Abstract
Caloric/dietary restriction (CR/DR) can promote longevity and protect against age-associated disease across species. The molecular mechanisms coordinating food intake with health-promoting metabolism are thus of significant medical interest. We report that conserved Caenorhabditis elegans microRNA-80 (mir-80) is a major regulator of the DR state. mir-80 deletion confers system-wide healthy aging, including maintained cardiac-like and skeletal muscle-like function at advanced age, reduced accumulation of lipofuscin, and extended lifespan, coincident with induction of physiological features of DR. mir-80 expression is generally high under ad lib feeding and low under food limitation, with most striking food-sensitive expression changes in posterior intestine. The acetyltransferase transcription co-factor cbp-1 and interacting transcription factors daf-16/FOXO and heat shock factor-1 hsf-1 are essential for mir-80(Δ) benefits. Candidate miR-80 target sequences within the cbp-1 transcript may confer food-dependent regulation. Under food limitation, lowered miR-80 levels directly or indirectly increase CBP-1 protein levels to engage metabolic loops that promote DR.
Author Summary
Dietary restriction, limitation of calorie intake with maintained vitamin and mineral support, can extend lifespan and protect against diseases of age across many species. Elaboration of molecular mechanisms that control dietary restriction in simple animal models may therefore inform on strategies to activate health-promoting metabolism to help address clinical challenges associated with aging and age-associated disease. We characterize a single Caenorhabditis elegans microRNA gene that keeps dietary restriction programs off when food is abundant. A mir-80 deletion exhibits beneficial features of dietary restriction regardless of food availability, including extended maintenance of mobility and cardiac-like muscle function later into life as well as lifespan extension. We identify three key longevity genes that are required for these benefits. We hypothesize that miR-80 is a core regulator by which diverse and intersecting metabolic pathways are coordinately regulated to respond to nutrient availability.
Introduction
The promotion of healthy aging is a goal of modern medicine, and simple interventions that protect against age-associated decline and disease are the dream of many in the general population. Genetics, environment, and stochastic factors all make substantial and complex contributions to healthspan. Single gene mutations that affect conserved pathways in model organisms can extend life and slow age-associated decline [1], [2]. Environmental factors such as diet can also have a profound effect on the quality of aging. For example, dietary restriction (DR), limitation of calorie intake with maintained vitamin and mineral support, can extend lifespan and protect against diseases of age across many species [3]. Elaboration of molecular mechanisms that control DR in simple animal models may thus inform on strategies to activate health-promoting metabolism to help address clinical challenges associated with aging.
In the nematode Caenorhabditis elegans, food limitation that results in lifespan extension can be introduced via several protocols [4], [5], [6], [7], [8], although the specific genetic requirements for longevity benefits of different DR regimens are not fully overlapping. For example, the transcription factor DAF-16/FOXO is dispensable for longevity induced in the feeding-impaired eat-2 mutant, whereas with a DR protocol in which bacterial food is diluted on plates, DAF-16/FOXO is essential for lifespan extension [4], [9]. Such observations most likely reflect highly complex regulatory loops that control the precise metabolic state.
microRNAs (miRNAs) can be metabolic regulators [10]. miRNAs are small, ∼22 nt non-coding RNAs that can bind to transcripts via partial sequence complementarity to down-regulate translation of those target mRNAs. Many miRNAs are conserved over their lengths or in the critical 5′ seed region, defining families across species [11], [12], [13]. Although the co-evolution of miRNAs and their targets is a complex process [14], some miRNA/target pairings have been molecularly and functionally conserved. For example, discovery of LET-7 miRNA regulation of target RAS in C. elegans [15] inspired anti-oncogenic therapies for mammalian lung tumors [16].
We took advantage of the powerful reagents for miRNA study in C. elegans [17], [18] and our previous characterization of a DR fluorimetric signature of endogenous gut fluorescence in these transparent nematodes (derived from lipofuscin+advanced glycation end products [19]) to identify miRNAs that might regulate DR. Here we report bantam-homolog miR-80 as a food-regulated miRNA that normally represses DR when food is abundant. Transcription factors DAF-16, HSF-1, and CBP-1 are required for mir-80(Δ) benefits. Of these, the cbp-1 transcript includes sequences that might be directly targeted by miR-80 to coordinate this circuit. Our data suggest an approach to metabolic activation of DR even under ad lib feeding that could inspire strategies for treating obesity, limiting age-associated disease, and promoting healthy aging.
Results
Deletion of microRNA-80 promotes system-wide healthy aging in C. elegans
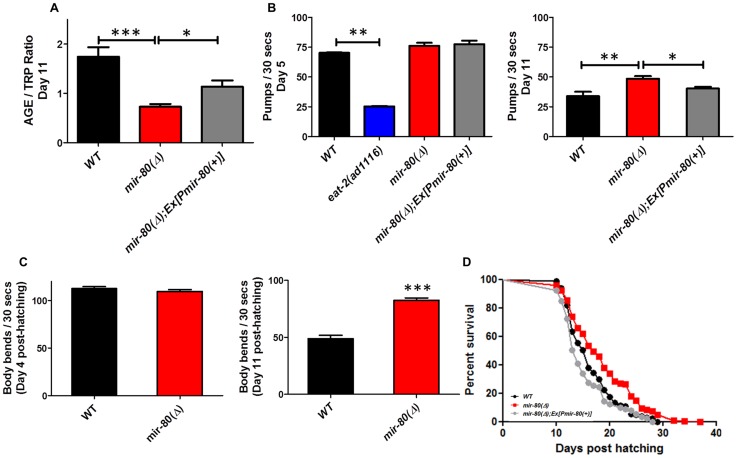
Our previous studies revealed that age pigment levels (lipofuscin+advanced glycation end products) inversely correlate with locomotory healthspan—low age pigment levels late in life are typical of animals that age gracefully and maintain strong locomotory vigor, whereas high age pigment levels are typical of same-chronological age animals that age poorly and appear decrepit [19]. Thus, to identify C. elegans miRNAs that might impact healthy aging, we screened available C. elegans mir deletion strains [20] for differences in autofluorescent age pigment levels in old animals. We found that mir-80(nDf53) [hereafter referred to as mir-80(Δ)], exhibits low age pigment fluorescence levels late in life compared to wild type (WT) animals (Fig. 1A, ∼58% lower, p<0.0005). The low age pigment phenotype of mir-80(Δ) is rescued by a transgene array harboring a mir-80(+) gene, confirming that the low age pigment phenotype is conferred by mir-80 deletion itself. Thus, late in adult life (∼2/3 through the WT lifespan), mir-80(Δ) mutants exhibit low age pigment accumulation typical of healthy aging animals.
Figure 1. mir-80(Δ) exhibits multiple features of healthy aging.
Fig. 1A. mir-80(Δ) has low intestinal age pigment levels compared to wild type during late adult life (day 11). We grew age-synchronized WT (black), mir-80(Δ) (red), and mir-80(Δ); Ex[Pmir-80(+)] (grey) under standard conditions (20°C, on E. coli OP50-1) and scored animals for age pigment levels using a fluorimeter (n = 100 per strain/trial; day 11, as counted from the hatch; mir-80(Δ) is nDf53; mir-80(+) rescue transgene is nEx1457 [18]). Age pigment fluorescence, which increases with age, is normalized to endogenous tryptophan fluorescence, which remains relatively constant with age [19], (AGE/TRP ratio ∼58% decreased in mir-80(Δ) vs. wild type). Graphs represent mean data from at least 3 independent trials. Data were compared using the One-way ANOVA followed by Newman-Keuls multiple comparison test, *** - p<0.0005, * - p<0.05; WT to Ex[Pmir-80(+)] rescue p<0.12. In the rescued strain, age pigment levels might not reach WT levels due to mosaicism of the extrachromosomal transgene, the mir-80 transgene dose, or “sponge” effects of overexpression. Fig. 1B. mir-80(Δ) maintains youthful pharyngeal pumping in late adulthood. We assayed age-synchronized WT (black), mir-80(Δ) (red), and mir-80(Δ); Ex[Pmir-80(+)] (grey, nEx1457 ([18])) for pharyngeal pumping rates on Day 5 (left) and Day 11 (right) (30 s interval, n = 10/trial, 3 trials). For day 5, we included the eat-2(ad1116) mutant (blue), impaired for pharyngeal pumping to ∼30% WT rate, as a negative control. In this assay we compared healthy appearing animals (most vigorous locomotion). Graph is of cumulative data from 3 independent trials. Data were compared using the One-way ANOVA followed by Newman-Keuls multiple comparison test. * - p<0.05; ** - p<0.005, *** - p<0.0005. mir-80(Δ) pumping rate is modestly higher than WT at day 5 (p = 0.023), but note that relative pumping differences at Day 5 are small compared to differences at Day 11 (∼44% increase). Fig. 1C. mir-80(Δ) maintains youthful swimming vigor in late adulthood. We assayed age-synchronized animals, WT (black), mir-80(Δ) (red), and mir-80(Δ); Ex[Pmir-80(+)] (grey) for swimming mobility at Day 5 and Day 11 post-hatching (n≥30, 3 independent trials are combined in presented data). Data were compared using 2-tailed Student's T-test, *** - p<0.0001. Although mir-80(Δ) and WT swim similarly in young adult life, mir-80(Δ) mutants better maintain swimming prowess late in life, ∼69% increased body bend rate. Fig. 1D. mir-80(Δ) mutants have increased mean and maximum lifespans. We assayed age-synchronized WT (black), mir-80(Δ) (red), and mir-80(Δ); Ex[Pmir-80(+)] (grey) animals grown under standard conditions (20°C, OP50-1) for viability (movement away from worm pick by gentle touch) at the indicated days. We initiated trials with relatively vigorous animals on day 9 from the hatch (10 animals per plate, ≥25 per strain per trial, 3 independent trials, which are combined here). Data from individual trials are shown in Fig. S1. Statistics were calculated using the Log-rank Test. mir-80(Δ) mutants exhibit a significant extension in lifespan as compared to WT (p<0.0001) and transgenic expression of mir-80(+) reversed the longevity increase (p<.0001).
