Abstract
Objective
To determine the pathogenesis of a patient born with congenital heart defects, who had appeared normal in prenatal screening.
Methods
In routine prenatal screening, G-banding was performed to analyse the karyotypes of the family and fluorescence in situ hybridization was used to investigate the 22q11.2 deletion in the fetus. After birth, the child was found to be suffering from heart defects by transthoracic echocardiography. In the following study, sequencing was used to search for potential mutations in pivotal genes. SNP-array was employed for fine mapping of the aberrant region and quantitative real-time PCR was used to confirm the results. Furthermore, other patients with a similar phenotype were screened for the same genetic variations. To compare with a control, these variations were also assessed in the general population.
Results
The child and his mother each had a region that was deleted in the beta-defensin repeats, which are usually duplicated in the general population. Besides, the child carried a SOX7-gene duplication. While this duplication was not detected in his mother, it was found in two other patients with cardiac defects who also had the similar deletion in the beta-defensin repeats.
Conclusion
The congenital heart defects of the child were probably caused by a SOX7-gene duplication, which may be a consequence of the partial haplotype of beta-defensin regions at 8p23.1. To our knowledge, this is the first congenital heart defect case found to have the haplotype of beta-defensin and the duplication of SOX7.
Introduction
The prevalence of congenital heart defects (CHD) has risen over the past few years, with a conservative estimate of 0.4∼5% of live births. Many changes may lead to cardiac malformations, including chromosomal abnormalities, gene mutations, copy number variations (CNV), or expression level changes [1]. One of the CHD hotspots is at 22q11.2, a deletion (occurring 1/4000 live births) which is already included in the prenatal diagnoses of cardiovascular anomalies using fluorescence in situ hybridization (FISH) [2]–[4]. Deletions or duplications in 8p23.1, caused by some formations of recurrent genomic rearrangement with unpredictable breakpoints, not only results in developmental delays, mental retardation and hypophrenia, but also have a close relationship with CHD [5]–[7]. GATA4 (next to 8p23.1) is thought to have a direct influence in cardiac morphogenesis and be a main cause of heart defects [8].
We describe here a different potential pathogenesis of CHD. A child with CHD had normal results of karyotype (G-banding), FISH (22q11.2) in prenatal screening and many pivotal genes like GATA4. We found that the child carries a partial haplotype of the beta-defensin region and a duplication of SOX7 which is normally underestimated in diagnoses.
Clinical Description
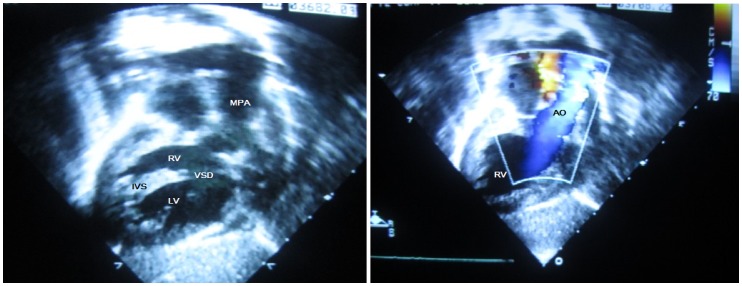
The child is currently a six-year-old male and was found to have a cardiac murmur during a health screening. He was diagnosed with complicated CHD when he was 4 months old. The symptoms included isolated dextrocardia, crisscross heart, double outlet of right ventricle (DORV), ventricular septal defect (VSD), atrial septal defect (ASD) and pulmonary hypertension (PH) (see Figure 1). Between the age of four months and six years old, he underwent Banding, Glenn and Fanton surgery and is under follow-up now. His mother had a cesarean section during labor due to the amniotic fluid II°contamination and the umbilical cord being wrapped around his neck. His Apar grade was 9/10 and weight was 3.6 kg. His parents were 29 when he was born; they are non-consanguineous and their karyotypes and cardiac morphology are normal. However, his sister (the proband) was diagnosed with tetralogy of Fallot (TOF) and died from anoxia when she was three years old. FISH results of the amniotic fluid were normal during his mother’s pregnancies, but she appeared to have the threat of miscarriages at early stages of these two pregnancies.
Figure 1. The echocardiography images of this case.
MPA: main pulmonary artery, RV: right ventricular, VSD: ventricular septal defect, IVS: inter ventricular septum, LV: left ventricular, AO: aortic artery.
Materials and Methods
Subjects
Samples from the child and his parents were collected from the clinic. 50 other patients with the similar defects (DORV, VSD, ASD, PH, TOF) were recruited from June 2008 to December 2009. None of them had a definite pathogenesis. 50 unrelated healthy Chinese people (of the Han ethnicity, like the patients) were enrolled as normal controls. All samples were collected in Xinhua Hospital and Shanghai Children’s Medical Center (SCMC). Diagnoses were confirmed by transthoracic echocardiography and karyotype, and extracardiac anomalies were also evaluated. Ethics committee of Xinhua Hospital specifically approved this study, and written informed consents were obtained from the participants or their parents. The individual in this manuscript has given written informed consent (as outlined in PLOS consent form) to publish these case detail.
Prenatal Screening: Karyotyping and FISH
The karyotypes were examined using G-banding at the 550 level. The peripheral blood (anticoagulation with heparin) and amniotic fluid were cultured in RPMI1640 culture medium with 20% calf serum (Invitrogen Gibco, USA) at 37°C in 5% CO2. Preparation of metaphase and conventional cytogenetics followed standard laboratory procedures. At least 20 banded metaphases with good chromosome separation were analyzed by experienced geneticists in each case. The commercially available locus specific probe kits N25 (D22S75) and TUPLE1 (HIRA) were purchased from Vysis (Downers Grove, IL, USA). Dual-color FISH was performed on metaphase spreads of the amniotic fluid cells, and 50 interphase nuclei were analyzed for the number of signals presented for each probe. Images were captured using an Olympus BX51 fluorescence microscope (Olympus, Japan). All of the screening was carried out in SCMC and followed the routine protocols [9].
