Summary
The NLRP3 inflammasome plays a major role in innate immune responses by activating caspase-1, resulting in secretion of interleukin (IL)-18 and IL-1β. Although cytosolic double-stranded RNA (dsRNA) and bacterial RNA are known to activate the NLRP3 inflammasome, the upstream sensor is unknown. We investigated the potential function of DExD/H-box RNA helicase family members (previously shown to sense cytosolic DNA and RNA to induce type 1 interferon responses) in RNA-induced NLRP3 inflammasome activation. Among the helicase family members tested, we found that targeting of DHX33 expression by short hairpin RNA efficiently blocked the activation of caspase-1 and secretion of IL-18/IL-1β in human macrophages that were activated by cytosolic poly I:C, reoviral RNA or bacterial RNA. DHX33 bound dsRNA via the helicase C domain. DHX33 interacted with NLRP3 and formed the inflammasome complex following stimulation with RNA. We, therefore, identified DHX33 as a cytosolic RNA sensor that activates the NLRP3 inflammasome.
Introduction
Recognition of cytosolic viral nucleic acids by specific pathogen recognition receptors (PRRs) is critical for the innate immune system to detect viral infection and mount anti-viral type 1 interferon (IFN) and inflammatory responses (Wilkins and Gale, 2010). During the past decade, four classes of sensors have been identified that sense cytosolic viral DNA and RNA and activate type-1 IFN responses: the DExD/H-box helicase family members RIG-I, MDA-5, LGP-2, DDX1, DDX41, DDX60, DHX9, DHX36 (Kato et al., 2006; Kim et al., 2010; Miyashita et al., 2011; Satoh et al., 2010; Yoneyama et al., 2004; Zhang et al., 2011a; Zhang et al., 2011b; Zhang et al., 2011c); IFI16 (a pyrin and HIN domain-containing protein) (Unterholzner et al., 2010); IFIT1 (interferon-induced protein with tetratricopeptide repeats 1) (Pichlmair et al., 2011); and LRRFIP1 (leucine-rich repeat flightless-interacting protein 1) (Yang et al., 2010). AIM2 (IFN-inducible absent in melanoma 2) was identified as a cytosolic DNA sensor that activates the ASC-containing inflammasome and triggers caspase-1-dependent IL-1β production (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009). Although cytosolic synthetic dsRNA, poly I:C (Kanneganti et al., 2006a; Rajan et al., 2010; Rintahaka et al., 2008), virus-derived dsRNA (Kanneganti et al., 2006a) and bacteria-derived RNA (Eigenbrod et al., 2012; Kanneganti et al., 2006b; Sander et al., 2011) were found to activate the NLRP3 inflammasome independently of known cytosolic RNA sensors, the upstream RNA sensors that activate the NLRP3 inflammasome have not been identified.
Inflammasomes are cytosolic multi-protein complexes that activate caspase-1. Activated caspase-1 processes pro-interleukin (IL)-18 and pro-IL-1β to their biologically mature secreted forms. IL-18 and IL-1β are pleiotropic proinflammatory cytokines and play pivotal roles in regulating innate immune responses in addition to instructing adaptive immune responses. The NLRP3 (also called cryopyrin, CIAS1 or NALP3) inflammasome recognizes various kinds of exogenous and endogenous danger signals. Once NLRP3 is activated by cytosolic stimuli, it starts to oligomerize and recruit the adaptor protein ASC (also called PYCARD), resulting in the cleavage of pro-caspase-1 to the active form of caspase-1 (Schroder and Tschopp, 2010). Recently, several groups reported an important role of mitochondria in NLRP3 inflammasome activation (Nakahira et al., 2011; Shimada et al., 2012; Zhou et al., 2011). Thioredoxin-interacting protein (TXNIP) was identified as an NLRP3-binding partner that triggers activation of the NLRP3 inflammasome in a mitochondrial reactive oxygen species (ROS)-sensitive manner by stimulation with monosodium urate crystals (MSU), R837, H2O2 and nigericin (Zhou et al., 2010; Zhou et al., 2011). The NLRP3 inflammasome can recognize diverse stimuli via the common mechanisms of mitochondrial damage. However, cytosolic nucleic acids do not induce mitochondrial depolarization (Shimada et al., 2012) and should be recognized specifically to distinguish non-self-pathogens from self. Thus, a cytosolic RNA sensor could act upstream of NLRP3 and interact with NLRP3 to initiate NLRP3 oligomerization, followed by recruitment of ASC.
Our laboratory has recently identified several members of the DExD/H-box helicase family as DNA and RNA sensors that induce type 1 IFN responses in dendritic cells (DCs) (Kim et al., 2010; Zhang et al., 2011a; Zhang et al., 2011b; Zhang et al., 2011c). We decided to systematically screen the 59 members of the DExD/H-box helicase superfamily for their potential functions in dsRNA-induced NLRP3 inflammasome activation. Here, we demonstrate that DHX33, a member of DExD/H-box helicase family, sensed cytosolic RNA and formed a complex with NLRP3 and ASC in human macrophages, resulting in the cleavage of caspase-1 and secretion of IL-18 and IL-1β. DHX33 as a cytosolic RNA sensor that trigger inflammasome activation may shed new light on infection-induced pathology and these findings may provide important information for infection-induced inflammatory diseases and self-RNA-induced autoimmune diseases.
Results
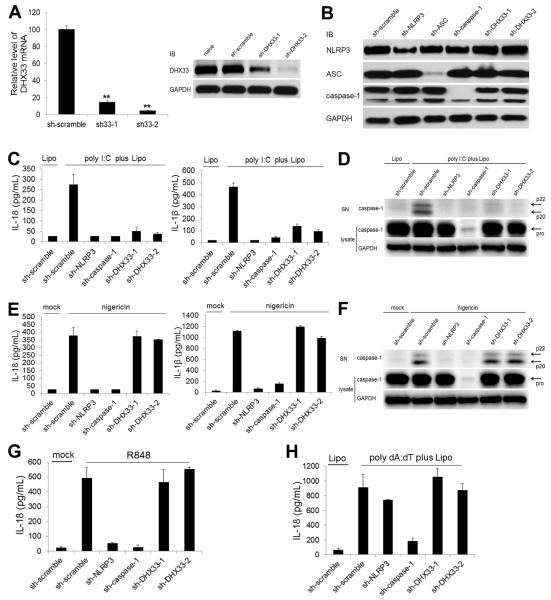
Targeting of DHX33 expression abolishes the activation of the NLRP3 inflammasome via cytosolic poly I:C
We first performed co-immunoprecipitation experiments in HEK293T cells over-expressing NLRP3 and one of helicases to determine whether any of the 59 members of the DExD/H helicase family could bind NLRP3. We found that DDX11, DHX29, DHX32, DHX33, DHX40 and DDX41 bound NLRP3 in this assay (Figure S1A available online). We then efficiently targeted the expression of each helicase mentioned above by short hairpin RNA (shRNA) in human THP-1 macrophages and screened for poly I:C-induced inflammasome activation. Pro-IL-18 was constitutively expressed before and after stimulation with poly I:C in THP-1 macrophages (Figure S1B), indicating that secretion of IL-18 depends on only inflammasome activation. We therefore analyzed IL-18 secretion by ELISA in the functional screening for the inflammasome activation. Among all potential NLRP3-binding helicases tested, we found that targeting of DHX33 expression led to the most substantial reduction in IL-18 secretion in THP-1 macrophages that were stimulated with poly I:C (Figure S1C). We therefore decided to focus on DHX33. We developed two distinct DHX33-targeting shRNA constructs that most efficiently targeted DHX33 expression without affecting expression of NLRP3, ASC, caspase-1 or other NLRP3-binding helicases in THP-1 cells (Figures 1A, 1B and S1D). shRNAs specific for NLRP3, ASC and caspase-1 only down-regulated the expression of the targeted protein (Figure 1B). Targeting DHX33 expression led to a substantial reduction in the secretion of IL-18 and IL-1β (Figure 1C) and the cleavage of caspase-1 (Figure 1D) that were induced by cytosolic poly I:C in THP-1 macrophages. We next investigated whether DHX33 was involved in other NLRP3 inflammasome activation pathways. Targeting of DHX33 expression did not block IL-18 and IL-1β secretion or cleavage of caspase-1 in THP-1 macrophages that were induced by nigericin and LPS+ATP, the known NLRP3 inflammasome activators through mitochondrial ROS (Figures 1E, 1F, S1E, and S1F) (Liao et al., 2012; Zhou et al., 2011). Targeting of DHX33 expression also did not block IL-18 and IL-1β secretion or cleavage of caspase-1 in THP-1 macrophages that were induced by a ligand of TLR7/8 (R848), a known NLRP3 inflammasome activator through mitochondria/TXNIP (Kanneganti et al., 2006b; Zhou et al., 2010). Similar experiments were conducted using poly dA:dT to examine cytosolic dsDNA-induced inflammasome activation. Targeting of DHX33 expression did not affect secretion of IL-18 induced by cytosolic poly dA:dT (Figure 1H). Therefore, DHX33 is specifically involved in cytosolic dsRNA-induced NLRP3 inflammasome activation.