To test if mir-80(Δ) mutants exhibited additional healthspan phenotypes, we next measured two indicators of maintained muscle integrity and function late into adult life–pharyngeal pumping rates and swimming vigor. Pharyngeal pumping is the mechanism by which food is pulled into the gut using specialized cardiac-like muscles. Pharyngeal pumping rates decline markedly with age, such that after the first week of life, feeding capacity is greatly diminished [21], a functional decline that tracks with physical changes in muscle integrity [22], [23], [24]. We find that pumping rates are significantly higher in mir-80(Δ) late in life (day 11) as compared to WT (44% increase), a phenotype reversed by a mir-80(+) transgene (Fig. 1B, right graph; p<0.005). Importantly, 5 day old WT and mir-80(Δ) (i.e., young adult; Fig. 1B, left graph) have similar pumping rates. Thus, mir-80(Δ) mutants are not simply hyper-activated for pumping, but rather maintain pumping function better late into life. We conclude that mir-80(Δ) exerts a positive effect on the quality of cardiac-like muscle aging.
As occurs with human skeletal muscle sarcopenia (the debilitating progressive loss of muscle mass and strength that accompanies aging across species), C. elegans body wall muscle deteriorates with age, featuring sarcomere loss [22], [24]. Physical decline is correlated with loss of locomotion vigor. We compared late-age swimming (body bend frequency) in WT and mir-80(Δ) to show that mir-80(Δ) mutants are significantly more vigorous swimmers in late adulthood (Fig. 1C right panel; 69% increase at day 11, p<0.0001). Early in adult life WT and mir-80(Δ) swim similarly (Fig. 1C, left panel). We conclude that mir-80(Δ) delays locomotory aging without altering young adult swimming behavior itself.
Given that mir-80(Δ) mutants exhibit several features of extended healthspan, we examined the longevity phenotype. We find that mir-80(Δ) mutants exhibit both mean and maximum healthspan extension, subject to mir-80(+) transgene rescue (Fig. 1D, p<0.0001, individual lifespan data in Fig. S1; average age increase at 75% mortality over all lifespan studies in this paper (13) was 24.1%+/−4.7%). Thus, deletion of mir-80 confers longevity.
In summary, mir-80(Δ) confers multiple features of extended adult healthspan late in life: lowered intestinal age pigment accumulation, maintained pharyngeal pumping capacity, increased swimming vigor, and lifespan extension. Because mir-80(Δ) does not exhibit notable defects in development ([20], and our observations), it appears that mir-80 has a predominant and focused impact on aging of the adult.
mir-80(Δ) mutants appear dietary-restriction constitutive
Spectral properties of age pigments
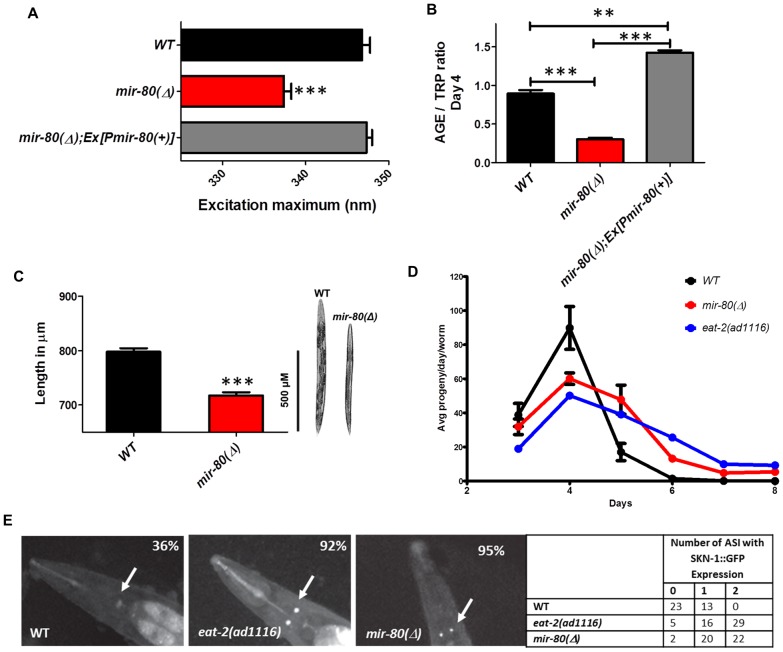
To ask whether mir-80(Δ) might act via a DR mechanism to extend healthspan and lifespan, we tested mir-80(Δ) mutants for phenotypic features of the DR state (Fig. 2). Our previous in vivo studies of fluorescent age pigments revealed that transparent C. elegans under DR have a unique fluorimetric “signature” that is distinct from spectral properties of both WT and long-lived mutants induced by other longevity pathways [19], [25], [26]. A spectrofluorimeter excitation/emission direct scan of WT reports a characteristic excitation maximum (Exmax) for age pigments at ∼345 nm. However, under all DR-inducing conditions we previously tested (multiple feeding-impaired mutants [27], liquid feeding of WT [7], [19], limiting bacterial concentrations for WT [4], complete removal of food [28], treatment with DR-mimetic drug metformin [25]), we noted a downward shift in Exmax. Thus, the age pigment Exmax shift indicates a DR-like state. We found that mir-80(Δ) consistently exhibits the DR Exmax shift despite growth in the presence of abundant food (Fig. 2A) and has low age pigment levels, even at young age (day 4, p<0.0005, Fig. 2B, about 66% lower in these studies), the latter of which also characteristic of DR mutants. Thus, mir-80(Δ) exhibits the spectral signature of DR despite the presence of food, consistent with mir-80(Δ) being a DR constitutive mutant.
Figure 2. mir-80(Δ) exhibits multiple characteristics typical of DR animals.
Fig. 2A. The mir-80(Δ) mutant exhibits the DR Exmax shift. We assayed age-synchronized WT (black), mir-80(Δ) (red), and mir-80(Δ); Ex[Pmir-80(+)] (grey) 4 day old animals grown under standard conditions (20°C, OP50-1). We used a spectrofluorimeter to scan transparent animals (n = 100 per strain/trial) to generate excitation/emission profiles as in [19]; wavelength of excitation at maximal fluorescence is indicated. Graphs represent mean data from at least 3 independent trials. Data were compared One-way ANOVA followed by Newman-Keuls multiple comparison test, *** - p<0.0005. mir-80(Δ) exhibits a significantly down-shifted Exmax (p<0.0005) as compared to wild type under conditions of abundant food, a feature unique to DR [19]. Fig. 2B. The mir-80(Δ) mutant exhibits low age pigment levels early in life, as occurs in C. elegans DR. We assayed age-synchronized WT (black), mir-80(Δ) (red), and mir-80(Δ); Ex[Pmir-80(+)] (grey) 4 day old animals grown under standard conditions (20°C, OP50-1). We scanned animals (n = 100 per strain) for fluorescence over a range of wavelengths, and normalized age pigment fluorescence (AGE) to tryptophan (TRP) fluorescence as in [19] for comparison. Graphs represent mean data from at least 3 independent trials. Data were compared using One-way ANOVA followed by Newman-Keuls multiple comparison test, *** - p<0.0005, ** - p<0.005. mir-80(Δ) exhibits low age pigment levels as compared to wild type (p<0.0005) early in life, which is true of all DR conditions previously tested (although not unique to DR). In these assays, levels were on average 66% lower in mir-80(Δ). Fig. 2C. mir-80(Δ) mutants are physically smaller than WT, typical of animals in DR. We measured age-synchronized WT (black) and mir-80(Δ) (red) 4 day old animals (examples at the right) grown under standard conditions (20°C, OP50-1) by imaging animals (WT n = 77, mir-80(Δ) n = 88) under DIC under low magnification. We measured using the segmented line tool in the ImageJ software by drawing a line across the length of the animal, and converted length in pixels to uM using a stage micrometer to assess image scale. We compared data using 2-tailed Student's T-test, *** - p<0.0005. mir-80(Δ) mutants are ∼10% shorter and look thinner than WT reared under the same conditions, typical of the scrawny appearance of animals in DR, example comparison on the right. Although size varies somewhat and is not as quantitative a measure as age pigment scores, we have used the scrawny appearance to identify likely mir-80(Δ) homozygotes in crosses. Fig. 2D. mir-80(Δ) mutants exhibit reduced fertility and an extended reproductive lifespan. We assayed egg production in age-synchronized WT (black), mir-80(Δ) (red), and DR mutant eat-2(ad1116) (blue) grown under standard conditions (20°C, OP50-1; parent n = 10, 3 independent trials). eat-2 is one trial so bars are not provided. Data were compared using 2-tailed Student's T-test. Early in adult life, mir-80(Δ) produce a reduced number of live births per day (33% decrease, p<0.05 for Day 3; 180% increase, p<0.001 Day 4–6) and exhibit a prolonged reproductive lifespan (through Day 8 for mir-80(Δ) as compared to WT Day 6, p<0.001). The constitutive DR mutant, eat-2 experiences a shift in reproductive lifespan (compared to WT, p<0.001) that is similar to mir-80(Δ). Fig. 2E. SKN-1-GFP, a molecular reporter of DR, is upregulated in mir-80(Δ) in the presence of food. SKN-1::GFP expression in the two ASI neurons is a molecular signal of some DR [7]. We constructed strains that included an integrated rescuing skn-1-gfp fusion gene expressed from the native skn-1 promoter, Is007[skn-1-gfp] [7], and measured at Day 7, 20°C, growth in OP50-1 (white arrows). WT animals show low levels of ASI expression (36% with very weak expression in only one ASI), while DR-constitutive eat-2(ad1116) animals display constitutive expression of SKN-1-GFP in the ASI neurons (92% in 1 or 2 neurons, strong expression). 95% of mir-80(Δ) have 1–2 ASIs expressing at this timepoint. These data support that mir-80(Δ) mutants are in DR even when reared in the presence of ample food.
