptide amphiphile) treated with PKA (2500 U) plus ATP (2 mM) and incubated at 30 °C for 24 hrs followed by dialysis. (c) TEM and (d) SEM: Phosphorylated particles subjected either to dialysis, or heat denaturation of PKA (20 min, 65 °C) prior to treatment with PP1 (2.5 U) for at 30 °C for 24 hrs. (e) DLS confirms increase and decrease of aggregate size in solution via phosphorylation and dephosphorylation respectively. (f) Phosphorylation and dephosphorylation were confirmed by radiolabelling the particles using [γ-32P] ATP. Heat denaturation and extended dialysis had no effect on M2 micelles alone. Note that no change is observed by DLS in the presence of ATP (2 mM) without addition of PKA.
Subsequent treatment of the phosphorylated micelles with PP1 for 24 hrs at 30 °C, following either dialysis (to remove ATP) or heat treatment at 65 °C (to denature the kinase), resulted in a reversion to the original size and morphology of particle (Figure 3c-e). Radiolabelling was conducted to confirm that phosphorylation and dephosphorylation occurred through the cycling process (Figure 3f). In this experiment, M2 was treated with PKA and radioactively labelled ATP. Following removal of excess ATP by dialysis, phosphorylation was observed with a scintillation counter. Results show that PKA successfully phosphorylated the particles, with the extent of phosphorylation by PKA estimated to be greater than 95% for M2 (see Supporting Information). Subsequent treatment with PP1 (again followed by dialysis) resulted in removal of the phosphate group. To establish that this process is indeed reversible, three cycles of phosphorylation/dephosphorylation were successfully performed and analyzed by radiolabeling and via DLS (Supporting Information, Figures 4S and 5S). Both M1 and M2 show the same reactivity patterns, but with M2 appearing to undergo a more complete reversible transition than M1 possibly because of the serine residue positioned further from the polymer backbone. This was confirmed via DLS analysis and is especially clear for the radiolabelling experiments (Figure 3f and Supporting Information). Together, these enzymatically-driven processes demonstrate the power of this approach to switching the morphology of a micellar particles via a selective biochemical reaction, not a change in bulk solution properties such as pH, or temperature.
To examine the role of site-specific, proteolytic cleavage on micelle morphology, M1 and M2 were treated with two cancer-associated proteases, MMP-2 and MMP-9, which were expected to have similar effects as they share a cleavage site. (Figure 4 and Supporting Information, Figure 6S). Reactions were performed on 20 μM solutions of micelles, (concentration is with respect to polymer-peptide amphiphile), for 24 hrs at 37 °C. TEM and SEM data showed no change in M1 morphology (Figure 4a-b), but DLS measurements indicated the formation of some larger aggregates in solution (Figure 4e). By contrast, a dramatic change in morphology (Figure 4c-d) and hydrodynamic diameter (Figure 4f) was observed upon treating M2 with MMP. SEM and TEM images both show evidence of the formation of an amorphous network upon peptide cleavage. The cleavage efficiency is estimated by HPLC analysis of the product to be approximately 21% (Supporting Information, Figure 7S), with product identity confirmed by MALDI-MS (Supporting Information, Figure 8S). No visible precipitate was formed in solution during this process. We infer from these results that the position of the cleavage site in the amphiphile plays a critical role in how the micelle responds to proteolysis and that complete shell cleavage is not necessary for phase transition. In particular, cleavage at sites more proximal to the polymer backbone leads to more dramatic morphological changes because of a larger change in peptide shell structure. Indeed, it is likely that the difference in the behaviour of M1 and M2 was further accentuated by the fact that proteolysis also affected the numbers of hydrophobic and hydrophilic residues. Cleavage of the M1 peptide removes three hydrophobic residues, while cleavage of the M2 peptide leads to the loss of all hydrophilic residues. Importantly, these results indicate a tunable relationship between peptide-sequence design and enzymatically directed morphology changes.
Figure 4.