After birth screening: Sequencing, SNP-array and quantitative real-time PCR (qPCR)
Peripheral blood samples were exsanguinated into an EDTA-anticoagulate tube. DNA was extracted using the QIAamp DNA Blood Midi Kit (Qiagen, Duesseldorf, Germany) by following the manufacturer’s instructions. Purified genomic DNA was resuspended in ddH2O for SNP-array analysis or in Tris-EDTA for other experiments, and DNA stocks were stored at −80°C.
Pivotal genes and regions with previously associated syndrome were identified through published articles. Multiple anomalies are influenced by the genes, including TBX1, TBX5, GATA4, GATA6, NKX2.5, SOX7 and FOG2 [10]. Information about the genes was searched from Build 36.1, which was released by the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) in March 2006. The primers concerning the sequencing were designed by our group using Primer 5. 0 software and listed in Table 1. PCR products were sequenced using the Sanger method on an ABI 3130 sequencer (Applied Biosystems). The sequence traces were aligned with the reference sequence in NCBI BLAST.
Table 1. Primers for sequencing TBX-1, TBX-5, GATA-4, GATA-6, SOX7, Nkx2.5 and Fog2.
| Gene | Primer-F (5′-3′) | Primer-R (5′-3′) |
| TBX1-1 | GGAGGAGCAGATGTCTCAGC | CCGGCTGCCTATACTCACTC |
| TBX1-2 | CCATGACGCCATAATCCTCT | TTGTGTTTTCTCCCCTTTGC |
| TBX1-3 | ACGCAGCTCTCGCATTTCT | GGCGGAGGATAGGTGTTAGG |
| TBX1-4 | GCCAAGCTCCCAGTTGAGTA | TCGCAGGTGCCTAAAGAGTT |
| TBX1-5 | GCAGCAGAGGGTTCAATCTC | TAGCCTCGCAGGGACTCTAA |
| TBX1-6 | AGTGACCCAGCCTCATCTTG | GTCTAAGCGGACCCACTGTC |
| TBX1-78 | CTTGGTGCGCTTCTCCTAAC | AGAGGGCCGAGGAGTGAG |
| TBX1-9A | GTTGGGAGATGCAGTCCTGT | TACTGGACAGCAGCACTTGG |
| TBX1-9B | GATGGTGTGTGAGGCTGATG | CTTGCATGCACACTTGACCT |
| TBX1-9C | GGCCAAGAGCCTTCTCTCC | ACTGGGGAACCGGATACTTC |
| TBX5-1 | CACTGAGTTATCGCATCCC | CACGAAGCCATTCTTATTT |
| TBX5-2 | GTGGGAGCTAGTTGGATAGGC | CGAACAAGATGCGGTTTGAC |
| TBX5-3 | CAAACTGCTCCCTCCTGT | AAGTAGATGGCAATACGCTA |
| TBX5-4 | ACTGTGGGTTCAAGTGGT | AGGATCTATCTTTCGCTCT |
| TBX5-5 | CCGCTTCCACGTTTCTCCAGG | TCTGAGCCTCCGCTTTCTCATCT |
| TBX5-6 | CTCACCTGGTGCGTGAACTGAA | GGTAGAGGCAGAAAGCGACGAAAG |
| TBX5-7 | AATGAAATCCCTGGCCCCTTTT | TTGTCCCCACCCCAGCACC |
| TBX5-8 | GAAGTGGTGGGTCCCGTTGA | TGGAGGGAGGTGCTGGGTTG |
| TBX5-9 | CTGGTTCAGCCACTCAGGAAATCT | CTCCAGCCTGGGTGATAGAGCA |
| TBX5-10A | TTGTATTCAGAATGGCGGTTAGGG | AAGTGAGCGGAGAAGTGCTGGTAG |
| TBX5-10B | TGCCCAGCCTAGAGGACATCAG | GGGGAGTAGCGTGAATGTGGC |
| TBX5-10C | CAAGGTCGCTGGATGCT | TTCGGCTTTCAGTAAACA |
| TBX5-10D | CCAACCTTCCAAACCTCCATCA | ACAACCTCTTCCTGTTTCCTCCAA |
| GATA4-1 | GACTCCCACAGGCCAGTCAG | GACAAGCAAAGGCGGAGAAG |
| GATA4-2 | ATTTGAAGCGTGGAAGAAGCAAC | CCTCGACAGGGCTCAAGACG |
| GATA4-3 | TTGTTTCTGTGCGCTCTAG | TCTCACCCACGTAATCCC |
| GATA4-4 | GAGTTAGGTGCCGTCACAGG | GGAAGAGGCCAGCAAAGTAG |
| GATA4-5 | CTTAGGTGTTGCCTTCTCG | TTTGCTGGGCTCTTCATC |
| GATA4-6 | GTTTGTCCCTGCCGCTGAT | GCTGCAAGTCCCACCCAGTA |
| GATA4-7A | GGTCATAGCCCTGGTTGTAT | AGGCTGTGCTGTGGTGG |
| GATA4-7B | CTGCATCCCTAATACCAAATC | AACCTCCCAGTGAAGACCA |
| GATA6-1 | CCGTCCCCTCCCCACCCTCTTT | GAGATCGCGCGAGGAGGAAGCA |
| GATA6-2A | TGGAGGCGAGGTAGCGTGCAG | AACTGAGCAGCAGCGAGCGGG |
| GATA6-2B | CTGAGCCCCTTCGCACCCGAG | CCTAGGGCGGGCTGGGAGAGT |
| GATA6-2C | CACCTGCAGGGGTCGGGCAGT | AAACAGGGCCCGAGTGGAGCA |
| GATA6-3 | CTACTGGGGCGCTCCGGGTGT | AGCGGGTGGGCGTTGGAACAG |
| GATA6-4 | TGGAGAAGAAACCAGGGATGA | TGCATTCAAATTTTTCACTTGAG |
| GATA6-56 | CGGCCGCCAAATTCTTTTA | AACCATAAAAAAATGATACCGATCT |
| GATA6-7 | TGGCCAGGGTCAGGTCAGTGG | GAGTGGCCCAAGCGCCCAGTT |
| SOX7-1 | CCGCTCTGAATCCTGGGCACC | ACTCCCTCCCTCCGTCCTCCTCC |