Figure 1. Targeting of DHX33 expression abolishes activation of the NLPR3 inflammasome induced by cytosolic dsRNA.
(A) DHX33 expression was analyzed by real-time PCR (left) and immunoblotting (IB) (right) in THP-1 cells that were naïve and expressing either scrambled shRNA (sh-scramble) or shRNA specific for DHX33 (sh-DHX33-1, sh-DHX33-2). GAPDH served as a loading control.
(B) NLRP3, ASC and caspase-1 expression was analyzed by IB in THP-1 macrophages expressing sh-scramble or shRNA specific for NLRP3 (sh-NLRP3), ASC (sh-ASC), caspase-1 (sh-caspase-1) or DHX33.
(C-F) THP-1 macrophages expressing shRNA were stimulated for 8 h with 5 μg/ml poly I:C delivered to the cytosol via Lipofectamine 2000 (Lipo) (C and D) or with 2 μM nigericin (E and F). Culture supernatants were analyzed for IL-18 and IL-β by ELISA (C and E) and for cleaved caspase-1 by IB (D and F). Cell lysates were analyzed by IB for pro-caspase-1 and GAPDH (D and F).
(G and H) IL-18 secretion was measured by ELISA in culture supernatants of THP-1 macrophages stimulated for 8 h with 5 μg/ml R848 (G) or with 5 μg/ml poly dA:dT plus Lipo (H).
Graph shows the mean ± SD of triplicate wells (**P<0.001 versus scramble). Data are representative of three independent experiments. See also Figure S1.
Viral dsRNA and bacterial RNA activate NLRP3 inflammasome
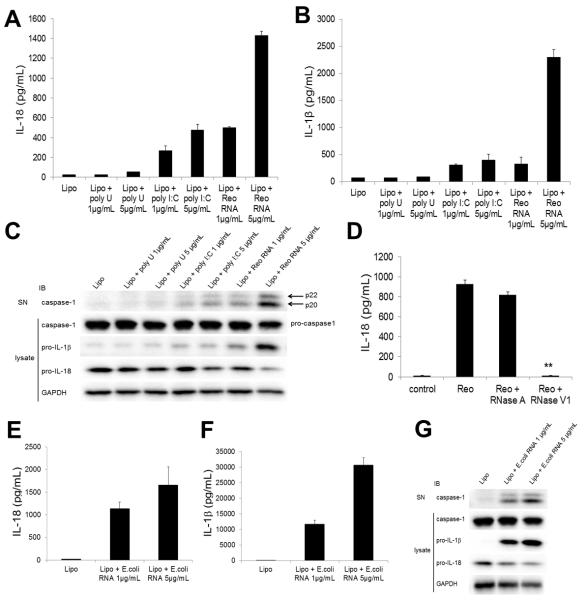
To further determine whether the presence of cytosolic dsRNA initiated activation of the NLRP3 inflammasome, we stimulated THP-1 macrophages with viral genomic dsRNA purified from reovirus. We found that it induced secretion of IL-18 (Figure 2A) and IL-1β (Figure 2B) and cleavage of caspase-1 (Figure 2C). Pretreatment of reoviral genomic RNA with RNase V1, which specifically degrades dsRNA, completely abolished the production of IL-18 (Figure 2D), indicating that the inflammasome activation in this system is mediated by dsRNA. In contrast to poly I:C, cytosolic poly U, a synthetic single-stranded RNA (ssRNA), did not induce inflammasome activation in THP-1 macrophages (Figures 2A-2C). In addition, as previously reported in murine macrophages and dendritic cells (Eigenbrod et al., 2012; Sander et al., 2011), cytosolic bacterial RNA from Escherichia coli (E. coli) induced IL-18 secretion (Figure 2E), IL-1β secretion (Figure 2F) and cleavage of caspase-1 (Figure 2G) in human THP-1 macrophages. Taken together, these results indicate that cytosolic RNA derived from both of virus and bacteria activate NLRP3 inflammasome in THP-1 macrophages.
Figure 2. Reoviral dsRNA and bacterial RNA activate caspase-1 and induce secretion of IL-18 and IL-1β.
(A-C) THP-1 macrophages were stimulated for 8 h with Lipo (control), poly U (1 or 5 μg/ml) plus Lipo, poly I:C (1 or 5 μg/ml) plus Lipo, or reoviral RNA (1 or 5 μg/ml) plus Lipo. Culture supernatants were analyzed for IL-18 (A) and IL-β (B) by ELISA and for cleaved caspase-1 by immunoblotting (IB) (C). Cell lysates were analyzed by IB for pro-caspase-1, pro-IL-1β, pro-IL-18 and GAPDH (C).
(D) THP-1 macrophages were stimulated for 8 h with Lipo (control) or 5 μg/ml reoviral RNA plus Lipo pretreated without or with RNase A or RNase V1. Culture supernatants were analyzed for IL-18 by ELISA.
(E-G) THP-1 macrophages were stimulated for 8 h with Lipo or bacterial RNA from E. coli (1 or 5 μg/ml) plus Lipo. Culture supernatants were analyzed for IL-18 (E) and IL-β (F) by ELISA and for cleaved caspase-1 by IB (G). Cell lysates were analyzed by IB for pro-caspase-1, pro-IL-1β, pro-IL-18 and GAPDH (G).
Graph shows the mean ± SD of triplicate wells (**P<0.001 versus reoviral RNA without pre-treatment).