Scrawny bodies
WT animals under DR appear thin and pale [29], [30]. We found that mir-80(Δ) had a somewhat scrawny and pale appearance and on average is ∼10% shorter in length than WT (p<0.0005, Fig. 2C). Thus, the physical appearance of mir-80(Δ) mutants resembles that of DR animals.
Reduced fecundity
Reduced fecundity is associated with DR across species [31]. We find that mir-80(Δ) exhibits a significant extension of reproductive period, producing progeny through Day 8 as compared to WT Day 6, p<0.001, Fig. 2D. In addition, there is a decrease in the number of live progeny laid per day by mir-80(Δ) (p<0.05 for Day 3 and p<0.001 for days 4–6), without a significant difference in the total number of surviving progeny (not shown), a pattern of progeny production similar to that of feeding-defective DR mutant eat-2 (Fig. 2D). We conclude that mir-80(Δ) exhibits reduced fecundity, similar to animals experiencing DR.
Hypersensitivity to a DR-mimetic drug
The anti-diabetes DR-mimetic drug metformin can induce a life-prolonging DR-like state in WT animals, but administering metformin to animals already in DR (e.g., the eat-2 mutant), leads to a reduced lifespan [25]. This metformin hypersensitivity in DR animals has been suggested to result from pushing DR metabolism into a deleterious starvation-like state [25], [32]. We found that although mir-80(Δ) is long-lived relative to WT under normal growth conditions (Fig. 1D), the mir-80(Δ) lifespan is decreased relative to WT in the presence of metformin (three individual trials and combined data in Fig. S2), similar to what occurs for DR mutant eat-2. Thus, like other DR strains, mir-80(Δ) is hypersensitive to metformin, consistent with mir-80(Δ) being in a DR constitutive state.
Molecular reporter of DR: SKN-1 expression in ASI neurons
SKN-1 is a transcription factor crucial for endodermal development and response to oxidative stress [33], that must also be expressed in the pair of chemosensory ASI neurons for the longevity outcomes of some DR regimens [7]. For example, SKN-1 appears continuously expressed in the ASI neurons in the constitutive DR eat-2(ad1116);Is007[skn-1-gfp] reporter strain, whereas this reporter is not highly expressed in wild type Is007[skn-1-gfp] animals that are grown under ad lib feeding conditions (Fig. 2E). We found that SKN-1-GFP is highly expressed in the ASI neurons in the well fed mir-80(Δ) mutant (Fig. 2E). At day 7, 95% mir-80(Δ) and 92% eat-2 mutants exhibited strong signals in 1–2 ASI neurons; but 36% of WT only express weak signal in at best one ASI.
Our data support that mir-80(Δ) induces molecular features of DR.
In summary, the mir-80(Δ) mutant has a lean body, reduced fecundity, hypersensitivity to metformin, and expresses both molecular and fluorimetric markers of DR despite growth in abundant food. Importantly, unlike the eat-2 mutant, pumping rates in mir-80(Δ) are normal in young animals and are actually enhanced relative to WT later in life (Fig. 1B). Thus, mir-80(Δ) does not physically reduce the ability to eat, but rather is likely to act further downstream to influence DR metabolism. Remarkably, then, deletion of a single miRNA gene can shift C. elegans metabolism into DR to promote healthy aging.
mir-80 expression is positively regulated by the presence of food
mir-80 is broadly expressed in well-fed animals
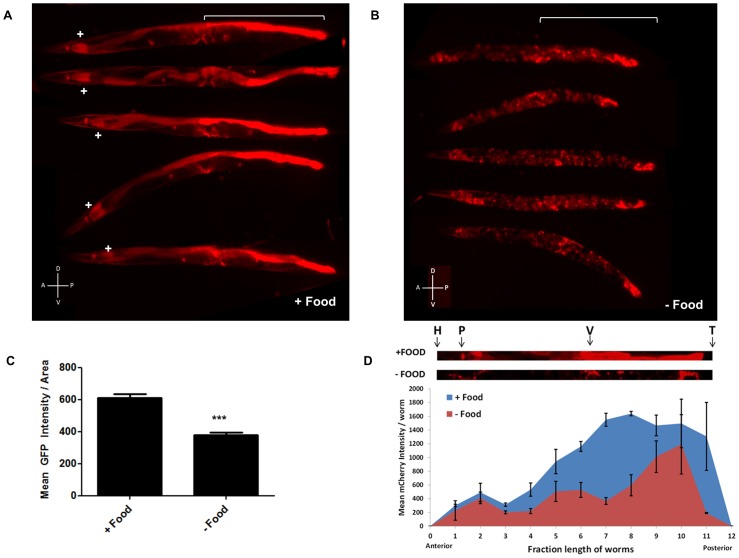
Previous deep sequencing studies indicate that miR-80 is a relatively high abundance miRNA expressed from late embryogenesis into adulthood [34], [35]. Somewhat paradoxically, two published transgenic lines of the same Pmir-80::GFP fusion transcriptional reporter (utilizing 1741 bp 5′ to mir-80 as promoter [17]), exhibit different cellular expression patterns, an observation we confirmed (Fig. S3A,B). To address this discrepancy, we constructed a new mCherry reporter that extended mir-80 5′ sequences up to the next annotated gene (1814 5′ bp, designated Pmir-80L). 5/5 extra-chromosomal transgenic lines of this reporter exhibit a broad cellular expression pattern, somewhat similar to the published extrachromosomal array line (vulva, hypodermis, body wall muscle, head neurons, tail neurons, excretory cell, dorsal/ventral nerve cord, and weaker expression in intestine [anterior or posterior] and pharynx). The Pmir-80L pattern is distinctive in exhibiting highest expression in the two most anterior gut cells and in posterior gut under high food conditions (Fig. 3A). Interestingly, mir-80 does not appear expressed in the ASI neurons in any lines (Fig. S4A,B) and thus miR-80 most likely acts non-cell autonomously to influence skn-1::GFP expression in the ASI neurons (Fig. 2E).
Figure 3. mir-80 expression is generally high in the presence of food, but low when food is lacking.
3A. Examples of expression of extrachromosomal bzEx207 [Pmir-80LmCherry] line grown in the presence of unlimited E. coli. Note that this transgenic line, typical of 4 lines that have the long mir-80 promoter region, exhibits substantial reporter expression in the first two cells of the intestine (indicated by white+sign) and in the posterior intestine (white bracket). Lower level expression is evident in several other tissues. Animals are adult day 6, but we find no bleed through of signals using red/green filter sets (Fig. S4B) so age pigments do not confound this analysis. 3B. Examples of expression of the bzEx207 [Pmir-80LmCherry] line grown in the presence of unlimited E. coli until young adulthood and then switched to no food for 48 hours. 6 day old adults are aligned with anterior to the left, posterior gut region indicated by white bracket. Most posterior gut fluorescence is markedly diminished, although expression in the anterior two intestinal cells, the central egg laying muscles, and the very posterior gut remains high. 3C. Quantitation of fluorescence signals for a mir-80 promoter fusion reporter line in food vs. food limitation. Fluorescence of overall bzEx207[Pmir-80LmCherry] line expression after 48 hrs on no-food plates. Food limitation in these studies was by dietary deprivation [28], but food dilution on solid NGM media [4] and food dilution in liquid media [4] induced similar changes in these lines (Fig. S4). Graph represents spectrofluorimeter measurements of fluorescence levels (whole body) for at least 50 animals per DR regimen. Pairwise comparisons were made using Two-tailed Students' T-test. *** - p<0.0005. Same exposure times were used for complementary panels. 3D. Analysis of food-regulated expression of pmir-80LmCherry expression along the nematode body implicates posterior intestinal regions as a major site of regulation. We compared pmir-80LmCherry signals in transgenic ZB3042 grown either in the presence of food (blue) or switched to no food for 24 hrs (red) (measured at day 4, n = 39). We used the ImageJ program to create a 25 pixel segmented line covering the animal and measured mean fluorescence intensity along the body, dividing the length into 12 equal bins and plotting the mean fluorescence intensity at each point. Representative animals are depicted above with the approximate body positions indicated (H = head, P = pharynx, V = vulva, T = tail). Note that although food regulation is apparent in most of the body, food-regulated expression changes in the regions of the mid- and posterior intestine are most dramatic. Error bars indicate standard error for each bin measurement.