| SOX7-2A | AGTTAGCCATACTGGTTAATTTCTC | CCTCAGTGGGCATGTTCC |
| SOX7-2B | GACGGCTCCTCTGCCACTCA | CCATCTCCTGCCTATTACTCCC |
| SOX7-2C | CTGTGGGACCCGTTGGTGT | CATGGCCTCCTCTGCCTTGT |
| NKX2.5-1 | GTGACACGAAACTGCTCATC | ACAACACCAGGCATCTTACA |
| NKX2.5-2A | AAGTCACCGTCTGTCTCCCTC | GCTCTGAACCGCATTCAAGTC |
| NKX2.5-2B | AAGCGCCGCAAGCTGAA | GGCCTCAATCCCTACGGTT |
| FOG2-1 | TCATCTCCGAACGTGAATCCG | TGGGCAATAATCCCACCAACTC |
| FOG2-2 | CGGATGTGGCATTATCT | TTACTCATGTCCCTCGA |
| FOG2-3 | GAGGGTGTGAATGTGAAAGAG | CAAGCAGAGGTAGCACTTTGG |
| FOG2-4 | GAGGTGGCTGCTGATAAAGTAC | GTTTCTGTCTAAATTCTGCGTAT |
| FOG2-5 | GGTTTGGGAGATTTAGTTG | AAGATATTAGTCAAGCCACTC |
| FOG2-6 | CATGAGAAGGTGCTATGGAC | GATGACGAGTTAGTGGGTG |
| FOG2-7 | AATGGACAGCAGCAAAT | CTGGAGCAACAGAAGAAAC |
| FOG2-8A | GAAAAGGTCCCTGTCATTC | CAGGTAGGCACATCTCATAC |
| FOG2-8B | CTACACGCCACGACCCT | CATCTTGTTTCAGTCCACC |
| FOG2-8C | GCTTCCTCAAATGGGTGT | CAGAGCCTGATTATCCAAGA |
| FOG2-8D | GTCACAATACAGAAAAGCAT | GGTGCCATTTGGAAACTA |
SNP-array (Illumina Omin1), which had an average resolution of 2. 5 kb, was performed following the manufacturer’s protocol for fine mapping of the potential aberrant regions in chromosomes. Data was analyzed using the Illumina Kayostudio software v1.3 (Build 36.1, CNV Plugin V3.0) in the recommended setting to identify only those regions larger than 75 kb comprising at least 50 contiguous markers. We deposited details of this experiment in dbVar (http://www.ncbi.nlm.nih.gov/dbvar) and got the accession number GSE48386 (NCBI GEO). The results were compared to cases in the Database of Genomic Variants (DGV, http://dgvbeta.tcag.ca/dgv/app/home ) and Online Mendelian Inheritance in Man (OMIM, http://www.ncbi.nlm.nih.gov/omim/) to distinguish common CNVs from likely causal CNVs. Segments that have a strong association with CHD were confirmed by qPCR.
Small portions of genomic DNA of healthy people were mixed to form a DNA pool serving as the normal control in qPCR. The gene COL1A1, which has few variations, was used as the control gene in qPCR. At least three selected genes in the targeted segments were searched in the UCSC Genome Browser (http://genome.ucsc.edu/). These genes were quantified to determine the copy number (CN), as were pivotal genes nearby. When the selected genes showed results different from the SNP-array, qPCR was trusted. When the same aberrant results were seen, the segments were next quantified in the genomic DNA of the parents and normal controls. They were considered to be common CNVs if detected in the normal controls. Otherwise, they were defined as potential CNVs if found to be parental, or causal CNVs if didn’t. These CNVs were inspected in Database of Chromosomal Imbalance and Phenotype in Humans using Ensembel Resources (DECIPHER, https://decipher.sanger.ac.uk/) and then verified in the samples of 50 patients we collected.
Results
Karyotyping, FISH and Sequencing
No obvious structural or numerical abnormalities were found on the metaphase spreads of the child and his parents by karyotype or FISH. Besides, the results of sequencing revealed no functional mutations in the coding sequences of TBX1, TBX5, GATA4, GATA6, NKX2.5, SOX7 and FOG2 genes.
Genotyping and CN Determination
SNP-array showed us the child carried only one copy in each beta-defensin gene cluster (DEFB) at chr8∶7230125–7342754 and chr8∶7677945–7835713. Another deletion was found in the olfactory receptor (OR) gene cluster (chr8∶11971611–12054845 and chr8∶12273531–12392405), which is much smaller and has few genes (see Figure 2).