DHX33 is required for NLRP3 activation induced by viral dsRNA and bacterial RNA
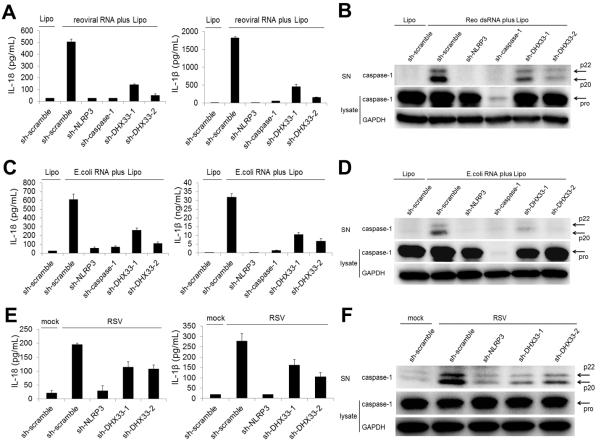
We examined whether DHX33 is involved in the NLRP3 inflammasome activation that is mediated by reoviral RNA. Targeting of DHX33, NLRP3, or caspase-1 expression led to a substantial reduction in secretion of IL-18 and IL-1β (Figure 3A) or cleavage of caspase-1 (Figure 3B) in THP-1 macrophages that were stimulated with cytosolic reoviral RNA. Similarly, secretion of IL-18 and IL-1β (Figure 3C) or cleavage of caspase-1 (Figure 3D) that were induced by bacterial RNA were abrogated by targeting of DHX33 expression. We next examined the involvement of DHX33 in inflammasome activation during live viral infection. We infected THP-1 macrophages with respiratory syncytial virus (RSV), which produces dsRNA during the replication process (Martinez and Melero, 2002). Targeting of NLRP3 or DHX33 expression substantially reduced the secretion of IL-18 and IL-1β (Figure 3E) or the cleavage of caspase-1 (Figure 3F) in THP-1 macrophages infected with RSV. In addition, RSV titers were higher in DHX33-gene targeting cells compared to scramble control cells (Figure S2), suggesting that DHX33 may play an important role in anti-viral defense. Thus, we conclude that DHX33 is critical for cytosolic RNA-induced inflammasome activation in human THP-1 macrophages.
Figure 3. DHX33 is required for NLRP3 inflammasome activation induced by reoviral dsRNA, bacterial RNA and live virus.
(A-F) THP-1 macrophages expressing shRNA were stimulated for 8 h with 5 μg/ml reoviral RNA with Lipo (A and B), or 5 μg/ml bacterial RNA from E. coli (C and D). THP-1 macrophages expressing shRNA were infected for 16 h with respiratory syncytial virus (RSV) (E and F). Culture supernatants were analyzed for IL-18 and IL-β (A, C, E) by ELISA and for cleaved caspase-1 by immunoblotting (IB) (B, D, F). Cell lysates were analyzed by IB for pro-caspase-1 and GAPDH (B, D, F).
Graph shows the mean ± SD of triplicate wells. Data are representative of three independent experiments. See also Figure S2.
DHX33 directly interacts with viral dsRNA via its helicase C domain
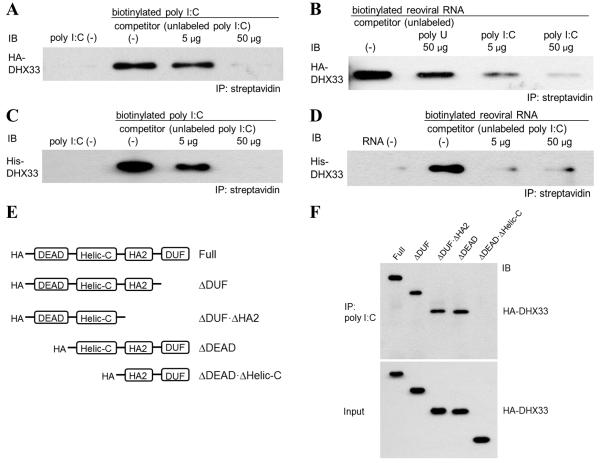
To determine whether DHX33 binds RNA, we performed co-precipitation experiments in which HA-tagged DHX33 (HA-DHX33), expressed and purified from HEK293T cells, was incubated with biotinylated poly I:C or biotinylated reoviral RNA, followed by precipitation with streptavidin beads. We found that HA-DHX33 bound biotin-labeled poly I:C and reoviral RNA (Figures 4A and 4B). With both dsRNA substrates, binding to DHX33 was blocked by unlabeled poly I:C. In addition, the interaction of DHX33 with reoviral RNA was not completely blocked by a high amount of unlabeled poly U (Figure 4B), indicating that DHX33 preferentially bind dsRNA, rather than ssRNA. To exclude the possibility that HA-DHX33 protein purified from HEK293T cells included other dsRNA-binding proteins that mediated the interaction between DHX33 and dsRNA, we repeated the co-immunoprecipitation experiments using histidine-tagged DHX33 (His-DHX33) that had been generated in E. coli. His-DHX33 was precipitated by both poly I:C and reoviral RNA, and binding was blocked by unlabeled poly I:C (Figures 4C and 4D). Therefore, DHX33 directly binds poly I:C and reoviral RNA. To map the regions of DHX33 that are required for dsRNA-binding, we conducted immunoprecipitation experiments with biotin-labeled poly I:C and truncated forms of HA-DHX33 (Figure S3A). We found that the helicase C (helic C) domain of DHX33 was required for binding to poly I:C (Figure 4E). In addition, we performed co-precipitation experiments using HA-DHX33 from HEK293T cells and biotinylated E. coli total RNA. E. coli RNA also pulled down HA-DHX33, and the binding was blocked by unlabeled E. coli RNA (Figure S3B).
Figure 4. DHX33 requires the helicase C domain for binding to dsRNA.
(A and B) HA-DHX33 was overexpressed in HEK293T cells and purified by anti-HA beads. Purified HA-DHX33 protein was incubated with biotinylated poly I:C (A) or biotinylated genomic reoviral RNA (B) in the absence (0 μg) or presence (A: 5 or 50 μg of poly I:C; B: 50 μg of poly U, 5 or 50 μg of poly I:C) of unlabeled competitor. Biotinylated poly I:C (A) or reoviral RNA (B) was precipitated by streptavidin beads, followed by immunoblotting (IB) for HA-DHX33.
(C and D) Recombinant His-DHX33, generated in E. coli, was incubated with biotinylated poly I:C (C) or biotinylated reoviral genomic RNA (D) in the absence (0 μg) or presence (5 or 50 μg) of unlabeled poly I:C as a competitor. Biotinylated dsRNA was immunoprecipitated by streptavidin beads, followed by IB for DHX33.
(E) Purified full-length and truncated HA-DHX33 proteins were incubated individually with biotinylated poly I:C. Biotinylated poly I:C was immunoprecipitated by streptavidin beads, followed by IB for HA-DHX33. DEAD, Asp-Glu-Ala-Asp domain; HELICc, helicase C-terminal domain; HA2, helicase-associated domain 2; DUF, domain of unknown function.
See also Figure S3.