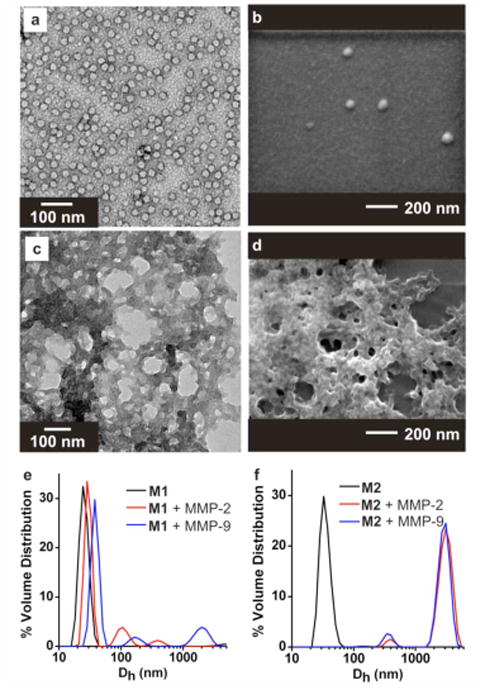
Response of micelles M1 and M2 to treatment with MMPs. (a) TEM M1 + MMP-2. (b) SEM M1 + MMP-2. (c) TEM M2 + MMP-2. (d) SEM M2 + MMP-2. (e-f) DLS shown for particles indicating changes in particle size upon MMP treatment as indicated. Micelles at 20 μM (with respect to polymer-peptide amphiphile), were incubated with MMPs (100 μU) at 37 °C for 24 hrs (Tris-HCL, 50 mM, pH 7.4).
Together these studies demonstrate the feasibility of designing enzymatically switchable micellar particle morphology. This was achieved by incorporating peptides as the hydrophilic block of a polymeric amphiphile. Indeed, in general biomolecules including proteins,9, 18 peptides,10, 19 and nucleic acids1k, 20 are attractive synthons for the development of supramolecular biomaterials21 because they are selective as substrates for enzymes, have inherently specific recognition properties, and consist of well-defined structural elements. It is anticipated that multi-enzyme responsive systems like those described here will provide a route toward materials capable of signalling specific patterns of multiple biochemical stimuli. Furthermore, the ability to program the nature of particle responses to disease-associated enzymes has broad implications for in vivo delivery and detection strategies where surface chemistry and morphology have critical roles in determining the targeting and pharmacokinetics of materials.
Supplementary Material
Acknowledgments
We gratefully acknowledge the support of this work by the University of California, a Camille & Henry Dreyfus Foundation New Faculty Award to N.C.G and DOD-AFOSR for a PECASE to N.C.G. We acknowledge use of the UCSD Cryo-Electron Microscopy Facility, which is supported in part by NIH grant 1S10 RR020016, a gift from the Agouron Institute, and UCSD funds provided to T.S.B. R.S.S, N.H.O and T.S.B were supported by NIH grants R37 GM-033050 and R01 AI-079095. We acknowledge the Nano-3 facilities at UCSD for use of their AFM instrument. The authors thank Prof. Simpson Joseph for kindly sharing his laboratory for radiolabeling studies. We acknowledge the support of the National Science Foundation under CHE-0741968.
Footnotes
Supporting Information Available: Experimental details including synthetic methods, materials characterization, enzymatic conditions, and procedures. This material is available free of charge via the internet at http://pubs.acs.org.