Figure 2. The copy number analysis of chromosome 8.
(A) Ideogram of chromosome 8. (B) Results of SNP-array integrated with CNV probes. Blue spots, B allele freq; Red line, smoothed Log R; Genes were annotated: Red, deletion; Black, normal; Green, duplication. Regions of REPD and REPP were annotated in brown bar.
QPCR was used to verify the CNs of DEFB104, SPAG11 and DEFB4 in the DEFB deletion. Given that chromosomal rearrangements frequently occur in this segment and likely affect adjacent segments, verification of genes like SOX7 and GATA4 is necessary. Interestingly, the CN of GATA4 was normal, while the CN of SOX7 was 2.5 times higher than that of the normal controls. Another primer-pair designed to quantify SOX7 CN showed the same result. Verification of some other nearby genes (TNKS, C8orf74, PINX1 for SOX7 and MTMR9, BLK, CSTBC for GATA4) to make the regions distinct in the child. They turned out to be normal except for MTMR9 which has no obvious relationship to heart defects [11]–[13]. Primers’ efficiencies in qPCR were tested qualified and the sequences were listed in Table 2. We detected the abnormal loci (SOX7, MTMR9, DEFB clusters, GATA4) in the child’s parents and other 100 samples (50 patients and 50 controls). The results showed that the deletion existed in the parents. Moreover, In other 2 patients there are duplications (SOX7) and deletions (DEFB clusters). Other ratios were normal, see Table 3.
Table 2. Primers of the genes in qPCR.
| Gene | Primer-F (5′-3′) | Primer-R (5′-3′) |
| COL1A1 | GGGGGAACAAGGCTGTCT | TCCTGGGGTTCAGACCAA |
| DEFB104 | AGCATTCTCTATCCCCCTCC | CATGCATAGGTGTTGGGACA |
| SPAG11 | AGAAGTCATCCTGGAGCACA | GTGACGGACGGGAGCAAT |
| DEFB4 | AGTTCTTACACGCTGTTTGC | AATCCGCATCAGCCACA |
| TNKS | TCAAAGCAAACCCATATTTTACTC | GCCAGTTAAAATAAAGCCATGTAG |
| C8orf74 | TCGCCATCTTGGACCTGA | TTTCCTTTGCTCGCTCTTT |
| SOX7 | GGGACATGGATCGCAATGAA | CAGCCAGGACGGAGATGAGG |
| SOX7-2 | GCGACTCTGGACAAGTCACATC | TTATCTCACCGAATCTTCACAACA |
| PINX1 | AGGTTCCAGTTCCAGGGTC | TTTGGGCTTCAGGGTGA |
| MTMR9 | CACCAAGCAGAAGTGGGAGG | TGCCCAGAAATGTTCCAAAC |
| BLK | CTGCTCATGGTCCTTCCTC | TTGGCAATGCTTCAGTGGT |
| GATA4 | GACATAATCACTGCGTAATCTTC | CTCCCTCCAGTCCCATCA |
| CSTBC | ATAACCAAGATGCTACC | CTTCTAGTTTGCTCTATACC |
Table 3. CNs of genes in the family, other patients and normal controls.
| Gene | Child | Mother | Father | Patient 1 | Patient 2 | Others |
| DEFB104 | 0.35 | 0.58 | 0.76 | 0.59 | 0.67 | n |
| SPAG11 | 0.37 | 0.49 | 0.75 | 0.54 | 0.65 | n |
| DEFB4 | 0.34 | 0.53 | 0.72 | 0.58 | 0.67 | n |
| TNKS | n | n | n | n | n | \ |
| C8orf74 | n | n | n | n | n | \ |
| SOX7 | 2.5 | n | n | 1.53 | 1.64 | n |
| PINX1 | n | n | n | n | n | \ |
| MTMR9 | 1.52 | n | n | n | n | n |
| BLK | n | n | n | n | n | \ |
| GATA4 | n | n | n | n | n | n |
| CSTBC | n | n | n | n | n | \ |
Left: Genes we detected. Right: CNs of the family, two patients, and other samples(50 controls and 48 patients); n: normal ratio comparing with the DNA pool. \: no data.
Discussion
The Repeat Regions
Many antimicrobial beta-defensin genes are located at 8p23.1, such as DEFB104, DEFB105, and DEFB106. The DEFB clusters are polymorphic in CN from 2 to 12 (4 is the average in the Chinese population), and the single copy has a frequency of 0. 2%∼0. 7% [14]. Rare report of single copy means that this CN haplotype may induce fatal diseases in early development [14]–[17]. Individual DEFB CN has been suggested as a genetic risk factor for psoriasis, ANCA-associated small vasculitis, Crohn’s disease and prostate cancer [5], [18], [19]. Two DEFB clusters, at 7.16–7.39 Mb and 7.67–7.89 Mb respectively, were separated by a gap containing the olfactory receptor (OR) gene cluster. The whole region is collectively named the REPD (distal repeat), and at a distance of 4.7 Mb away is another smaller repeat unit, the REPP (proximal repeat) [14], [16], [20]–[22]. S. Giglio reported that haploinsufficiency of the region between WI-8372 (6.36–6.57 Mb) and D8S1825 (8.86–9.06 Mb) was associated with congenital heart defects using short-tandem repeat (STR) analysis [23], which suggested that the REPD may be related to CHD. However, Chen found that the microdeletion (chr8∶7227000–7916187) was insufficient to induce heart defects [24].