Reconstitution of shRNA-resistant DHX33 rescues inflammasome activation in DHX33-gene targeting cells
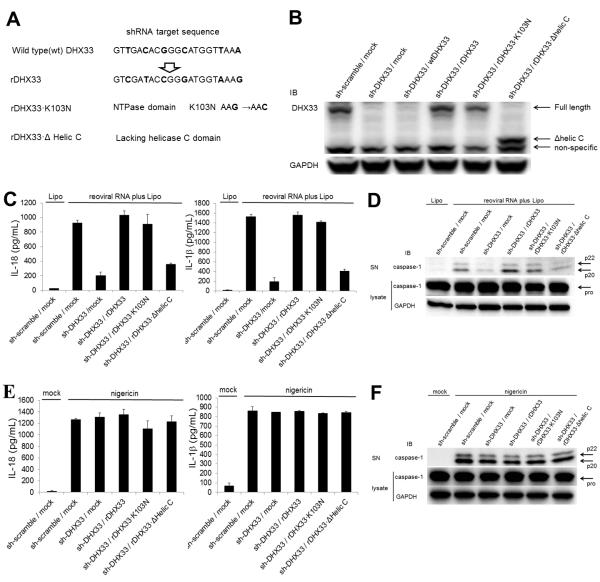
To exclude the possibility of off-target effects of DHX33-shRNA, we reconstituted DHX33 expression in DHX33-gene targeting cells. Since the shRNA-targeting sequence was in the translated region, transfection of wild type DHX33 could not reconstitute DHX33 expression in DHX33-gene targeting cells (Figures 5A and 5B). Therefore, an shRNA-resistant DHX33 (rDHX33) was generated by substitution of 6 nucleotides within the targeting sequence (without changing the amino acid sequence) (Figure 5A). In addition, we generated two forms of rDHX33 with either a dead NTPase domain that lacked ribosomal RNA synthesis activity (K103N) or with a helic C domain deletion that lacks the ability to bind dsRNA (Δhelic C) (Figure 5A). Transfection of these rDHX33 constructs into DHX33-gene targeting cells could reconstitute DHX33 expression successfully (Figure 5B). Reconstitution of rDHX33 rescued the ability of DHX33-gene targeting cells to secret IL-18 and IL-1β or to activate caspase-1 in THP1 macrophages stimulated with dsRNA, indicating that DHX33-shRNA has no off-target effects (Figures 5C and 5D). Reconstitution of rDHX33 with the K103N mutation also rescued the ability of DHX33-gene targeting cells to secret of IL-18 and IL-1β or to activate caspase-1 in THP1 macrophages that were stimulated with dsRNA. This indicates that the function of DHX33 in dsRNA-induced inflammasome activation is independent of the rRNA synthesis function, which requires NTPase domain (Figures 5C and 5D). However, reconstitution of rDHX33 with the Δhelic C mutation failed to rescue inflammasome activation induced by dsRNA, indicating that dsRNA binding to the helic C domain is critical for inflammasome activation (Figures 5C and 5D).
Figure 5. Reconstitution of DHX33 in DHX33-gene targeting cells rescues inflammasome activation induced by cytosolic dsRNA.
(A and B) The shRNA-resistant DHX33 with 6-nucleotide substitution in the 21mer targeting sequence was constructed without changing amino acids (rDHX33). Using this rDHX33 construct, an NTPase dead mutant (rDHX33·K103N) and a helic C domain-deleted mutant (rDHX33·Δhelic C) were generated (A). The wild type DHX33 (wtDHX33) and shRNA-resistant DHX33 constructs were transfected into DHX33-gene targeting THP-1 cells, followed by immunoblotting (IB) for DHX33 (B).
(C-F) Cells were stimulated for 8 h with 5 μg/ml reoviral RNA plus Lipo (C and D) or 2 μM nigericin (E and F). Culture supernatants were analyzed for IL-18 and IL-β by ELISA (C and E) and for cleaved caspase-1 by IB (D and F). Cell lysates were analyzed by IB for pro-caspase-1 and GAPDH (D and F).
Graph shows the mean ± SD of triplicate wells.
DHX33 binds NLRP3 via the DEAD domain of DHX33 and the NACHT domain of NLRP3
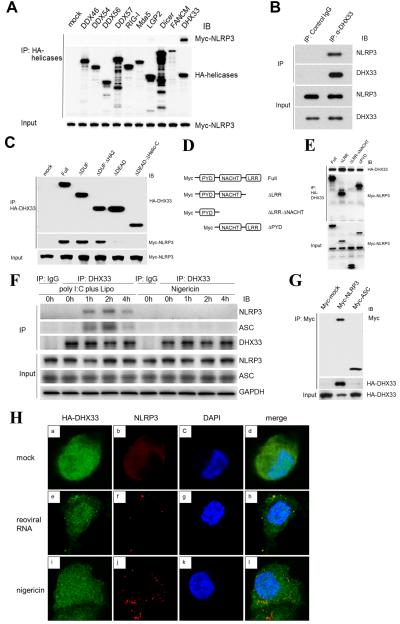
HEK293T cells were co-transfected with a vector encoding Myc-NLRP3 plus a vector encoding HA-tagged helicases. Lysates of transfected cells were immunoprecipitated with anti-HA antibody. We show the precipitation of DDX46, DDX54, DDX57, RIG-I, MDA-5, LGP-2, Dicer, FANCM and DHX33 in Figure 6A. DHX33 pulled down NLRP3, but the other tested helicases here, including cytosolic RNA sensors RIG-I and Mda-5 did not. In addition, we showed that Myc-NLRP3 pulled down HA-DHX33 (Figure S1A); thus, we confirmed the binding of DHX33 to NLRP3 in the HEK293T cells overexpression system. To see the specificity of the binding between DHX33 and NLRP3, we tested the binding between DHX33 and other NLR (nucleotide-binding domain and leucine-rich repeat containing) family proteins NOD1/NOD2. The binding affinities of NOD1 and NOD2 to DHX33 were much weaker than that of NLRP3 to DHX33 (Figure S4A). To determine whether DHX33 directly binds NLRP3, we conducted immunoprecipitation experiments using a mixture of His-DHX33 and GST-NLRP3, each expressed and purified from E. coli. We found that pull-down of recombinant DHX33 co-precipitated recombinant NLRP3, indicating that DHX33 directly binds NLRP3 (Figure 6B). To map the site of DHX33 required for NLRP3-binding, we overexpressed full length or truncated forms of HA-DHX33 with Myc-NLRP3 in HEK293T cells, followed by precipitation with anti-HA beads. We found that the Asp-Glu-Ala-Asp (DEAD) domain of DHX33 was required to interact with NLRP3 (Figure 6C). Likewise, we generated truncated forms of Myc-NLRP3 (Figure S4B) and expressed each of them in HEK293T cells along with HA-DHX33 to identify the binding site of NLRP3 to DHX33 by co-immunoprecipitation. We found that the NACHT domain of NLRP3 was required for NLRP3:DHX33 interaction (Figure 6D). Taken together, these results indicate that DHX33 directly binds NLRP3 through the DEAD domain of DHX33 and the NACHT domain of NLRP3.
Figure 6. DHX33 forms the inflammasome complex via direct binding to NLRP3.