References
- (1).(a) Torchilin VP. Coll Surf B. 1999;16:305–319. [Google Scholar]; (b) Shanmugananda Murthy K, Ma Q, Clark CG, Remsen EE, Wooley KL. Chem Commun. 2001:773–774. [Google Scholar]; (c) Hamley IW. Angew Chem, Int Ed. 2003;42:1692–1712. doi: 10.1002/anie.200200546. [DOI] [PubMed] [Google Scholar]; (d) Alarcon CdlH, Pennadam S, Alexander C. Chem Soc Rev. 2005;34:276–285. doi: 10.1039/b406727d. [DOI] [PubMed] [Google Scholar]; (e) Gothelf KV, LaBean TH. Org Biomol Chem. 2005;3:4023–4037. doi: 10.1039/b510551j. [DOI] [PubMed] [Google Scholar]; (f) Nayak S, Lyon LA. Angew Chem, Int Ed. 2005;44:7686–7708. doi: 10.1002/anie.200501321. [DOI] [PubMed] [Google Scholar]; (g) Petrak K. Drug Disc Today. 2005;10:1667–1673. doi: 10.1016/S1359-6446(05)03698-6. [DOI] [PubMed] [Google Scholar]; (h) Vasir JK, Reddy MK, Labhasetwar VD. Curr Nanosci. 2005;1:47–64. [Google Scholar]; (i) Hawker CJ, Wooley KL. Science. 2005;309:1200–1205. doi: 10.1126/science.1109778. [DOI] [PubMed] [Google Scholar]; (j) Mastrobattista E, van der Aa Marieke AEM, Hennink Wim E, Crommelin Daan JA. Nat Rev Drug Discov. 2006;5:115–121. doi: 10.1038/nrd1960. [DOI] [PubMed] [Google Scholar]; (k) Na K, Sethuraman VT, Bae YH. Anti-Canc Agents Med Chem. 2006;6:525–535. doi: 10.2174/187152006778699068. [DOI] [PubMed] [Google Scholar]; (l) Alemdaroglu FE, Herrmann A. Org Biomol Chem. 2007;5:1311–1320. doi: 10.1039/b617941j. [DOI] [PubMed] [Google Scholar]; (m) Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]; (n) Smith D, Pentzer EB, Nguyen ST. Pol Rev. 2007;47:419–459. [Google Scholar]; (o) Ganta S, Devalapally H, Shahiwala A, Amiji M. J Cont Release. 2008;126:187–204. doi: 10.1016/j.jconrel.2007.12.017. [DOI] [PubMed] [Google Scholar]; (p) Du J, O'Reilly RK. Soft Matter. 2009;5:3544–3561. [Google Scholar]; (q) Meng F, Zhong Z, Feijen J. Biomacromolecules. 2009;10:197–209. doi: 10.1021/bm801127d. [DOI] [PubMed] [Google Scholar]
- (2).(a) Zhu L, Anslyn EV. Angew Chem, Int Ed. 2006;45:1190–1196. doi: 10.1002/anie.200501476. [DOI] [PubMed] [Google Scholar]; (b) Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]; (c) Engvall E, Perlmann P. Immunochem. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- (3).(a) Sawyers CL. Nature. 2008;452:548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]; (b) Vartak DG, Gemeinhart RA. J Drug Targ. 2007;15:1–20. doi: 10.1080/10611860600968967. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Raffetto JD, Khalil RA. Biochem Pharm. 2008;75:346–359. doi: 10.1016/j.bcp.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Kessenbrock K, Plaks V, Werb Z. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Liotta LA, Tryggvason K, Garbisa S, Hart I, Foltz CM, Shafie S. Nature. 1980;284:67–68. doi: 10.1038/284067a0. [DOI] [PubMed] [Google Scholar]; (f) Davies B, Waxman J, Wasan H, Abel P, Williams G, Krausz T, Neal D, Thomas D, Hanby S, Balkwill F. Cancer Res. 1993;53:5365–5369. [PubMed] [Google Scholar]; (g) MacDougall JR, Bani MR, Lin Y, Muschel RJ, Kerbel RS. Br J Cancer. 1999;80:504–512. doi: 10.1038/sj.bjc.6690385. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) MacDougall JR, Bani MR, Lin Y, Rak J, Kerbel RS. Cancer Res. 1995;55:4174–4181. [PubMed] [Google Scholar]; (j) Jinga DC, Blidaru A, Condrea I, Ardeleanu C, Dragomir C, Szegli G, Stefanescu M, Matache C. J Cell Mol Med. 2006;10:499–510. doi: 10.1111/j.1582-4934.2006.tb00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Maatta M, Santala M, Soini Y, Turpeenniemi-Hujanen T, Talvensaari-Mattila A. Acta Obstet Gynecol Scand. 2010;89:380–384. doi: 10.3109/00016340903555990. [DOI] [PubMed] [Google Scholar]