Pmir-80 reporter expression is down-regulated in the absence of food
Since the genetic elimination of mir-80 results in constitutive DR phenotypes (Fig. 2, Fig. S2), we hypothesized that mir-80 expression might be reduced when food is limited. We examined multiple mir-80 transcriptional reporters for expression level 48 hours after shift from abundant food to no food (Fig. 3B–D, Fig. S3). We find that for all reporters examined, expression for mir-80 is significantly lower in the absence of food. Furthermore, food limitation by alternative diet regimens is also associated with general down-regulation of mir-80 reporter expression (Fig. S5). For the broadly expressing transgenes, it appeared that overall expression in many cells was down-regulated, although the changes in the mid and posterior intestine have the largest differential, ∼4–10× Pmir-80L::mCherry level changes food/no food (Fig. 3D). We confirmed general down-regulation in the absence of food by deep sequence analysis of miR-80: overall expression levels food/no food are 1.5 increased (data not shown, p-value<0.05). However, we emphasize that not all cells exhibit miR-80 down-regulation: expression in two anterior-most and two posterior-most gut cells, and vulval muscle expression appear maintained, and possibly enhanced, in no food. We conclude that mir-80 expression can be modulated by the presence of food: in most cells, mir-80 expression is relatively high in the presence of food and is reduced when food is limiting. The broad mir-80 expression pattern suggests a potential role for miR-80 in global regulation of metabolism; although dramatic posterior intestinal regulation raises the possibility that major changes in this tissue could provide the most critical influence on organism-wide regulation (see Discussion).
Transcription factors implicated in DR metabolism are required for mir-80(Δ)-associated fluorimetric features
To identify genes required for mir-80(Δ)-regulated DR, we used RNAi to knockdown genes previously implicated in DR lifespan benefits, hypothesizing that genes required for mir-80(Δ) DR should be needed for the Exmax shift and low age pigment levels typical of multiple DR states. Of the 18 genes we screened, we found that RNAi knockdown of transcription factors daf-16/FOXO, heat shock transcription factor hsf-1, and CREB binding protein homolog cbp-1 modulated both the Exmax shift and low age pigment levels of mir-80(Δ) (Tables S1, S2, Figs. 4A,B; 5A,B; 6A,B).
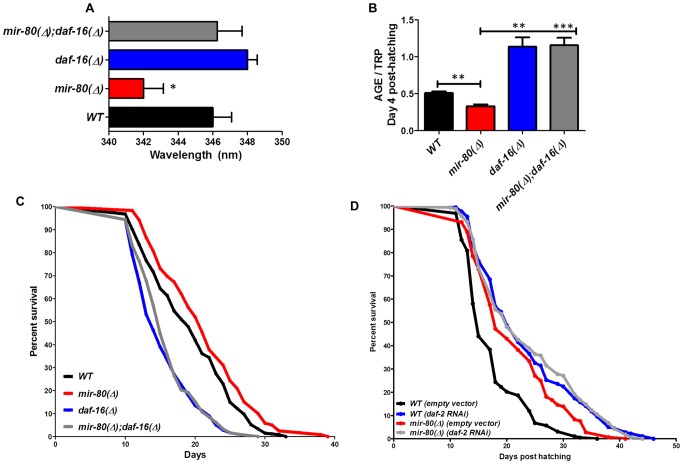
Figure 4. daf-16/FOXO is needed for the fluorimetric DR signature and longevity phenotypes of mir-80(Δ).
Fig. 4A. Transcription factor daf-16/FOXO is required for the Exmax shift phenotype in mir-80(Δ). We reared age-synchronized animals under standard growth conditions (20°C, OP50-1) and measured age pigment spectral properties at Day 4 (50 animals per strain) for WT (black bar), mir-80(Δ) (red), daf-16(Δ) allele mgDf50 (blue), and mir-80(Δ);daf-16(Δ) double mutant (grey). The same color coding is used for panels 4A–4D. We recorded Exmax as the highest peak detected by the Datamax software package suite (Horiba Scientific). Graphs represent mean data from at least 3 independent trials. Data were compared using 2-tailed Student's T-test. mir-80(Δ) compared to WT * - p<0.05; mir-80(Δ);daf-16(Δ) double mutant compared to WT, ns. Deletion of daf-16 reverses the Exmax shift phenotype of mir-80(Δ). Fig. 4B. daf-16/FOXO is required for low age pigment levels in mir-80(Δ). We grew age-synchronized animals under standard conditions (20°C, OP50-1) and measured total age pigment fluorescence, normalized to total tryptophan fluorescence as in [19] (Day 4, 50 animals per trial). Graphs represent mean data from at least 3 independent trials. Error bars represent ±S.E.M. Data were compared using 2-tailed Student's T-test. *** - p<0.0005, ** - p<0.005. The low age pigment accumulation phenotype of mir-80(Δ) is reversed in the mir-80(Δ);daf-16(Δ) double mutant on day 4 (shown here) as well as on day 9 (data not shown). Fig. 4C. daf-16 is required for the lifespan extension of mir-80(Δ). We grew age-synchronized animals under standard conditions (20°C, OP50-1). At day 9, we placed 10 healthy animals per plate, ≥40 per strain per trial, and we scored viability as movement away from pick touch on the indicated days. The graphs represent data combined from 3 independent trials. Statistics are calculated using the Log-rank Test. The mir-80(Δ);daf-16(Δ) double mutant is suppressed for the longevity phenotype of mir-80(Δ) (p<0.0001). We did not, however, observe dramatic overall changes in nuclear localization of DAF-16::GFP +/− mir-80 (data not shown). Fig. 4D. mir-80(Δ) lifespan can be further extended by daf-2(RNAi). We placed age-synchronized mir-80(Δ) L1 larvae (Day 1) on empty vector control (pL4440) or daf-2 RNAi plates under standard conditions (20°C). At day 9, we placed 10 healthy animals per plate, ≥40 per strain per trial, and we scored viability as movement away from pick touch at the indicated days. The graphs represent data combined from 3 independent trials. Statistics are calculated using the Log-rank Test. daf-2(RNAi) increases the lifespan of mir-80(Δ) vector control (p<0.005), but additive effects for mir-80(Δ)+daf-2(RNAi) above the daf-2(RNAi) level are not observed (p = 0.98). Note that data from these experiments also provide a general sense of how mir-80(Δ) compares to daf-2 for lifespan extension; roughly we find mir-80(Δ) effects are slightly less than half those of daf-2(rf), see Table S4 for exact data from individual trials.
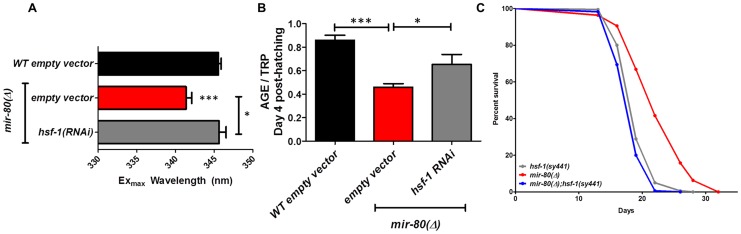
Figure 5. hsf-1 is needed for the fluorimetric DR signature and longevity phenotypes of mir-80(Δ).
Fig. 5A. hsf-1(RNAi) in the mir-80(Δ) background reverses the DR Exmax shift. We grew age-synchronized animals under standard RNAi feeding conditions (20°C, HT115) and measured age pigments at Day 4 (50 animals per RNAi clone). We recorded Exmax as the highest peak detected by the Datamax software package suite (Horiba Scientific). Black bar, WT+ empty vector RNAi; red bar, mir-80(Δ)+empty vector RNAI; grey bar, mir-80(Δ)+hsf-1(RNAi). Graphs represent cumulative data from 3 independent trials. Error bars represent ±S.E.M. Data were compared using 2-tailed Student's T-test (** p<0.001). Note that hsf-1(RNAi) treatment of WT does not change Exmax (data not shown). Fig. 5B. hsf-1(RNAi) in the mir-80(Δ) background partially counters the low age pigment level phenotype of mir-80(Δ). We grew age-synchronized animals under standard conditions (20°C, HT115) and measured total age pigment fluorescence, normalized to total tryptophan fluorescence as in [19] (Day 4 post-hatching, 50 animals per RNAi clone). Black bar, WT+ empty vector RNAi; red bar, mir-80(Δ)+empty vector RNAi; grey bar, mir-80(Δ)+hsf-1(RNAi). Graphs represent cumulative data from 3 independent trials. Error bars represent ±S.E.M. Data were compared using 2-tailed Student's T-test (*** p<0.0001, * p<0.05 compared to mir-80(Δ) empty vector). Note that hsf-1(RNAi) treatment of WT does not change age pigment scores at day 4 (data not shown). Fig. 5C. hsf-1 is required for mir-80(Δ)-induced longevity. We grew age-synchronized animals under standard conditions with low levels of FUDR to prevent progeny production (20°C, OP50-1, 50 uM FuDR). At day 9, we placed 10 healthy animals per plate, ≥40 per strain per trial, and we scored viability as movement away from pick touch at the indicated days. The graphs represent data combined from 3 independent trials. Statistics are calculated using the Log-rank Test. Error bars indicate ± S.E.M. The mir-80(Δ); hsf-1(sy441) double mutant is shorter lived than mir-80(Δ) (p<0.0001). Because RNAi knockdown is inefficient the nervous system (see [59]), the profound effects of hsf-1(RNAi) suggest that critical hsf-1 and mir-80 regulation occurs outside of the C. elegans nervous system.