(A) HEK293T cells were co-transfected with a vector encoding Myc-NLRP3 plus a vector encoding an HA-DExD/H family member (DDX46, DDX54, DDX56, DDX57, RIG-I, Mda-5, LGP-2, Dicer, FANCM or DHX33) or a pCMV-HA empty vector (mock). HA-tagged helicases were immunoprecipitated, followed by immunoblotting (IB) for Myc-NLRP3 and HA-helicases.
(B) A mixture of the purified GST-NLRP3 and His-DHX33 proteins generated in E. coli was immunoprecipitated with anti-DHX33 antibody or isotype control IgG, followed by IB for NLRP3 and DHX33.
(C) HEK293T cells were co-transfected with a vector encoding Myc-NLRP3 along with a vector encoding a full-length or truncated HA-DHX33 (as in Figure 4E). HA-tagged helicases were immunoprecipitated, followed by IB for HA-DHX33 and Myc-NLRP3.
(D) HEK293T cells were co-transfected with a vector encoding HA-DHX33 along with a vector encoding full-length or truncated Myc-NLRP3. HA-DHX33 was immunoprecipitated followed by IB for HA-DHX33 and Myc-NLRP3.
(E) THP-1 macrophages were stimulated for 0, 1, 2 and 4 h with poly I:C plus Lipo or nigericin. Immunoprecipitation was performed by anti-DHX33 antibody or isotype control IgG, followed by IB for DHX33, NLRP3, ASC and GAPDH.
(F) HEK293T cells were co-transfected with a vector encoding HA-DHX33 along with a pCMV-Myc empty vector (Myc-mock), or a vector encoding Myc-NLRP3 or Myc-ASC. Myc-tagged proteins were immunoprecipitated, followed by IB for Myc and HA-DHX33.
(G) THP-1 macrophages transfected with HA-DHX33 were stimulated for 2 h with Lipo (mock) (a-d), reoviral RNA plus Lipo (reoviral RNA) (e-h) or nigericin (i-l) and analyzed for the localization of HA-DHX33 (red) (a, e, i), NLRP3 (green) (b, f, j), nucleus marker DAPI (blue) (c, g, k), and merge (d, h, l) using confocal microscopy. See also Figure S4.
DHX33 forms the inflammasome complex with NLRP3 following stimulation with cytosolic RNA
We next investigated the presence of endogenous DHX33-NLRP3-ASC complexes in THP-1 macrophages (that were resting or stimulated with cytosolic poly I:C or nigericin) by immunoprecipitation with anti-DHX33 antibody. While DHX33-NLRP3-ASC complexes were undetectable in resting THP-1 macrophages, those complexes were detected following poly I:C-stimulation (Figure 6E). This kinetics pattern was similar to the interaction between TXNIP and NLRP3 (Zhou et al., 2010). Alternatively, neither NLRP3 nor ASC were co-precipitated with DHX33 after stimulation with nigericin (Figure 6E). To examine whether DHX33 was able to bind ASC directly, we co-transfected vectors encoding HA-DHX33 and either Myc-ASC or Myc-NLRP3 into HEK293T cells that endogenously expressed neither NLRP3 nor ASC. In the co-immunoprecipitation assay, ASC did not bind to DHX33 (Figure 6F). In addition, an in-vitro binding assay using recombinant DHX33 and ASC protein generated in E. coli did not show the interaction of these molecules (Figure S4C). Therefore, DHX33 may bind ASC indirectly through NLRP3 in dsRNA-activated THP-1 macrophages. To characterize the distribution of DHX33 in cellular compartments, we analyzed its subcellular localization by confocal microscopy in THP-1 monocytes and THP-1 macrophages that had been transfected with a vector encoding HA-DHX33. DHX33 was detected mainly in the nucleus of THP-1 monocytes and was diffusely distributed in both the nucleus and cytosol of THP-1 macrophages (Figure S4D). We observed that NLRP3 redistributed from a diffused expression pattern (Figures 6Gb and S4Eb) into distinct spots after stimulation with both reoviral RNA and nigericin (Figures 6Gf, 6Gj, S4Ef and S4Ej). Some bright DHX33 spots were observed in reoviral RNA-stimulated cells (Figures 6Ge and S4Ee) but not in nigericin-stimulated cells (Figures 6Gi and S4Ei). These DHX33 bright dots were found to colocalize with NLRP3 dots (Figures 6Gh and S4Eh). Therefore, DHX33 mediates inflammasome complex formation after stimulation with cytosolic RNA.
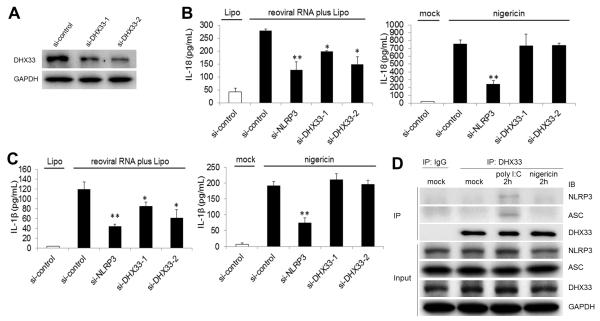
DHX33 is involved in cytosolic RNA-induced NLRP3 inflammasome activation in human primary monocyte-derived macrophages
We next examined the role of DHX33 in human primary monocyte-derived macrophages (MDM). We targeted DHX33 expression in MDM through the use of small interfering RNA (siRNA) (Figure 7A) without affecting expression levels of the other NLRP3-binding helicases (Figure S5A). Although DHX33 expression was not completely abolished, secretion of IL-18 (Figure 7B) and IL-1β (Figure 7C) induced by reoviral RNA was significantly reduced in DHX33-siRNA cells (P<0.05). On the other hand, IL-18 (Figure 7B) and IL-1β (Figure 7C) secretion induced by nigericin was not altered in DHX33-gene targeting cells. In addition, the production of TNF-α induced by reoviral RNA was normal in DHX33-gene targeting cells (Figure S5B). We next examined the inflammasome complex formation by immunoprecipitation in MDM. Pull-down of DHX33 co-precipitated NLRP and ASC in MDM stimulated with reoviral RNA but not in resting cells or in nigericin-treated cells (Figure 7D). Therefore, DHX33 is required for the activation of the NLRP3 inflammasome induced by cytosolic RNA in human primary MDM.
Figure 7. DHX33 is involved in cytosolic RNA-induced NLRP3 inflammasome activation in human monocyte-derived macrophages.
(A) DHX33 expression was analyzed by immunoblotting (IB) in human primary monocyte-derived macrophages (MDM) transfected with control siRNA (si-control) or siRNA specific for DHX33 (si-DHX33-1, si-DHX33-2).
(B and C) MDM transfected with control siRNA or DHX33 siRNA were stimulated for 8 h with 5 μg/ml reoviral RNA plus Lipo (left) or 2 μM nigericin (right). Culture supernatants were analyzed for IL-18 (B) and IL-β (C) by ELISA.
(D) MDM were stimulated without (mock) or with poly I:C plus Lipo or nigericin for 2 h. Immunoprecipitation was performed by anti-DHX33 antibody or isotype control IgG, followed by IB for DHX33, NLRP3, ASC and GAPDH.
Graph shows the mean ± SD of triplicate wells (*P<0.05, **P<0.01 versus scramble siRNA). See also Figure S5.