Recombination and Heart Defects
Florida reported some kinds of 8p23.1 nonrandom recombination that was consistently of maternal origin [25]. For three of the described recombinations, inv dup (8p), der (8p) and del (8p), Giglio considered REPD and REPP as the substrates for the formation of non-allelic homologous recombinations. Common 8p23 polymorphic inversion has a frequency in general populations of 25.6% in European [21] and 34% in Japanese [22]. However, inv dup (8p) was suggested as an independent risk factor for abnormal recombinations, which lead to several diseases, including mental retardation, facial dysmorphisms, brain defects and some other syndromes [7], [25]–[27]. Studies mentioning heart defects, always refer to the transcription factors SOX7 and GATA4, which are located between REPD and REPP in the polymorphic inversion region. Both deletion and duplication of these genes are associated with recombinations [28]–[31]. That is to say, the recombinations would highly increase the mutation risk of SOX7 and/or GATA4, thereby inducing heart defects [21]. In clinic, when a patient with heart defects due to abnormal 8p23 was found to be carrying a duplication of SOX7 and GATA4, the dosage of GATA4 was always defined as likely the most responsible cause [32], [33]. While SOX7 alterations have been seldom reported in patients with heart defects, SOX7 is a tumor suppressor in many organs [34], [35]. Whether CN variation of SOX7 (without changes in GATA4) leads to heart defects in patients needs to be further studied.
SOX7 in Heart Development
SOX7, which belongs to the family of proteins equipped with SRY-type HMG boxes, was first identified in Xenopus and in mouse [36], [37]. SOX7 has the ability to select, bind and bend DNA chains, and interact with partner proteins (MEF2C, β-catenin) and growth factor signaling pathways (VEGFs). Previous research demonstrated that SOX7 was widespread and played essential functions during cardiovascular development in zebrafish, frogs mice and human, while SOX18 has the same role in cardiogenesis, allowing it to substitute when SOX7 is insufficient [38], [39]. Researchers injected SOX7 RNAs into the animal caps of Xenopus cell and found it would enhance the expression of MHCa, TBX5 and regulate the Xnr genes and Nkx2.5. With the increasing of SOX7 RNAs in the later stage when SOX7 should have disappeared, the Xenopus showed much more defective embryos [40]–[44]. Besides, Wnt11 inhibits canonical Wnt signaling and acts through the protein kinase C (PKC) and jun kinase (JnK) to induce cardiogenesis [39], [40], [45].
In the experiments in mouse embryonic stem cells, SOX7 was found to dictate cell fate of cardiovascular progenitors: Flk-1+ progenitors with increased expression levels of SOX7 are associated with a vascular phenotype. However, Flk-1+ progenitors with decreased expression levels of SOX7 are associated with a cardiogenic phenotype [46], [47]. Some researches directly pointed out that SOX7 is only transiently expressed at the onset of hematopoietic differentiation, and the sustained expression will block the specification and maturity [48], [49]. In addition, researches in which SOX7 expression vectors were transferred to the Human embryonic stem cells proved that SOX7 over-expression will up-regulated the expression of GATA4 and GATA6 [50], [51]. Besides, with the rising expression of SOX7, cardiac differentiation would be significantly reduced [52]. SOX7 over-expression was normally described as a destroyer in the differentiations of cells or model animals but without clinic records. This is mainly because suitable specimens were difficult to find, likely because SOX7 expresses at a very early stage before we can detect it, and serious neonatal defects can rise from SOX7 up-regulation.
Our Case
The abnormal region in the child’s chromosome 8 is a deletion of repeat regions (1 copy) and duplication of SOX7 (5 copies). The small regions and only symptom suggest the specific loci related to heart development.
Repeat regions: When compared to the normal controls, the DEFB CNs in this family showed much lower ratios of 1/3 (child) and 1/2 (mother) and 2/3 (father). That means the CN of the parents are as low as 2-0 and 3-1. As a result, the child carries only one copy (1-0), which has never been previously reported in the Chinese population [14]. DEFB CNs in two other patients were also lower than ordinary, at 1/2 and 3/5, respectively. Although this is not enough evidence to argue the DEFB clusters are the causes of CHD, it is noticeable that 94.4% of patients carrying interstitial deletions have cardiac-malformation children [53]. The loss of the entire REPP and most of the REPD means that recombinations happened before. REPP (Losing or gain) in DGV may won’t lead to diseases, while recombinations in DECIPHER usually include large segments of duplication or deletion which have brought about many diseases (Table 4). Unusually, the affected range in our case was so limited (shorter than 65 kb) that it was not detected by SNP-array and may cause just the symptom of heart defects.
SOX7: When defects are found, the stage of heart development usually has passed already. As it is difficult to detect the precise time of expression, we have to analyze genotype-phenotype correlations. The symptoms of the child (DROV, ASD, VSD, PH, etc.), his elder sister (TOF) and another patient (TOF) may due primarily to three genes located downstream of SOX7 (GATA4, TBX5, Nkx2. 5), but only GATA4 can cause all of the symptoms [10], [53]–[55]. Without an abnormality in GATA4 (sequence and CN), SOX7 became the most likely candidate gene in our case. Duplication of SOX7 alone did not exist in the 50 healthy normal controls, the general population in DGV and patients in DECIPHER. However, it is the only common abnormality in the child and two patients when REPP seems merely has the potential to impact the recombinations (Table 4).