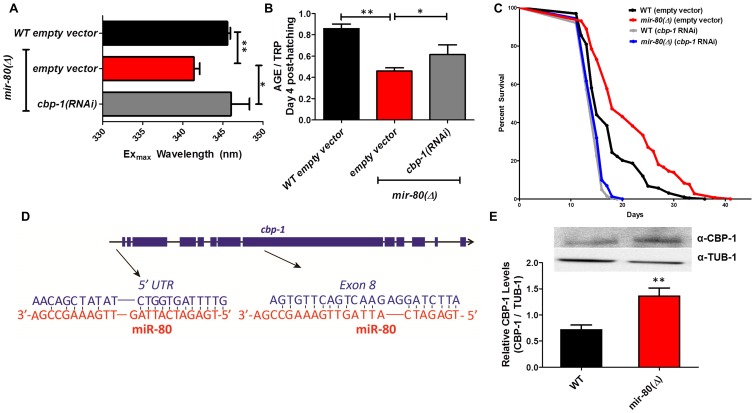
Figure 6. CBP-1 is critical for mir-80(Δ) healthspan benefits, and is a candidate direct binding target of miR-80.
Fig. 6A. cbp-1(RNAi) in the mir-80(Δ) background reverses the DR Exmax shift. We grew age-synchronized animals under standard RNAi feeding conditions (20°C, HT115) and measured age pigments at Day 4 (50 animals per RNAi clone). We recorded Exmax as the highest peak detected by the Datamax software package suite (Horiba Scientific). Graphs represent cumulative data from 3 independent trials. Error bars represent ±S.E.M. Data were compared using 2-tailed Student's T-test (** p<0.001, * p≤0.055 compared to mir-80(Δ) empty vector). cbp-1(RNAi) Exmax is comparable to that of ad lib wild type (p = 0.729). Note that cbp-1(RNAi) treatment of WT does not change Exmax (data not shown), so this effect is specific to the DR signature of mir-80(Δ). Fig. 6B. cbp-1(RNAi) in the mir-80(Δ) background partially reverses low age pigment levels. We grew age-synchronized animals under standard conditions (20°C, HT115) and measured total age pigment fluorescence at day 4 (50 animals per RNAi clone), normalized to total tryptophan fluorescence as in ref. [19]. Graphs represent cumulative data from 3 independent trials. Error bars represent ±S.E.M. Data were compared using 2-tailed Student's T-test (** p<0.05, * p<0.1 compared to mir-80(Δ)+empty vector RNAi). Note that cbp-1(RNAi) treatment of WT induces modest reduction of age pigment levels (p = 0.01, data not shown). Fig. 6C. mir-80(Δ) longevity is dependent on cbp-1. We placed age-synchronized L1 larvae on empty vector control (pL4440) plates under standard conditions (20°C) until Day 4 (day 1 of adult life) at which time animals were moved to either empty vector control (L4440) or cbp-1(RNAi) plates. At day 9, we placed 10 healthy animals per plate (≥40 per strain per trial), and we scored viability as movement away from pick touch on the indicated days. The graphs represent data combined from 3 independent trials. Statistics are calculated using the Log-rank Test. cbp-1(RNAi) decreases the lifespan of mir-80(Δ) (p<0.0001 compared to vector control. Because RNAi knockdown is inefficient the nervous system (see [59]), the profound effects of cbp-1(RNAi) suggest that critical cbp-1/mir-80 regulation occurs outside of the C. elegans nervous system. Fig. 6D. The cbp-1 transcript includes two predicted binding sites for miR-80. Exon structure of cbp-1 is indicated by thick blue bars, introns in thin black lines (see WormBase for details). The rna22 algorithm [10], which searches for target sites outside the 3′UTR, predicts that miR-80 binds cbp-1 within the 5′ UTR and within exon 8. The potential alignments of miR-80 (red) to C. elegans cbp-1 (blue) sequences are indicated. Note that the seed match to the exon 8 region is a perfect 10 bp match for C. elegans, and that the target sequence is conserved in mouse and human CBP1 (see Fig. S7). Fig. 6E. Endogenous CBP-1 protein levels are increased in 7 day old mir-80(Δ) mutants. We grew age-synchronized animals under standard conditions (20°C, OP50-1) and extracted total protein at Day 7 (100 animals per strain) for Western blot analysis (top). Graphs represent CBP-1 levels for each strain normalized to own TUB-1 levels. Error bars represent ±S.E.M. Data were compared using 2-tailed Student's T-test (** p<0.005). The graphs represent data combined from 3 independent trials. We noted that during young adulthood, native levels of CBP-1 seemed comparable to WT in mir-80(Δ), suggesting that additional regulatory controls are exerted on CBP-1 expression levels in development or early adulthood.
DAF-16/FOXO is required for fluorimetric indicators of DR age pigment and lifespan extension in mir-80(Δ)
Transcription factor daf-16/FOXO, an important modulator of longevity through insulin signaling, is also critical for lifespan extension benefits of serial dilution of bacteria on plates (sDR) and peptone dilution on plates (pDR) [4]. We found that the mir-80(Δ);daf-16(Δ) double mutant was reversed for both the DR Exmax shift (Day 4, Fig. 4A, p<0.05) and the low age pigment levels (Fig. 4B, p<0.005) that are characteristic of mir-80(Δ). Moreover, the mir-80(Δ);daf-16(Δ) double mutant had a short lifespan, similar to that of daf-16(Δ) (Fig. 4C). These data identify DAF-16 as an required regulator of the fluorimetric DR signature and longevity benefits in the mir-80(Δ) background. Our in silico analyses did not identify candidate miR-80 target sites in the daf-16 transcript, suggesting an indirect role in the mir-80(Δ)-regulated DR pathway.
To address the relationship of mir-80(Δ) and the insulin signaling pathway further, we compared longevity phenotypes of mir-80(Δ) and mir-80(Δ) treated with daf-2 RNAi, which targets the C. elegans insulin receptor (Fig. 4D). We find that mir-80(Δ) lifespan can be further extended by daf-2(RNAi) (p<0.005). The additive effects of mir-80(Δ)+daf-2(RNAi) knockdown suggest that healthspan and longevity benefits of mir-80(Δ) may be conferred in part by a daf-2-independent pathway. However, the fact that mir-80(Δ) does not further extend daf-2(RNAi) lifespan (p<.98) is also consistent with a model in which miR-80 partially down-regulates the insulin pathway, and that daf-2(RNAi) reflects a stronger activation of the DAF-16-dependent transcriptional response, more toward an optimal healthspan signaling strength. Regardless of the details of pathway overlap, our data are definitive in establishing that daf-16/FOXO is needed for fluorimetric properties and longevity outcomes of mir-80(Δ).
hsf-1 deficiency eliminates multiple mir-80(Δ) healthspan phenotypes
hsf-1 regulates the expression of many heat-inducible target genes, modulates longevity, and is required for lifespan extension conferred by bacterial food deprivation [36] and dietary deprivation [5], [28]. In the mir-80(Δ) background, hsf-1(RNAi) reverses the Exmax shift that typifies DR (p<0.06, Table S1, Fig. 5A) and partially restores 4 day age pigment levels (p<0.05 compared to mir-80(Δ)+empty vector RNAi, Table S2, Fig. 5B). hsf-1(RNAi) does not affect Exmax or age pigment levels in WT (data not shown). To determine if hsf-1 is also required for mir-80(Δ) longevity, we examined survival curves for the mir-80(Δ);hsf-1(sy441) double mutant. We find that disruption of hsf-1 eliminates the lifespan extension conferred by mir-80(Δ) (Fig. 5C). We conclude that hsf-1 is required for both mir-80(Δ)-induced fluorimetric features that typify DR and for mir-80(Δ)-induced longevity. Consistent with a role for hsf-1 in mir-80(Δ)-induced benefits, HSF-1 target gene hsp-16.2 transcripts are elevated in the mir-80(Δ) mutant (Fig. S6). In silico analyses did not reveal candidate miR-80 target sites in the hsf-1 transcript, suggesting indirect regulation in the mir-80(Δ)-induced DR pathway.
The CREB-binding protein CBP-1 is required for mir-80(Δ)-dependent changes in DR fluorimetric indicators and for mir-80(Δ)-dependent longevity
C. elegans histone acetyltransferase transcriptional coactivator homolog cbp-1 is required for lifespan extension via at least three different DR regimens (growth in axenic media, growth in diluted bacteria in liquid media, and the eat-2 feeding-impaired model). In the bDR regimen, cbp-1 deficiency has been shown to disrupt expression of daf-16 and hsf-1 target genes [32], [37] and thus the action of two transcription factors that influence mir-80(Δ) benefits has been mechanistically linked to CBP-1 in DR. We find that cbp-1(RNAi) in the mir-80(Δ) mutant reverses the DR-associated Exmax shift (p<0.05 +/− RNAi; Fig. 6A), and increases age pigment levels in day 4 animals (p<0.09, +/− RNAi, Fig. 6B) (cbp-1(RNAi) does not affect Exmax but modestly reduces age pigment levels in WT (data not shown)). Thus, cbp-1 activity plays a role in mir-80(Δ) regulation of age pigments, and appears generally needed for mir-80(Δ) DR metabolism. Consistent with a contribution to DR benefits, we find that the lifespan extension conferred by mir-80(Δ) depends strongly on cbp-1 (p<0.003; Fig. 6C).