- (4).(a) Israelachvilli JN, Mitchell DJ, Ninham BW. J Chem Soc, Faraday Trans 2. 1976;72:1525–1568. [Google Scholar]; (b) Tanford C. The Hydrophobic Effect: Formation of Micelles and Biological Membranes. Second. John Wiley & Sons, Inc; New York: 1980. [Google Scholar]; (c) Matsen MW, Bates FS. Macromolecules. 1996;29:7641–7644. [Google Scholar]; (d) Discher BM, Won YY, Ege DS, Lee JCM, Bates FS, Discher DE, Hammer DA. Science. 1999;284:1143–1146. doi: 10.1126/science.284.5417.1143. [DOI] [PubMed] [Google Scholar]; (e) Nagarajan R. Langmuir. 2002;18:31–38. [Google Scholar]; (f) Jain S, Bates FS. Science. 2003;300:460–464. doi: 10.1126/science.1082193. [DOI] [PubMed] [Google Scholar]; (g) Smart T, Lomas H, Massignani M, Flores-Merino MV, Perez LR, Battaglia G. Nano Today. 2008;3:38–46. [Google Scholar]
- (5).(a) Zhang L, Yu K, Eisenberg A. Science. 1996;272:1777–1779. doi: 10.1126/science.272.5269.1777. [DOI] [PubMed] [Google Scholar]; (b) Bendejacq D, Ponsinet V, Joanicot M. Langmuir. 2005;21:1712–1718. doi: 10.1021/la048983r. [DOI] [PubMed] [Google Scholar]; (c) LaRue I, Adam M, Pitsikalis M, Hadjichristidis N, Rubinstein M, Sheiko SS. Macromolecules. 2006;39:309–314. [Google Scholar]; (d) Buetuen V, Liu S, Weaver JVM, Bories-Azeau X, Cai Y, Armes SP. React Funct Pol. 2006;66:157–165. [Google Scholar]; (e) Lee HI, Wu W, Oh JK, Mueller L, Sherwood G, Peteanu L, Kowalewski T, Matyjaszewski K. Angew Chem, Int Ed. 2007;46:2453–2457. doi: 10.1002/anie.200604278. [DOI] [PubMed] [Google Scholar]; (f) Ishihara Y, Bazzi HS, Toader V, Godin F, Sleiman HF. Chem Euro J. 2007;13:4560–4570. doi: 10.1002/chem.200601423. [DOI] [PubMed] [Google Scholar]; (g) Rijcken CJF, Soga O, Hennink WE, van Nostrum CF. J Cont Rel. 2007;120:131–148. doi: 10.1016/j.jconrel.2007.03.023. [DOI] [PubMed] [Google Scholar]; (h) Nakayama M, Okano T. Macromolecules. 2008;41:504–507. [Google Scholar]; (i) Sundararaman A, Stephan T, Grubbs RB. J Am Chem Soc. 2008;130:12264–12265. doi: 10.1021/ja8052688. [DOI] [PubMed] [Google Scholar]; (j) Klaikherd A, Nagamani C, Thayumanavan S. J Am Chem Soc. 2009;131:4830–4838. doi: 10.1021/ja809475a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Roy D, Cambre JN, Sumerlin BS. Chem Commun. 2009:2106–2108. doi: 10.1039/b900374f. [DOI] [PubMed] [Google Scholar]; (l) Wang YC, Tang LY, Li Y, Wang J. Biomacromolecules. 2009;10:66–73. doi: 10.1021/bm800808q. [DOI] [PubMed] [Google Scholar]; (m) Moughton AO, O'Reilly RK. Chem Commun. 2010;46:1091–1093. doi: 10.1039/b922289h. [DOI] [PubMed] [Google Scholar]; (n) Agut W, Brulet A, Schatz C, Taton D, Lecommandoux S. Langmuir. 2010;26:10546–10554. doi: 10.1021/la1005693. [DOI] [PubMed] [Google Scholar]; (o) Chien MP, Rush AM, Thompson MP, Gianneschi NC. Angew Chem Int Ed. 2010;49:5076–5080. doi: 10.1002/anie.201000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).(a) Amir RJ, Zhong S, Pochan DJ, Hawker CJ. J Am Chem Soc. 2009;131:13949–13951. doi: 10.1021/ja9060917. [DOI] [PubMed] [Google Scholar]; (b) Azagarsamy MA, Sokkalingam P, Thayumanavan S. J Am Chem Soc. 2009;131:14184–14185. doi: 10.1021/ja906162u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wang Y, Xu H, Zhang X. Adv Mat. 2009;21:1–16. [Google Scholar]
- (8).For a review of enzyme-responsive materials in general see: Ulijn RV. J Mater Chem. 2006;16:2217–2225.