Prenatal screening: Since screening for CHD will improve the quality of public health services and reduce health care costs, it has been taken seriously in many countries [56]–[58]. The most common methods of routine CHD prenatal screening, including echocardiographic examination, may reduce the incidence of CHD but still leave the pathogenesis unexplained [59], [60]. Normal karyotype and negative results of FISH make genetic counseling difficult. Prenatal molecular diagnosis for CHD mainly aimed at mutations of the pivotal genes (such as GATA4, GATA6, TBX5 and so on) and CNVs of 22q11. Some MLPA kits (http://www.mlpa.com/) were launched to detect the CNVs of the genes and chromosomes (but without any SOX7 probes) [9], [61]. CGH array and SNP array have also been performed as a comprehensive method to analyze the genetics of heart defects, but the probe numbers and false negative results are issues that should be taken seriously. In our study, however, we found three patients with SOX7 duplication in an average-sized-patient group. That there are few reports about SOX7 duplication in CHD patients may be because of being overlooked in diagnosis. Copy number of SOX7, as a single locus for CHD and a reference locus for GATA4, has the potential to be a potential hotspot in the future.
Table 4. Comparison among the subjects’ genotypes and symptoms among samples.
| Sample | Cardiac defects | REPD | SOX7 | GATA4 |
| Father | None | − | n | n |
| Mother | None | − | n | n |
| Child | DORV,ASD,VSD,PS,etc | − | + | n |
| patient 1 | TOF | − | + | n |
| patient 2 | SV | − | + | n |
| general people in DGV | none | −/+ | n | n |
| patients in DECIPHER | ASD,VSD,PS,etc | −/+ | −/+ | −/+ |
+: duplication, −: deletion, n: normal. Losing or gain in the whole segments leads to diseases. REPD variations may not leads to diseases but impacts SOX7 which is related to CHD.
Conclusion
Our study suggests that the loss of REPD or nearby regions may raise the risk of heart diseases by impacting the following genes, such as SOX7. To the best of our knowledge, we provide the first published evidence that the duplication of SOX7 has a strong association with heart defects using clinical specimens. Further research should be carried out to clarify and confirm the SOX7-CHD mechanism. Updated guidelines including SOX7 are needed for heart defects in prenatal screening.
Funding Statement
The project was funded by the National Natural Science Foundation of China (81070135/H0204) (http://www.nsfc.gov.cn/); National Basic Research Program of China (2010CB529500) (http://www.most.gov.cn/); Innovative Research Team of Shanghai Municipal Education Commission the Shanghai University Innovation Team (Phase I): Pediatrics (http://www.shmec.gov.cn/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Pierpont ME, Basson CT, Benson DW Jr, Gelb BD, Giglia TM, et al. (2007) Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 115: 3015–3038. [DOI] [PubMed] [Google Scholar]
- 2. Momma K (2010) Cardiovascular anomalies associated with chromosome 22q11.2 deletion syndrome. Am J Cardiol 105: 1617–1624. [DOI] [PubMed] [Google Scholar]
- 3. Schinke M, Izumo S (2001) Deconstructing DiGeorge syndrome. Nat Genet 27: 238–240. [DOI] [PubMed] [Google Scholar]
- 4. Momma K (2007) Cardiovascular anomalies associated with chromosome 22q11.2 deletion. Int J Cardiol 114: 147–149. [DOI] [PubMed] [Google Scholar]
- 5. Yu S, Fiedler S, Stegner A, Graf WD (2010) Genomic profile of copy number variants on the short arm of human chromosome 8. Eur J Hum Genet 18: 1114–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pabst B, Arslan-Kirchner M, Schmidtke J, Miller K (2003) The application of region-specific probes for the resolution of duplication 8p: a case report and a review of the literature. Cytogenet Genome Res 103: 3–7. [DOI] [PubMed] [Google Scholar]
- 7. Giorda R, Ciccone R, Gimelli G, Pramparo T, Beri S, et al. (2007) Two classes of low-copy repeats comediate a new recurrent rearrangement consisting of duplication at 8p23.1 and triplication at 8p23.2. Hum Mutat 28: 459–468. [DOI] [PubMed] [Google Scholar]
- 8. Devriendt K, Matthijs G, Van Dael R, Gewillig M, Eyskens B, et al. (1999) Delineation of the critical deletion region for congenital heart defects, on chromosome 8p23.1. Am J Hum Genet 64: 1119–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu YJ, Wang J, Xu R, Zhao PJ, Wang XK, et al. (2011) Detecting 22q11.2 deletion in Chinese children with conotruncal heart defects and single nucleotide polymorphisms in the haploid TBX1 locus. BMC Med Genet 12: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nemer M (2008) Genetic insights into normal and abnormal heart development. Cardiovasc Pathol 17: 48–54. [DOI] [PubMed] [Google Scholar]
- 11. Yanagiya T, Tanabe A, Iida A, Saito S, Sekine A, et al. (2007) Association of single-nucleotide polymorphisms in MTMR9 gene with obesity. Hum Mol Genet 16: 3017–3026. [DOI] [PubMed] [Google Scholar]
- 12. Goh XY, Rees JR, Paterson AL, Chin SF, Marioni JC, et al. (2011) Integrative analysis of array-comparative genomic hybridisation and matched gene expression profiling data reveals novel genes with prognostic significance in oesophageal adenocarcinoma. Gut 60: 1317–1326. [DOI] [PubMed] [Google Scholar]