Sequences within the cbp-1 transcript may be direct binding targets of miR-80
Interestingly, of the three transcription factors required for mir-80(Δ) healthspan, cbp-1 is the only one for which the transcript is predicted to include potential miR-80 miRNA target sequences (Fig. 6D). One candidate miR-80 binding site is present in the cbp-1 5′UTR, and another is present within exon 8. To test whether direct CBP-1 regulation might be a mechanism by which miR-80 controls metabolic state, we constructed translational reporters in which the cbp-1 promoter drives expression of a GFP that includes either no candidate miR-80 binding sites (NBS) or both the 5′UTR and the exon 8 candidate binding sites (5+8BS) (Fig. S7A).
We compared GFP expression levels of these constructs in ad lib fed animals +/− mir-80, with a focus on the posterior gut region in which mir-80 regulation is most dramatic. We find that the NBS construct is not regulated by miR-80(Δ) (Fig. S7B, left panel); whereas the 5+8BS construct is expressed at a higher level in the absence of mir-80 (Fig. S7B, right panel). Although rigorous testing in native context will be required to validate cpb-1 as a direct miR-80 target, our data suggest binding sites in the cbp-1 transcript may contribute to cbp-1 inhibition by miR-80 when levels are high in food.
If mir-80 represses cbp-1 translation, then we would expect higher levels of CBP-1 protein in mir-80(Δ) animals. We measured CBP-1 protein levels using anti-CBP antibodies against human CREBBP for WT and mir-80(Δ) mutants (day 7). We find that CBP-1 protein levels are significantly increased in mir-80(Δ) mutants compared to WT (p<0.05, Fig. 6E). Thus, in whole animal context, mir-80(Δ) is associated with increased CBP-1 protein.
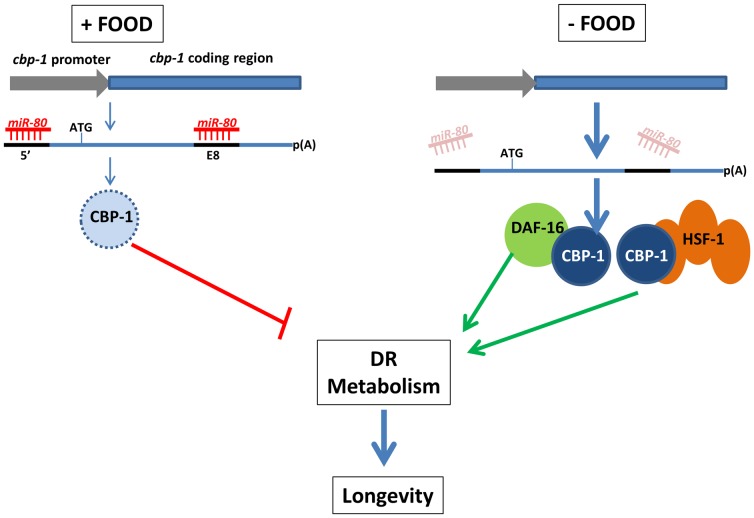
Our data are consistent with a model in which in the presence of food, cbp-1 is translationally repressed by binding of miR-80 to target sites within the cbp-1 transcript (Fig. 7). When food is lacking, miR-80 levels drop, translational repression of cbp-1 is relieved, and CBP-1+DAF-16+HSF-1-mediated transcriptional changes induce DR within the animal. Interestingly, the human CREBBP transcript might be targeted by miR-80 family members or another miRNA homologous to the exon 8 site (Fig. S7C), suggesting miRNAs could exert a conserved role in DR metabolic regulation that might be harnessed in the future to promote healthy metabolism with anti-aging applications.
Figure 7. A model for miR-80 regulation of DR metabolism.
In adults, when food is abundant, mir-80 is expressed at a high level, and miR-80 binds to metabolic and signaling targets to down-regulate their expression. The cbp-1 transcript, which includes two potential binding sites for miR-80, one in the 5′ UTR and one in exon 8 (exons thick dark blue lines, promoter lighter blue), and is essential for mir-80(Δ) benefits, is one candidate target (light blue represents relatively low CBP-1 concentration in food). When food is limiting, miR-80 levels drop, and translational repression of cbp-1 could be relieved (dark blue circle represents higher concentration CBP-1). The CBP-1 protein associates with DAF-16 and HSF-1 to promote expression of genes required for DR metabolism and longevity. Note that although cbp-1 is essential for mir-80(Δ) DR benefits, direct targeting remains to be proved and it is likely that additional targets help modulate the DR state. Since we cannot rule out that daf-16, hsf-1, and cbp-1 disruptions make animals too generally sick to gain mir-80(Δ) benefits, alternative models are possible.
Discussion
Deletion of a single C. elegans miRNA, mir-80, induces systemic healthy aging—improving cardiac muscle-like and skeletal-muscle-like maintenance and function later into life, limiting age-associated accumulation of lipofuscin-like material in the gut, and extending lifespan. Our data indicate that miR-80 acts as a negative regulator of metabolic loops that promote DR metabolism when nutrients are scarce. Acetyltransferase CBP-1 acts together with DAF-16/FOXO and HSF-1 to promote healthy metabolism in this regulatory circuit. Sequences within the Cecbp-1 transcript, the protein product of which increases in DR nematodes ([32], Fig. 6E) and in hypoglycemic mouse [38], may serve as direct targets of miR-80 down-regulation when food is abundant. Similarities between miR-80/target features in nematodes and mammals raise the possibility that miRNA manipulation of related DR metabolic loops in humans might be recruited to promote healthy aging.
mir-80 is expressed broadly and is regulated by food availability
mir-80 is an abundant, widely-expressed miRNA, and thus might be involved in global regulation of metabolism coordinated across tissues. Indeed, multiple mir-80 reporters indicate broad cellular expression and regulation by E. coli food availability. However, not all tissues/cells reflect similar magnitudes of regulation, with the largest fold food-induced change in expression in posterior intestine (Fig. 3D). The dramatic gut regulation raises the possibility that intestinal cells, well-positioned to monitor nutrient uptake, might play the most critical role in metabolic sensing and control. We speculate that miR-80 level changes in intestinal cells might initiate body-wide signaling via gut secretion of insulins and other hormones, analogous to human gastrointestinal tract and adipose tissue hormonal signaling to hypothalamus [39]. Because mir-80(Δ) induces skn-1::GFP expression in the ASI neurons (previously suggested to be similar to hypothalamic neurons [7]) but mir-80 is not expressed in ASI neurons (Fig. S4A,B), relief of miR-80 repression under food limitation could act upstream of ASI skn-1 induction via a gut-to-neuron signaling relationship.
DAF-16, HSF-1 and CBP-1 transcription factors are needed for mir-80(Δ)-induced healthspan benefits in a likely complex regulatory circuit
The requirement for daf-16, hsf-1, and cbp-1 in mir-80(Δ) DR is interesting in multiple regards. First, DAF-16/FOXO and HSF-1 can each individually bind to CBP-1 in nematodes and mammals (C. elegans DAF-16 and CBP-1; mammalian FOXO3A and CBP [40]; mammalian HSF-1 and CBP1 [41]), underscoring their capacity to co-regulate transcription. Second, previous work identified C. elegans daf-16 and hsf-1 as required for the CBP-1-dependent bDR lifespan extension [37]. In the bDR study, cbp-1(RNAi) blocked expression of DAF-16 and HSF target genes sod-1 and sip-1, respectively, rather than blocking transcriptional induction of daf-16 and hsf-1 that accompanies bDR. These data suggest that the CBP-1 cofactor couples and modifies transcriptional outputs of DAF-16- and HSF-1-dependent longevity pathways under bDR conditions, a model likely to apply for mir-80(Δ)-induced DR.
Although our study focused on DR genes that have most dramatic impact on the age pigment DR signature, we emphasize that our data support that additional genes contribute in a complex network to regulate age pigment phenotypes in mir-80(Δ). For example, knockdown of either AMPK subunit encoded by the C. elegans genome, aak-1 or aak-2, can alter age pigment levels (Tables S1 and S2), but not Exmax shift, suggesting separate regulation of lipofuscin content and levels. We thus anticipate that our data just touches the surface of a large interrelated network of metabolic genes and processes that are regulated by miR-80.
Could miR-80 directly target the cbp-1 transcript to regulate protein levels?
We fully expect that miR-80 regulates dietary restriction by binding to multiple target transcripts. An interesting candidate target, however, is the cbp-1 gene itself, which we have shown to be critical for mir-80(Δ)-induced DR benefits. The potential cbp-1 target sites for miR-80 binding are unusual, being situated in the 5′ UTR and within a highly conserved exon. Interestingly, the 5′ UTR sequences in cbp-1 are perfectly conserved in C. brenneri, C. briggsae, and C. remanei (though not in C. japonica) and the exon 8 site is somewhat conserved among all (Fig. S7D). Exon targeting by miRNAs is common in plants [42] and has been demonstrated for mammalian transcription factors Nanog, Oct4, and Sox2 [43], [44], fly DICER [45], and is now predicted in many additional genes after algorithm refinements that consider coding sequences [45], [46].