- (9).For protein-polymer amphiphiles see: Velonia K, Rowan AE, Nolte RJM. J Am Chem Soc. 2002;124:4224–4225. doi: 10.1021/ja017809b.; Dirks AJ, Nolte RJM, Cornelissen JJLM. Adv Mater. 2008;20:3953–3957. [Google Scholar]
- (10).For examples of peptide-amphiphile assemblies see: Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187.; (b) Chécot F, Lecommandoux S, Gnanou Y, Klok HA. Angew Chem, Int Ed. 2002;41:1339–1343. doi: 10.1002/1521-3773(20020415)41:8<1339::aid-anie1339>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]; (c) Loewik DWPM, van Hest JCM. Chem Soc Rev. 2004;33:234–245. doi: 10.1039/b212638a. [DOI] [PubMed] [Google Scholar]; (d) Bull SR, Guler MO, Bras RE, Meade TJ, Stupp SI. Nano Lett. 2005;5:1–4. doi: 10.1021/nl0484898. [DOI] [PubMed] [Google Scholar]; (e) Rodriguez-Hernandez J, Lecommandoux S. J Am Chem Soc. 2005;127:2026–2027. doi: 10.1021/ja043920g. [DOI] [PubMed] [Google Scholar]; (f) Kuo SW, Lee HF, Huang CF, Huang CJ, Chang FC. J Poly Sci A. 2008;46:3108–3119. [Google Scholar]; (g) Robson-Marsden H, Korobko AV, van Leeuwen ENM, Pouget EM, Veen SJ, Sommerdijk NAJM, Kros A. J Am Chem Soc. 2008;130:9386–9393. doi: 10.1021/ja800254w. [DOI] [PubMed] [Google Scholar]; (h) Chen CL, Zhang P, Rosi NL. J Am Chem Soc. 2008;130:13555–13557. doi: 10.1021/ja805683r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Cui H, Webber MJ, Stupp SI. Biopolymers. 2009;94:1–18. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Versluis F, Marsden HR, Kros A. Chem Soc Rev. 2010;39:3434–3444. doi: 10.1039/b919446k. [DOI] [PubMed] [Google Scholar]; (k) Pashuck ET, Cui H, Stupp SI. J Am Chem Soc. 2010;132:6041–6046. doi: 10.1021/ja908560n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).(a) Kellermann M, Bauer W, Hirsch A, Schade B, Ludwig K, Boettcher C. Angew Chem, Int Ed. 2004;43:2959–2962. doi: 10.1002/anie.200353510. [DOI] [PubMed] [Google Scholar]; (b) Schade B, Ludwig K, Boettcher C, Hartnagel U, Hirsch A. Angew Chem, Int Ed. 2007;46:4393–4396. doi: 10.1002/anie.200604784. [DOI] [PubMed] [Google Scholar]; (c) Parry AL, Bomans PHH, Holder SJ, Sommerdijk NAJM, Biagini SCG. Angew Chem, Int Ed. 2008;47:8859–8862. doi: 10.1002/anie.200802834. [DOI] [PubMed] [Google Scholar]; (d) Kato T, Goodman RP, Erben CM, Turberfield AJ, Namba K. Nano Lett. 2009;9:2747–2750. doi: 10.1021/nl901265n. [DOI] [PubMed] [Google Scholar]
- (12).Maller JL, Kemp BE, Krebs EG. Proc Natl Acad Sci U S A. 1978;75:248–251. doi: 10.1073/pnas.75.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wera S, Hemmings BA. Biochem J. 1995;311:17–29. doi: 10.1042/bj3110017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).(a) Pontrello JJ, Allen MJ, Underbakke ES, Kiessling LL. J Am Chem Soc. 2005;127:14536–14537. doi: 10.1021/ja053931p. [DOI] [PubMed] [Google Scholar]; (b) Li Y, Akiba I, Harrisson S, Wooley KL. Adv Func Mater. 2008;18:551–559. [Google Scholar]; (c) Theato P. J Poly Sci A. 2008;46:667–6687. [Google Scholar]; (d) Wiss KT, Theato P. J Poly Sci A. 2010;48:4758–4767. [Google Scholar]; (e) Pauly AC, Theato P. J Poly Sci A. 2011;49:211–224. [Google Scholar]