Discussion
In this study, we have identified DHX33 as a sensor for viral dsRNA that has the ability to directly bind NLRP3 and activate the NLRP3-ASC inflammasome. This inflammasome mediates caspase-1 cleavage and processing as well as secretion of mature IL-18 and IL-1β. DHX33 bind dsRNA via the helicase C domain. Following recognition of dsRNA, DHX33 interacted with NLRP3 via the DEAD domain of DHX33 and the NACHT domain of NLRP3, leading to the recruitment of ASC. This DHX33:NLRP3 interaction at the protein level and clustered colocalization within the cytosol could be induced only by dsRNA stimulation, but not by another NLRP3 inflammasome activator, nigericin. Together with the finding that shRNA-resistant DHX33 requires the presence of an intact dsRNA binding domain (the helic C domain) to rescue inflammasome activation in DHX33-gene targeting THP1 macrophages, these data indicate that DHX33 is a specific dsRNA sensor for NLRP3 inflammasome activation. A key question is whether shRNA for DHX33 has any off-target effect that may be important for the NLRP3 inflammasome activation. Our reconstitution experiments using shRNA-resistant DHX33 concluded that DHX33-shRNA used in our study has no such off-target effect. Another important question is whether the effect of DHX33 in dsRNA-induced NLRP3 inflammasome activation is due to the function of DHX33 in rRNA synthesis. We performed reconstitution experiments using shRNA-resistant DHX33 that contained a dead NTPase domain, whose normal function is critical for rRNA synthesis. These data demonstrated that NLRP3 inflammasome activation in THP-1 macrophages induced by cytosolic dsRNA is independent of the NTPase domain. It is not uncommon that most cytosolic DNA or RNA sensors, such as RIG-I, also have multiple functions in cell proliferation, apoptosis and development (Besch et al., 2009; Jiang et al., 2011; Liu and Gu, 2011; Zhang et al., 2008).
We therefore demonstrated that DHX33 represents a receptor that links the pattern recognition of viral dsRNA to NLRP3 inflammasome activation. The DExD/H-box helicase superfamily has at least 59 members. Previous studies have demonstrated that members of this family including RIG-I, MDA-5, LGP-2, DDX1, DDX41, DDX60, DHX9 and DHX36 play critical roles in sensing viral DNA/RNA and triggering anti-viral type 1 IFN responses (Kato et al., 2006; Kim et al., 2010; Miyashita et al., 2011; Satoh et al., 2010; Yoneyama et al., 2004; Zhang et al., 2011a; Zhang et al., 2011b; Zhang et al., 2011c). Interestingly, different helicases appear to use different signaling adaptor molecules for the activation of the type-1 IFN pathway. For example, RIG-1, MDA-5 and DHX9, which recognize dsRNA, use MAVS (IPS-1) (Kawai et al., 2005; Zhang et al., 2011c); DDX1-DDX21-DHX36, which recognizes dsRNA, uses TRIF (Zhang et al., 2011a); and DDX41, a dsDNA sensor, uses STING (Zhang et al., 2011b) to activate the type-1 IFN pathway. Here, we demonstrate that DHX33, a RNA sensor, uses NLRP3 to activate the secretion of proinflammatory cytokines. These data suggest that the DExD/H-box helicase superfamily is highly utilized by the innate immune system for detecting viral infection, which activates different type 1 IFN pathways and the inflammasome pathway.
It has been suggested that members of NLR family, including NAIP (NLR family, Apoptosis Inhibitory Protein), NOD1, NOD2, NLRC4, NLRP1 and NLRP3, are cytosolic pattern recognition receptors that link inflammatory responses to pathogen-associated molecular patterns (PAMPs) and host-derived molecular patterns (danger-associated molecular patterns, DAMPs) (Bryant and Fitzgerald, 2009; Franchi et al., 2009). However, it is still not clear whether NLRs directly interact with PAMPs or DAMPs. Our study suggests that NLRP3 functions as a signaling adaptor rather than a sensor, and is used by the RNA sensor DHX33 to activate the inflammasome pathway. Two distinct bacterial proteins, flagellin and PrgJ, have been shown to activate the NLRC4 inflammasome (Franchi et al., 2006; Miao et al., 2006; Miao et al., 2010). Kofoed et al. reported that NAIP5/6 and NAIP2, specifically recognize flagellin and PrgJ, respectively. Then, NAIP5/6 and NAIP2 commonly bind to NLRC4 and forms inflammasome complexes (Kofoed and Vance, 2011). Thus, DHX33 or NAIPs provide specificity for ligands and interact with adapter NLRP3 or NLRC4, triggering subsequent inflammasome activation.
Recent studies demonstrated that bacterial messenger RNA could activate NLRP3 inflammasome (Kanneganti et al., 2006b; Sander et al., 2011). We demonstrated in this study that DHX33 also senses bacteria-derived RNA and is responsible for NLRP3 inflammasome activation in human macrophages. How DHX33 distinguishes viral/bacterial RNA from self-RNA deserves further investigation.
Viral and bacterial infections trigger inflammasome activation that contributes to infection-associated inflammatory diseases and autoimmune diseases (Davis et al., 2011; Martinon et al., 2009; Shaw et al., 2011). The identification of AIM2 as a dsDNA sensor and DHX33 as a dsRNA sensor that trigger inflammasome activation may shed new light on infection-induced pathology. These findings may provide important information for the development of new therapies for infection-induced inflammatory diseases and self-DNA/RNA-induced autoimmune diseases.
Experimental Procedures
Cell culture
THP-1 cells, a human acute monocytic leukemia cell line, were maintained in RPMI-1640 medium containing 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine and 50 μM β-mercaptoethanol. HEK293T cells and Vero cells were maintained in DMEM medium with 10% (vol/vol) heat-inactivated FBS.
Isolation of reoviral genomic RNA
Vero cells were infected with reovirus serotype 3 (American Type Culture Collection) at a multiplicity of infection of 0.1 PFU/cell. The supernatant was collected when cells showed maximal cytopathic effect from viral infection, centrifuged at 2500 rpm for 30 min, and then passed through 0.45 μm filters. The filtered supernatant was centrifuged at 21,000 rpm for 120 min. The viral pellet was suspended in RLT Buffer (Qiagen) and then RNA was extracted using RNeasy kit with DNase I treatment (Qiagen).
Plasmids
cDNA encoding the open reading frames of DExD/H-box RNA helicases, NLRP3, NOD1, NOD2 or ASC was subcloned into pCMV-HA or pCMV-Myc vectors (Clontech). Based on pCMV-HA-full length DHX33 and pCMV-Myc-full length NLRP3, truncated forms of the HA-DHX33 and Myc-NLRP3 genes were generated by inverse PCR methods using the primers shown in Table S1. For reconstitution of DHX33, DHX33 cDNA was subcloned into pCDH vector (System Biosciences). Based on pCDH-wild type DHX33 (wtDHX33), genes encoding shRNA-resistant DHX33 (rDHX33), an NTPase domain-dead mutant (K103N) and the deletion mutant of helic C domain (Δhelic C) were generated by inverse PCR methods using the primers shown in Table S1.