Ideally, we could test direct miR-80 targeting in vivo by manipulation of a cbp-1 transgene, +/− candidate miR-80 binding sites. Technical challenges, including the long length of the cbp-1 gene/cDNA, as well as an apparent exquisite sensitivity of CBP-1 activity levels for health and viability [47], [48], precluded direct study. Our studies of expression of a GFP transgene flanked by the 5′ UTR and the exon 8 sites from cbp-1 supported that miR-80 can down-regulate artificial construct expression in posterior gut. Although not definitive proof of direct targeting, these data, together with our findings that CBP-1 protein levels are elevated in DR (Fig. 6E; DR induction of CBP-1 also reported in [32], [37]) and miR-80 levels drop in DR (Fig. 3, Fig. S3, S5) are consistent with a model in which miR-80 mediates DR regulation by directly effecting CBP-1 levels (Fig. 7). Even if miR-80 effects are indirect, it is clear that cbp-1 is critical for mir-80(Δ)-induced age pigment and lifespan changes. Given that cbp-1 plays a role in dietary restriction associated with growth in axenic medium, growth on diluted bacteria, and eat-2 feeding impairment [37] and intersects with the insulin pathway for lifespan extension [37], and that we have noted mir-80 expression regulation under bacterial dilution and dietary deprivation, and a partial engagement of the insulin signaling pathway in mir-80(Δ)-induced longevity (Fig. 4D), the miR-80/CBP-1 regulatory loop may constitute a core mechanism by which diverse and intersecting metabolic pathways are coordinately regulated to respond to nutrient availability.
Might the conserved miR-80 microRNA family regulate metabolism across species?
C. elegans miR-80 family
The most conserved mir-80 family members encoded in the C. elegans genome are mir-80, mir-58, mir-81 and mir-82 [12], [18]. We did not find an Exmax shift in mir-58(Δ) or in the double mir-81(Δ) mir-82(Δ) mutant (data not shown) and thus mir-80 is the sole family member that can be deleted to induce the DR Exmax shift. Interestingly, however, the quadruple mutant mir-80; mir-58; mir-81-82 has a very small body size, more severe than the scrawny body type we documented for mir-80(Δ) (Fig. 2C), which can be rescued by a mir-80 high copy number transgene [18] suggesting some functional redundancy among mir-80 family members.
Drosophila melanogaster
mir-80 is homologous to the miRNA bantam in Drosophila melanogaster (Fig. S7E), well studied for roles in developmental growth and cell death regulation [49], [50], [51], [52], and more recently implicated in regulation of the core circadian clock [53], neuronal dendritic growth regulation by epithelia [51], and regulation of ecdysone/insulin interplay that influences body size [54]. Our data raise the question of whether developmental processes or circadian clocks might be sensitive to metabolic state, or whether modulation of metabolism might be used to regulate growth.
Human
To date, there are 3 identified human miRNAs closely related to miR-80: hsa-mir-450b-3p; hsa-mir-556-5p, and hsa-mir-3689a-5p (Fig. S7E), none of which have been well studied for function in mammalian biology. The human hCBP coding sequences corresponding to Cecbp-1 exon 8 have sufficient homology to human miR-80 family members to raise the possibility of analogous interaction and conserved regulatory mechanism. We do note, however, that hCBP coding sequences in the Ce exon 8-homologus region have a stronger match to hsa-mir-136 (more distantly related to Cemir-80) (Fig. S7C). If the negative regulatory interaction between miR-80 (or specific miRNAs) and DR targets is conserved, disruption of human regulatory miRNAs might be exploited to promote healthy aging.
Materials and Methods
We grew C. elegans under standard conditions [55] at 20°C, on E. coli strain OP50-1 or HT115 for RNAi. Age-synchronized cultures were prepared by bleach treatment egg preparation, with hatch counted as day 1. Note all presented data represent 3 independent combined trials; error bars +/− SEM. Protocol details are provided in Figure Legends and Supplemental Methods. Note all presented data in this paper represent at least 3 independent combined trials; error bars +/− SEM; we counted age with the egg hatch corresponding to day 0.
Strains and plasmids
A detailed list of strains is included as Table S3.
We grew C. elegans under standard conditions [55] at 20°C unless otherwise indicated. The food sources we used were E. coli strain OP50-1 or HT115 for RNAi feeding experiments (Caenorhabditis Genetics Center, University of Minnesota, Twin Cities, MN, USA). To generate synchronized cultures, we bleached gravid adults and starved L1 progeny. The wild-type strain was var. Bristol N2 [55]. The mir-80(nDf53) allele breakpoints are 5′- tgctttcgatgtctatactctc -3′ and 5′-tctggcgaacgaaatgagt-3′, encompassing part of the promoter region, the entire precursor sequence and ∼300 bp downstream. We genotyped mir-80(Δ) by PCR using primer pairs mir80Out-F (5′- ttcgtcgccatcaacacacg-3′)+mir80Out-R (5′- gagcgcggatagatatacagtcag-3′) that flank the deletion and mir80Flank-F (5′- caacaacgatgtgaatgctcgtc-3′)+mir80Flank-R (5′- ctcgcacacggacggactgcc-3′) that bind internal to nDf53. We worked with a 6× outcrossed line. The mir-80 deletion mutant does not exhibit gross developmental phenotypes ([20]; our observations). Developmental timing, L1 nuclei numbers, early adult locomotion, pumping rates, defecation rates, amphid neuron dye filling, and dauer entry/exit behaviors are within wild type ranges in mir-80(Δ), supporting that mir-80 does not contribute an essential role in development and basic function. Thus, mir-80 deletion primarily impacts adult maintenance and DR phenotypes.
For the Pmir-80L:mCherry transcriptional reporter, we amplified the mir-80 promoter using primers 5′-cgagatgagaagtaagaagagtgg-3′ and 5′-tccgtgtgcgagagagtgagcgag-3′ and cloned into the Pmec4::mCherry plasmid vector at the start codon of mCherry from [56] using the In-fusion cloning kit (Clontech Inc). The resulting plasmid was injected at 50 ng/ul into wild type animals along with a rol-6 co-injection marker (100 ng/ul) to generate extrachromosomal transgenic lines ZB3039-ZB3043.
For the binding site test constructs (Fig. S7A), we amplified the cbp-1 promoter (4.4 kb until start codon) using primers 5′-gACTAGTc tcttcc atgtcg gtttaa gcgcgg aaacgg tttttt aaa-3′ and 5′-tcccCCCGGGggga caatta gtagaa aaatgt atatat ttgac-3′ containing Spe1 and Xma1 restriction sites, respectively. This product was introduced within the Spe1-Xma1 digested pKS(-) vector. The mir-80 target sites were incorporated within primers that amplified the GFP coding sequence using the primer sites (outlined below) and introduced at the Xma1 site of the above cloned Pcbp-1 vector: NBS: 5′- tccccccgggatgagtaaaggagaagaacttttcactgg-3′+5′-cggggtaccctatagttcatccatgccatgtgtaatccc-3′; 5+8 BS: 5′- tccccccgggaacagctatatctggtgatttgatgagtaaagaagaag-3′+5′-cggggtacctaagatcctcttgactgaacacttcatagttcatccatgcc-3′
Measurement of fluorescent age pigments
We grew age-synchronized animals (see above) under standard conditions (20°C, OP50-1) and scanned animals (n≥50 per strain) for age pigment accumulation (Day 4, Day 9, Day 11) using a Fluorolog 3 spectroflorimeter as in Gerstbrien et al. [19]. All graphs represent mean data from at least 3 independent trials. For Exmax determination at Day 4, we used Datamax software (Horiba Scientific) to identify the peak excitation value. The peak for tryptophan fluorescence was also analyzed to normalize scores, as TRP levels do not change markedly with age.
qRT-PCR experiments for assaying gene expression changes
We synchronized strains by alkaline bleaching [57] and placed synchronized L1 larvae (Day 1) on NGM plates seeded with OP50-1 bacteria. On Day 4 or day 7, we moved approximately half the animals to plates containing OP50-1 with 50 uM FUdR. We used the other half for total RNA extraction using TRIZOL as described below. ∼1.5 ug of total RNA was used for cDNA synthesis using the Invitrogen SuperScript III cDNA synthesis kit and OligoDT primers to synthesize cDNA from all poly-adenylated RNA. We used 100 ng of cDNA to measure gene expression levels using the standard curve approach. Standard curves were generated from wild type cDNA by utilizing multiple dilutions of cDNA (1000, 100, 10, 1, 0.1, 0.01 ng) and probing for expression levels of the house-keeping gene, actin (act-1). Primers used were act1RT-F (5′- ttactctttcaccaccaccgctga-3′) and act1RT-R (5′- tcgtttccgacggtgatgacttgt -3′) for act-1, ama1RT-F (5′- cctacgatgtatcgaggcaaa-3′) and ama1RT-F (5′- cctccctccggtgtaataatg-3′) for ama-1, hsf1RT-F (5′-tagtaatggcagagatgcgtgcga-3′) and hsf1RT-R (5′- tggctgcatgacagagacgagaaa-3′) for hsf-1 and hsp16.2RT-F (5′- atggaacgccaatttgctccagtc-3′) and hsp16.2RT-R (5′- tccttggattgatagcgtacgacc-3′) for hsp-16.2. We plotted Ct values obtained from amplification for target DR genes against this standard curve to determine transcript levels.
Quantification of GFP fluorescence from mir-80 target site constructs
We synchronized strains (refer Fig. S7A,B, and Table S3) by alkaline bleaching [57] and placed synchronized L1 larvae (Day 1) on NGM plates seeded with OP50-1 bacteria. On Day 4, 100 mCherry(+) animals were picked and GFP fluorescence was measured in the spectrofluorimeter at 488 nm excitation and 511 nm emission. We measured fluorescence using ImageJ with a region-of-interest (ROI) that included the entire length of the body (using the line tool) and then plotted a histogram of the mean intensity along the length of the line.