- (15).(a) Yan X, Dryden KA, Tang J, Baker TS. J Struct Biol. 2006;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yan X, Sinkovits RS, Baker TS. J Struct Biol. 2006;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).(a) Pedersen JS. J Chem Phys. 2001;114:2839–2846. [Google Scholar]; (b) Castelletto V, Hamley IW. Curr Opinion Coll Int Sci. 2002;7:167–172. [Google Scholar]; (c) Gohr K, Schaertl W, Willner L, Pyckhout-Hintzen W. Macromolecules. 2002;35:9110–9116. [Google Scholar]; (d) Jorge M. J Mol Struct. 946:88–93. [Google Scholar]; (e) Yoshii N, Okazaki S. Chem Phys Lett. 2006;425:58–61. [Google Scholar]
- (17).(a) Signarvic RS, DeGrado WF. J Mol Biol. 2003;334:1–12. doi: 10.1016/j.jmb.2003.09.041. [DOI] [PubMed] [Google Scholar]; (b) Sonoda T, Nogami T, Oishi J, Murata M, Niidome T, Katayama Y. Bioconj Chem. 2005;16:1542–1546. doi: 10.1021/bc049710a. [DOI] [PubMed] [Google Scholar]
- (18).(a) van Hest JCM, Tirrell DA. Chem Commun. 2001:1897–1904. doi: 10.1039/b105185g. [DOI] [PubMed] [Google Scholar]; (b) Padilla JE, Colovos C, Yeates TO. Proc Natl Acad Sci U S A. 2001;98:2217–2221. doi: 10.1073/pnas.041614998. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Salgado EN, Faraone-Mennella J, Tezcan FA. J Am Chem Soc. 2007;129:13374–13375. doi: 10.1021/ja075261o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).(a) MacEwan SR, Chilkoti A. Biopolymers. 2010;94:60–77. doi: 10.1002/bip.21327. [DOI] [PubMed] [Google Scholar]; (b) Zelzer M, Ulijn RV. Chem Soc Rev. 2010;39:3351–3357. doi: 10.1039/c0cs00035c. [DOI] [PubMed] [Google Scholar]; (c) Aili D, Stevens MM. Chem Soc Rev. 2010;39:3358–3370. doi: 10.1039/b919461b. [DOI] [PubMed] [Google Scholar]; (d) Loewik DWPM, Leunissen EHP, van den Heuvel M, Hansen MB, van Hest JCM. Chem Soc Rev. 2010;39:3394–3412. doi: 10.1039/b914342b. [DOI] [PubMed] [Google Scholar]; (e) Chen CL, Rosi NL. Angew Chem, Int Ed. 2010;49:1924–1942. doi: 10.1002/anie.200903572. [DOI] [PubMed] [Google Scholar]
- (20).(a) Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]; (b) Storhoff JJ, Mirkin CA. Chem Rev. 1999;99:1849–1862. doi: 10.1021/cr970071p. [DOI] [PubMed] [Google Scholar]; (c) Aldaye FA, Palmer AL, Sleiman HF. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]; (d) He Y, Ye M, Su C, Zhang AE, Ribbe W, Jiang, Mao C. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]; (e) Ofir Y, Samanta B, Rotello VM. Chem Soc Rev. 2008;37:1814–1825. doi: 10.1039/b712689c. [DOI] [PubMed] [Google Scholar]; (f) Gu H, Chao J, Xiao SJ, Seeman NC. Nat Nanotechnol. 2009;4:245–248. doi: 10.1038/nnano.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Langer R, Tirrell DA. Nature. 2004;428:487–492. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