- 13. Guo L, Martens C, Bruno D, Porcella SF, Yamane H, et al. (2013) Lipid phosphatases identified by screening a mouse phosphatase shRNA library regulate T-cell differentiation and Protein kinase B AKT signaling. Proc Natl Acad Sci U S A 110: E1849–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hollox EJ (2008) Copy number variation of beta-defensins and relevance to disease. Cytogenet Genome Res 123: 148–155. [DOI] [PubMed] [Google Scholar]
- 15. Zhou XJ, Cheng FJ, Lv JC, Luo H, Yu F, et al. (2012) Higher DEFB4 genomic copy number in SLE and ANCA-associated small vasculitis. Rheumatology (Oxford) 51: 992–995. [DOI] [PubMed] [Google Scholar]
- 16. Abu Bakar S, Hollox EJ, Armour JA (2009) Allelic recombination between distinct genomic locations generates copy number diversity in human beta-defensins. Proc Natl Acad Sci U S A 106: 853–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hollox EJ, Barber JC, Brookes AJ, Armour JA (2008) Defensins and the dynamic genome: what we can learn from structural variation at human chromosome band 8p23.1. Genome Res 18: 1686–1697. [DOI] [PubMed] [Google Scholar]
- 18. Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, et al. (2008) Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet 40: 23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Taudien S, Groth M, Huse K, Petzold A, Szafranski K, et al. (2010) Haplotyping and copy number estimation of the highly polymorphic human beta-defensin locus on 8p23 by 454 amplicon sequencing. BMC Genomics 11: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hollox EJ, Armour JA, Barber JC (2003) Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am J Hum Genet 73: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Giglio S, Broman KW, Matsumoto N, Calvari V, Gimelli G, et al. (2001) Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet 68: 874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sugawara H, Harada N, Ida T, Ishida T, Ledbetter DH, et al. (2003) Complex low-copy repeats associated with a common polymorphic inversion at human chromosome 8p23. Genomics 82: 238–244. [DOI] [PubMed] [Google Scholar]
- 23. Giglio S, Graw SL, Gimelli G, Pirola B, Varone P, et al. (2000) Deletion of a 5-cM region at chromosome 8p23 is associated with a spectrum of congenital heart defects. Circulation 102: 432–437. [DOI] [PubMed] [Google Scholar]
- 24. Chen CP, Wang TH, Chen YJ, Chang TY, Liu YP, et al. (2007) Prenatal diagnosis of Fryns syndrome associated with a microdeletion at 8p23.1. Prenat Diagn 27: 967–969. [DOI] [PubMed] [Google Scholar]
- 25. Floridia G, Piantanida M, Minelli A, Dellavecchia C, Bonaglia C, et al. (1996) The same molecular mechanism at the maternal meiosis I produces mono- and dicentric 8p duplications. Am J Hum Genet 58: 785–796. [PMC free article] [PubMed] [Google Scholar]
- 26. Seltmann M, Harrington P, Ponder BA (2000) A case of inv dup(8p) with early onset breast cancer. J Med Genet 37: 70–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiao B, Zhang JM, Ji X, Jiang WT, Hu J, et al. (2011) [Two cases of partial trisomy 8p derived from paternal reciprocal translocation or maternal insertion translocation: clinical features and genetic abnormalities]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 28: 247–250. [DOI] [PubMed] [Google Scholar]
- 28. Ballarati L, Cereda A, Caselli R, Selicorni A, Recalcati MP, et al. (2011) Genotype-phenotype correlations in a new case of 8p23.1 deletion and review of the literature. Eur J Med Genet 54: 55–59. [DOI] [PubMed] [Google Scholar]
- 29. Tomita-Mitchell A, Mahnke DK, Struble CA, Tuffnell ME, Stamm KD, et al. (2012) Human gene copy number spectra analysis in congenital heart malformations. Physiol Genomics 44: 518–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han X, Zhang JM, Jiang WT, Hu Q, Tao J (2010) [Cytogenetic and molecular genetic study of a case with 8p inverted duplication deletion syndrome]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 27: 361–366. [DOI] [PubMed] [Google Scholar]
- 31. Barber JC, Maloney V, Hollox EJ, Stuke-Sontheimer A, du Bois G, et al. (2005) Duplications and copy number variants of 8p23.1 are cytogenetically indistinguishable but distinct at the molecular level. Eur J Hum Genet 13: 1131–1136. [DOI] [PubMed] [Google Scholar]
- 32. Barber JC, Bunyan D, Curtis M, Robinson D, Morlot S, et al. (2010) 8p23.1 duplication syndrome differentiated from copy number variation of the defensin cluster at prenatal diagnosis in four new families. Mol Cytogenet 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barber JC, Maloney VK, Huang S, Bunyan DJ, Cresswell L, et al. (2008) 8p23.1 duplication syndrome; a novel genomic condition with unexpected complexity revealed by array CGH. Eur J Hum Genet 16: 18–27. [DOI] [PubMed] [Google Scholar]
- 34. Li B, Ge Z, Song S, Zhang S, Yan H, et al. (2012) Decreased expression of SOX7 is correlated with poor prognosis in lung adenocarcinoma patients. Pathol Oncol Res 18: 1039–1045. [DOI] [PubMed] [Google Scholar]
- 35. Guo L, Zhong D, Lau S, Liu X, Dong XY, et al. (2008) Sox7 Is an independent checkpoint for beta-catenin function in prostate and colon epithelial cells. Mol Cancer Res 6: 1421–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shiozawa M, Hiraoka Y, Komatsu N, Ogawa M, Sakai Y, et al. (1996) Cloning and characterization of Xenopus laevis xSox7 cDNA. Biochim Biophys Acta 1309: 73–76. [DOI] [PubMed] [Google Scholar]