Lentivirus production and infection
The targeting sequences of shRNAs for human DHX33, NLRP3, ASC, and caspase-1 are the following (5′ to 3′); sh-DHX33-1: CATTTCCTTTAGAACCCAAAT; sh-DHX33-2: GTTGACACGGGCATGGTTAAA; sh-NLRP3: GCGTTAGAAACACTTCAAGAA; sh-ASC: CGGAAGCTCTTCAGTTTCACA; sh-caspase-1: CTACAACTCAATGCAATCTTT. A PLKO.1 vector encoding shRNA for a scrambled (Sigma) or a specific target molecule (Open Biosystems or Sigma) was transfected into HEK293T cells together with psPAX2, a packaging plasmid, and pMD2.G, an envelope plasmid, for producing viral particles using Lipofectamine 2000 (Life Technologies). Similarly, pCDH vector encoding wtDHX33, rDHX33, rDHX33·K103N or rDHX33· Δhelic C was transfected into HEK293T cells together with pPACKH1™ Packaging Plasmid mix (System Biosciences) using Lipofectamine 2000. Culture supernatants were harvested 24 h and 48 h after transfection, then centrifuged at 2200 rpm for 15 min. THP-1 cells were infected with collected supernatants containing lentiviral particles in the presence of 4 μg/ml polybrene (Sigma). After 48 h of culture, lentiviral-infected cells were selected by 5 μg/ml puromycin (Invivogen). Gene targeting efficiency of each shRNA-targeted molecule and expression level of reconstituted DHX33 were detected by real-time PCR and/or immunoblot analysis.
Stimulation of THP-1 macrophages
THP-1 cells were differentiated to macrophages with 60 nM phorbol 12-myristate 13-acetate (PMA; Sigma) for 16 h, and cells were cultured for an additional 48h without PMA. Differentiated cells were stimulated for 8 h in 96 well-plates with 1.0 or 5.0 μg/ml poly I:C (Invivogen) plus Lipofectamine 2000, 1.0 or 5.0 μg/ml reoviral genomic RNA plus Lipofectamine 2000, 1.0 or 5.0 μg/ml poly U (Invivogen) plus Lipofectamine 2000, 5.0 μg/ml poly dA:dT (Sigma), 5.0 μg/ml R848, or 2 μM nigericin. Cells were also stimulated with 1 ng/ml LPS for 8 h, followed by treatment with 5 mM ATP for 2 h. Culture supernatants were collected for measurement of cytokines and cleavage of caspase-1. Cells were harvested for real-time PCR or immunoblot analysis.
Stimulation of human monocyte-derived macrophages
Buffy coats of blood samples from healthy donors were used for isolation of peripheral blood monocytes. Monocytes were isolated using RosetteSep™ Human Monocyte Enrichment Cocktail (STEMCELL Technologies) and were differentiated into macrophages in RPMI-1640 medium containing 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), 2 mM L-glutamine and 10 ng/ml M-CSF (Life Technologies) for 10 days. Control siRNA or DHX33 siRNA (Dharmacon) was transfected into macrophages using HiPerFect Transfection Reagent (Qiagen). The target sequences for DHX33 siRNA are as follows; si-DHX33-1, CUACUAGAGUCUCAGAUGA; si-DHX33-2, CCAAAGGGCUAUCGCAAAG. Forty-eight hours after siRNA transfection, cells were stimulated for 8 h in 96 well-plates with 5.0 μg/ml reoviral RNA plus Lipofectamine 2000 or 2 μM nigericin.
Viral infection
THP-1 macrophages were infected with RSV (B strain; Advanced Biotechnologies Inc) in Opti-MEM® reduced-serum medium (Life Technologies). Following adsorption for 2 h at 37°C, cells were washed with regular culture medium and the infection was continued in regular culture medium for the indicated time periods.
Enzyme-linked immunosorbent assay (ELISA)
The concentrations of IL-18, IL-1β and TNF-α in culture supernatants were measured by ELISA kit (IL-18, MBL International; IL-1β, BD Biosciences; TNF-α, R&D).
Immunoblot analysis
Proteins were probed with a primary antibody as follows: polyclonal rabbit anti-DHX33 (sc-137424; Santa Cruz), monoclonal mouse anti-NLRP3 (Cryo-2) (ALX-804-881-C100; Enzo Life Sciences), monoclonal anti-ASC (D086-3; MBL international), monoclonal rabbit anti-caspase-1 (D7F10) (3866; Cell signaling), monoclonal mouse anti-IL-1β (sc-52770; Santa Cruz), monoclonal mouse anti-IL-18 (sc-13602; Santa Cruz) and monoclonal mouse anti-GAPDH (G9295; Sigma).
Expression and purification of recombinant proteins
For the generation in a mammalian system, a PCMV-HA plasmid encoding human DHX33 was transfected into HEK293T cells. The expressed HA-DHX33 protein was purified by using the HA-tagged Protein Purification Kit (MBL International). For the generation in a bacterial system, the cDNA sequence encoding human DHX33 was subcloned into the pRSET vector (Life Technologies) to encode a histidine (His) fusion. The plasmid was transformed into E. coli BL21 (DE3) pLysS (Life Technologies), which was treated with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The His-DHX33 protein was then purified using HisTrap FF (GE Healthcare Life Sciences). GST-tagged NLRP3 recombinant protein and GST-tagged ASC recombinant protein generated in E. coli were obtained from Novus.
In vitro RNA binding assay
Poly I:C, reoviral RNA and E. coli total RNA were labeled with biotin using the RNA 3′ end Biotinylation Kit (Thermo Scientific). Purified DHX33 protein was incubated with biotinylated RNA in the buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.3% (vol/vol) Nonidet-P40, 5 mM EDTA and 10% (vol/vol) glycerol). Biotin-labeled RNA was precipitated with High Capacity NeutrAvidin Agarose (Thermo Scientific). For the confirmation of DHX33-binding specificity to RNA, immunoprecipitation was done in the absence or presence of competitors (unlabeled poly I:C, poly U or E. coli total RNA).
Co-immunoprecipitation assays using transfected HEK293T cells
pCMV vectors encoding Myc-tagged cDNA and HA-tagged cDNA were co-transfected into HEK293T cells using Lipofectamine 2000. Cells were lysed 24 h after transfection in the buffer (50 mM Tris, pH 7.5, 300 mM NaCl, 1% (vol/vol) Triton-X, 5 mM EDTA and 10% (vol/vol) glycerol). Lysates were immunoprecipitated by anti-HA agarose (Sigma) or by anti-Myc agarose (Sigma).
Co-immunoprecipitation assays using stimulated THP-1 cells and monocyte-derived macrophages
THP-1 cells (1×107) were differentiated with PMA, and then treated for 0, 1, 2, or 4 h with 5.0 μg/ml poly I:C plus Lipofectamine 2000 or 2 μM nigericin. Similarly, monocyte-derived macrophages were treated for 2 h with or without 5.0 μg/ml poly I:C plus Lipofectamine 2000 or 2 μM nigericin. Stimulated cells were resuspended in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% (vol/vol) Nonidet-P40, 5 mM EDTA and 10% (vol/vol) glycerol). Lysates were immunoprecipitated with control rabbit IgG or anti-DHX33 antibody (NB100-2581; Novus) using Protein-G Sepharose (Thermo Scientific).