- 37. Taniguchi K, Hiraoka Y, Ogawa M, Sakai Y, Kido S, et al. (1999) Isolation and characterization of a mouse SRY-related cDNA, mSox7. Biochim Biophys Acta 1445: 225–231. [DOI] [PubMed] [Google Scholar]
- 38. Francois M, Koopman P, Beltrame M (2010) SoxF genes: Key players in the development of the cardio-vascular system. Int J Biochem Cell Biol 42: 445–448. [DOI] [PubMed] [Google Scholar]
- 39. Takash W, Canizares J, Bonneaud N, Poulat F, Mattei MG, et al. (2001) SOX7 transcription factor: sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Res 29: 4274–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang C, Basta T, Klymkowsky MW (2005) SOX7 and SOX18 are essential for cardiogenesis in Xenopus. Dev Dyn 234: 878–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang C, Basta T, Fawcett SR, Klymkowsky MW (2005) SOX7 is an immediate-early target of VegT and regulates Nodal-related gene expression in Xenopus. Dev Biol 278: 526–541. [DOI] [PubMed] [Google Scholar]
- 42. Fawcett SR, Klymkowsky MW (2004) Embryonic expression of Xenopus laevis SOX7. Gene Expr Patterns 4: 29–33. [DOI] [PubMed] [Google Scholar]
- 43. Grepin C, Nemer G, Nemer M (1997) Enhanced cardiogenesis in embryonic stem cells overexpressing the GATA-4 transcription factor. Development 124: 2387–2395. [DOI] [PubMed] [Google Scholar]
- 44. Gove C, Walmsley M, Nijjar S, Bertwistle D, Guille M, et al. (1997) Over-expression of GATA-6 in Xenopus embryos blocks differentiation of heart precursors. EMBO J 16: 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pandur P, Lasche M, Eisenberg LM, Kuhl M (2002) Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature 418: 636–641. [DOI] [PubMed] [Google Scholar]
- 46. Yamauchi F, Okada M, Kato K, Jakt LM, Iwata H (2007) Array-based functional screening for genes that regulate vascular endothelial differentiation of Flk1-positive progenitors derived from embryonic stem cells. Biochim Biophys Acta 1770: 1085–1097. [DOI] [PubMed] [Google Scholar]
- 47. Nelson TJ, Chiriac A, Faustino RS, Crespo-Diaz RJ, Behfar A, et al. (2009) Lineage specification of Flk-1+ progenitors is associated with divergent Sox7 expression in cardiopoiesis. Differentiation 77: 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gandillet A, Serrano AG, Pearson S, Lie ALM, Lacaud G, et al. (2009) Sox7-sustained expression alters the balance between proliferation and differentiation of hematopoietic progenitors at the onset of blood specification. Blood 114: 4813–4822. [DOI] [PubMed] [Google Scholar]
- 49. Costa G, Mazan A, Gandillet A, Pearson S, Lacaud G, et al. (2012) SOX7 regulates the expression of VE-cadherin in the haemogenic endothelium at the onset of haematopoietic development. Development 139: 1587–1598. [DOI] [PubMed] [Google Scholar]
- 50. Seguin CA, Draper JS, Nagy A, Rossant J (2008) Establishment of endoderm progenitors by SOX transcription factor expression in human embryonic stem cells. Cell Stem Cell 3: 182–195. [DOI] [PubMed] [Google Scholar]
- 51. Paige SL, Osugi T, Afanasiev OK, Pabon L, Reinecke H, et al. (2010) Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One 5: e11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bauwens CL, Song H, Thavandiran N, Ungrin M, Masse S, et al. (2011) Geometric control of cardiomyogenic induction in human pluripotent stem cells. Tissue Eng Part A 17: 1901–1909. [DOI] [PubMed] [Google Scholar]
- 53. Wat MJ, Shchelochkov OA, Holder AM, Breman AM, Dagli A, et al. (2009) Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A 149A: 1661–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Futaki S, Hayashi Y, Emoto T, Weber CN, Sekiguchi K (2004) Sox7 plays crucial roles in parietal endoderm differentiation in F9 embryonal carcinoma cells through regulating Gata-4 and Gata-6 expression. Mol Cell Biol 24: 10492–10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wolf M, Basson CT (2010) The molecular genetics of congenital heart disease: a review of recent developments. Curr Opin Cardiol 25: 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Olney RS, Botto LD (2012) Newborn screening for critical congenital heart disease: essential public health roles for birth defects monitoring programs. Birth Defects Res A Clin Mol Teratol 94: 965–969. [DOI] [PubMed] [Google Scholar]
- 57. Slodki M, Szymkiewicz-Dangel J, Tobota Z, Seligman NS, Weiner S, et al. (2012) The Polish National Registry for Fetal Cardiac Pathology: organization, diagnoses, management, educational aspects and telemedicine endeavors. Prenat Diagn 32: 456–460. [DOI] [PubMed] [Google Scholar]
- 58. Sharland G (2012) Fetal cardiac screening and variation in prenatal detection rates of congenital heart disease: why bother with screening at all? Future Cardiol 8: 189–202. [DOI] [PubMed] [Google Scholar]
- 59. Li H, Meng T, Shang T, Guan YP, Zhou WW, et al. (2007) Fetal echocardiographic screening in twins for congenital heart diseases. Chin Med J (Engl) 120: 1391–1394. [PubMed] [Google Scholar]
- 60. Yang XY, Li XF, Lu XD, Liu YL (2009) Incidence of congenital heart disease in Beijing, China. Chin Med J (Engl) 122: 1128–1132. [PubMed] [Google Scholar]
- 61. Mademont-Soler I, Morales C, Soler A, Clusellas N, Margarit E, et al. (2012) MLPA: a prenatal diagnostic tool for the study of congenital heart defects? Gene 500: 151–154. [DOI] [PubMed] [Google Scholar]