Quantitative real-time PCR
RNA was isolated with the RNeasy Kit (Qiagen) and used to synthesize cDNA with the iScript cDNA Synthesis Kit (Bio-Rad). iTaq SYBR Green Supermix with ROX (Bio-Rad) was used for quantitative PCR with the primers shown in Table S2. RPL13A or GAPDH was used as a housekeeping control to normalize the amounts of cDNA between each sample. The amount of the helicase cDNA relative to the amount of RPL13A cDNA was measured as −ΔCT = CT·RPL13A − CT·helicase. The ratio of helicase mRNA copies relative to RPL13A mRNA copies was defined as 2-ΔCT × K (K: constant).
Confocal microscopy
THP-1 cells expressing HA-DHX33 were differentiated with PMA on coverslips for 48h. Cells were stimulated with reoviral RNA plus Lipofectamine 2000, 2 μM nigericin, or Lipofectamine 2000 alone. Then, the stimulated cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% saponin. After blocking with 10% goat serum (Sigma), cells were incubated with monoclonal mouse IgG1 anti-NLRP3 antibody (ALX-804-818-C100; Enzo Life Sciences) overnight at 4°C, followed by treatment with anti-mouse IgG1 antibody conjugated with Alexa Fluor 555 (A21127; Life Technologies). After blocking with mouse IgG1 (eBioscience), cells were stained with Alexa Fluor 488-conjugated monoclonal mouse anti-HA antibody (2350; Cell Signaling). Slides were mounted with ProLongGold-DAPI mounting media (Life Technologies) and were analyzed with a confocal microscope.
Statistical analysis
Data were analyzed for statistical significance by two-tailed student’s t-test or Welch’s t-test.
Supplementary Material
Highlights.
Gene targeting of DHX33 abolishes cytosolic RNA-induced inflammasome activation.
DHX33 binds viral double stranded-RNA directly via its helicase C domain.
DHX33 binds NLRP3 via the DEAD domain of DHX33 and the NACHT domain of NLRP3.
DHX33 forms inflammasome complex with NLRP3 after stimulation with cytosolic dsRNA.
Acknowledgements
The authors thank M. Wentz (MD Anderson Cancer Center) and C. Harrod (Baylor Institute for Immunology Research) for critical reading of the manuscript and all colleagues in our laboratory for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Besch R, Poeck H, Hohenauer T, Senft D, Hacker G, Berking C, Hornung V, Endres S, Ruzicka T, Rothenfusser S, Hartmann G. Proapoptotic signaling induced by RIG-I and MDA-5 results in type I interferon-independent apoptosis in human melanoma cells. The Journal of clinical investigation. 2009;119:2399–2411. doi: 10.1172/JCI37155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant C, Fitzgerald KA. Molecular mechanisms involved in inflammasome activation. Trends in cell biology. 2009;19:455–464. doi: 10.1016/j.tcb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature immunology. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annual review of immunology. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenbrod T, Franchi L, Munoz-Planillo R, Kirschning CJ, Freudenberg MA, Nunez G, Dalpke A. Bacterial RNA mediates activation of caspase-1 and IL-1beta release independently of TLRs 3, 7, 9 and TRIF but is dependent on UNC93B. Journal of immunology (Baltimore, Md.: 1950) 2012;189:328–336. doi: 10.4049/jimmunol.1103258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nature immunology. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunological reviews. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang LJ, Zhang NN, Ding F, Li XY, Chen L, Zhang HX, Zhang W, Chen SJ, Wang ZG, Li JM, et al. RA-inducible gene-I induction augments STAT1 activation to inhibit leukemia cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1897–1902. doi: 10.1073/pnas.1019059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, Nunez G. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. The Journal of biological chemistry. 2006a;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006b;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature immunology. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, Liu YJ. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao PC, Chao LK, Chou JC, Dong WC, Lin CN, Lin CY, Chen A, Ka SM, Ho CL, Hua KF, et al. Lipopolysaccharide/adenosine triphosphate-mediated signal transduction in the regulation of NLRP3 protein expression and caspase-1-mediated interleukin-1beta secretion. Inflammation research: official journal of the European Histamine Research Society. 2012 doi: 10.1007/s00011-012-0555-2. [DOI] [PubMed] [Google Scholar]
- Liu F, Gu J. Retinoic acid inducible gene-I, more than a virus sensor. Protein & cell. 2011;2:351–357. doi: 10.1007/s13238-011-1045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I, Melero JA. A model for the generation of multiple A to G transitions in the human respiratory syncytial virus genome: predicted RNA secondary structures as substrates for adenosine deaminases that act on RNA. The Journal of general virology. 2002;83:1445–1455. doi: 10.1099/0022-1317-83-6-1445. [DOI] [PubMed] [Google Scholar]
- Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annual review of immunology. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nature immunology. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita M, Oshiumi H, Matsumoto M, Seya T. DDX60, a DEXD/H box helicase, is a novel antiviral factor promoting RIG-I-like receptor-mediated signaling. Molecular and cellular biology. 2011;31:3802–3819. doi: 10.1128/MCB.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nature immunology. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, Burckstummer T, Stefanovic A, Krieger S, Bennett KL, et al. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nature immunology. 2011;12:624–630. doi: 10.1038/ni.2048. [DOI] [PubMed] [Google Scholar]
- Rajan JV, Warren SE, Miao EA, Aderem A. Activation of the NLRP3 inflammasome by intracellular poly I:C. FEBS letters. 2010;584:4627–4632. doi: 10.1016/j.febslet.2010.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintahaka J, Wiik D, Kovanen PE, Alenius H, Matikainen S. Cytosolic antiviral RNA recognition pathway activates caspases 1 and 3. Journal of immunology (Baltimore, Md.: 1950) 2008;180:1749–1757. doi: 10.4049/jimmunol.180.3.1749. [DOI] [PubMed] [Google Scholar]
- Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science (New York, N.Y.) 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Muller M, Blander JM. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature. 2011;474:385–389. doi: 10.1038/nature10072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:1512–1517. doi: 10.1073/pnas.0912986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Shaw PJ, McDermott MF, Kanneganti TD. Inflammasomes and autoimmunity. Trends in molecular medicine. 2011;17:57–64. doi: 10.1016/j.molmed.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, et al. Oxidized Mitochondrial DNA Activates the NLRP3 Inflammasome during Apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, et al. IFI16 is an innate immune sensor for intracellular DNA. Nature immunology. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins C, Gale M., Jr. Recognition of viruses by cytoplasmic sensors. Current opinion in immunology. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nature immunology. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature immunology. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zhang NN, Shen SH, Jiang LJ, Zhang W, Zhang HX, Sun YP, Li XY, Huang QH, Ge BX, Chen SJ, et al. RIG-I plays a critical role in negatively regulating granulocytic proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10553–10558. doi: 10.1073/pnas.0804895105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kim T, Bao M, Facchinetti V, Jung SY, Ghaffari AA, Qin J, Cheng G, Liu YJ. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011a;34:866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature immunology. 2011b;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Lu N, Facchinetti V, Liu YJ. DHX9 Pairs with IPS-1 To Sense Double-Stranded RNA in Myeloid Dendritic Cells. Journal of immunology (Baltimore, Md.: 1950) 2011c;187:4501–4508. doi: 10.4049/jimmunol.1101307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nature immunology. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.