Western blot analysis for detection of CBP-1 protein
We synchronized strains by alkaline bleaching [57] and placed synchronized L1 larvae (Day 1) on NGM plates seeded with OP50-1 bacteria. On Day 7, 250 animals were placed in 50 ul of RIPA buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% Sodium deoxycholate, 2.5 mM beta-glycerophosphate, 1 mM Na3VO4)+Protein sample buffer and heated at 95°C for 15 mins. 25 ul of samples was loaded onto a MiniProtean TGX gradient gel (4–20%, Bio-Rad) and transferred onto PVDF membrane following separation. Membrane was blocked using 5% non-fat milk in PBST buffer for 1 hour. Membrane was then incubated with CBP-1 and TUB-1 antibodies (Santa Cruz Biotechnology) at 1∶500 and 1∶4000 dilutions respectively in 2% non-fat milk overnight at 4°C. Protein bands were detected using the ECL reagent (Invitrogen) using horseradish peroxidase conjugated secondary antibodies (Jackson ImmunoResearch Labs) at 1∶10,000 dilutions. Band intensities were calculated using ImageJ [58].
Supporting Information
Individual lifespan data for lifespan analysis of mir-80(Δ). We grew age-synchronized WT (black), mir-80(Δ) (red) or the mir-80(+) (grey) under standard plate conditions (200C, OP50-1). At day 9, we placed 10 healthy animals per plate, and we scored viability as movement away from pick touch on the indicated days. Statistics were calculated using the OASIS software. Details are presented in Table format. Three additional trials that did not include the rescued strain, as well as other trials featured in the text, showed similar trends, on the order of 10–30% lifespan extension. Note the some trials with mir-80 transgene rescue suggest that overexpression of mir-80 may be deleterious, and that in a small minority of trials, we did not see life extension although the culture always trended in that direction.
(TIF)
The mir-80(Δ) mutant exhibits hypersensitivity to the DR-mimetic drug metformin, similar to DR mutant eat-2. We grew age-synchronized WT (black), mir-80(Δ) (red) or the eat-2 mutant (blue) under standard plate conditions supplemented with 50 mM metformin (20°C, OP50-1). At day 9, we placed 10 healthy animals per plate, ≥40 per strain per trial, and we scored viability as movement away from pick touch on the indicated days. The upper left graph (A) represents data combined from 3 independent trials, which are presented individually in the other panels (B-D). Statistics are calculated using the Log-rank Test. Error bars indicate ± S.E.M. Metformin reduces lifespan for mir-80(Δ) and eat-2 as compared to WT.
(TIF)
Published mir-80::GFP reporters are regulated by food availability. Fig. S3A. Example of expression in the Ex[Pmir-80::GFP] line VL211 ([17], wwEx18) grown in the presence of unlimited E. coli. Animals are at day 6 from the hatch for all images in this figure. + indicates anterior. Fig. S3B. Excretory duct cell in the Is[Pmir-80::GFP] in line VT1492 ([17], maIs196) grown in the presence of unlimited E. coli. White arrow indicates the fluorescent cell that is well labeled in this integrated line, tentatively identified as the excretory duct cell. . Example of expression of Ex[Pmir-80::GFP] in line VL211 grown in the presence of unlimited E. coli to young adulthood and then switched to no food for 48 hours. Fig. S3D. Example of Is[Pmir-80::GFP] line VT1492 excretory duct cell (white arrow) grown in the presence of unlimited E. coli to young adulthood and then switched to no food for 48 hours. Fig. S3 E,F. Quantitation of fluorescence signals for mir-80 promoter fusion reporter lines in food vs. food limitation. (E) Fluorescence of overall Ex[Pmir-80::GFP] line VL211, (F) excretory duct cell Is[Pmir-80::GFP] line VT1492; after 48 hrs on no-food plates. Food limitation in these studies was by dietary deprivation [5], but food dilution on solid NGM media [8] and food dilution in liquid media [37] induced similar changes in these lines (see Fig. S5). Graphs represent measured fluorescence levels (whole body for E; cell region for F) for at least 50 animals per DR regimen. Error bars represent mean intensity ± S.E.M. Pairwise comparisons were made using Two-tailed Students' T-test. *** - p<0.0005, ** - p<0.005. Same exposure times were used for complementary panels.
(TIF)
A,B. Two different transcriptional reporters for mir-80 fail to co-localize with ASI sensory neurons, so up-regulation of skn-1::GFP expression in those neurons in DR may be a non-autonomous consequence of miR-80 activity. The expression of transcription factor skn-1 in the two head ASI sensory neurons can be necessary for DR lifespan extension benefits [7], and we have shown that mir-80(Δ) increases the expression of a skn-1::GFP reporter in the ASI neurons (Fig. 2E). Thus, an important mechanistic question is whether miR-80 is present in the ASI neurons where it might cell-autonomously affect skn-1 expression. To test for mir-80 expression in ASI neurons, we took advantage of the fact that ASI neuron endings are open to the environment and can uptake fluorescent dye from their surroundings. We used a dye-filling assay to label the amphid sensory neurons in the Pmir-80 fluorescent reporter lines to test for co-expression (Pmir-80::GFP with red DiO, and Pmir-80L::mCherry with yellow DiI). We reared animals under standard conditions (ad lib OP50-1, 20°C). We labelled {A} VL211 expressing Pmir-80::GFP with red DiO, and reciprocally, {B} strain ZB3042 containing bzEx207[Pmir-80L::mCherry] with yellow fluorescent DiI, using a standard protocol that enables the amphid neurons that are open to the environment (ASI, ADL, ASK, AWB, ASH and, ASJ) to dye-fill. For both approaches, we never observed co-label of the mir-80 reporter with any amphid neurons (n = 30 per reporter); white arrows indicate ASI in representative images. Thus, although mir-80(Δ) influences skn-1::GFP expression in ASI neurons in older animals (Fig. 2E), expression data suggest miR-80 does not act cell autonomously in ASI neurons to exert this regulation. C. Absence of fluorescent bleed-through in through the GFP/DAPI filter sets in the Pmir-80L::mCherry lines Pmec-4::GFP (top row) and Pmir-80::mCherry (bottom row) lines were age-synchronized via alkaline bleaching and plated on standard NGM containing OP50-1 as food. Day 4 animals were imaged using 3 filter sets – DIC (left), FITC (to image GFP, middle) and Texas Red (to image mCherry, right). Touch neurons labeled with GFP in the Pmec-4::GFP strain are clearly and distinctly visible using the FITC channel (white arrows) and are absent in the Texas Red channel. Conversely, mCherry signal from a transcriptional reporter of mir-80 is clearly visible in the TexasRed channel (white arrows) but is absent in the FITC channel. Thus, our images of mCherry in adult intestine is unlikely to inadvertently score age pigment fluorescence.
(TIF)
mir-80 is regulated by food availability under multiple food restriction conditions and as assayed with multiple reporters. We grew strains either under abundant food conditions or using the dietary limitation protocol indicated. Graphs represent measured fluorescence levels from areas indicated for 2 trials of at least 50 animals per DR regimen (except liquid DR for VL211, 17 worms as the strain bagged frequently), measured 48 hours after dietary limitation. Error bars represent Mean Intensity ± S.E.M. Pairwise comparisons were made using Two-tailed Students' T-test, ** - p<0.0005, ** - p<0.005. a) overall pmir-80GFP line VL211 expression, food dilution on solid NGM media. b) overall pmir-80GFP line VL211 expression, food dilution in liquid media. c) excretory duct cell pmir-80GFP line VT1492 expression, food dilution on solid NGM media. d) excretory duct cell pmir-80GFP line VT1492 expression, food dilution in liquid media. e) overall fluorescence pmir-80LmCherry expression for 4 independently derived lines, in abundant food or under dietary deprivation. In individual trials, 4/5 tested lines exhibited significant differences; one line was not regulated in this direction (not shown).
(TIF)
Relative HSF-1 target gene hsp-16.2 transcripts are elevated in mir-80(Δ), but HSF-1 transcript levels are maintained. Temperature-induced HSF-1 activation occurs post-translationally, via formation of an active trimer from inactive monomers that persist under basal conditions [60]. To address the question of whether mir-80(Δ) mutants exhibit increased HSF-1 transcriptional activity compared to WT, we used qRT-PCR to measure transcript levels of HSF-1 target gene hsp-16.2 [61], +/− mir-80. Our data suggest that loss of mir-80 indirectly upregulates HSF-1 activity to increase expression of HSF-1-dependent target genes. A. Transcriptional expression of HSF-1 target gene hsp-16.2 is elevated in mir-80(Δ). We grew age-synchronized WT and mir-80(Δ) under standard conditions of abundant food (20°C, OP50-1) and harvested animals at Day 4 for total RNA isolation. We normalized raw qPCR scores for hsp-16.2 (an hsf-1 target) to ama-1. Graphs represent cumulative data from 3 independent trials with 3 technical replicates per trial. Error bars represent ±S.E.M. for RAW transcript gene values normalized to RAW ama-1 levels. Normalized transcript levels of hsp-16.2 are elevated in mir-80(Δ), * p<0.1 (2-tailed Student's T-test). B. hsf-1 transcript levels (day 4) are not changed by the absence of miR-80. We grew age-synchronized WT animals in abundant food and harvested animals at Day 4 for RNA isolation. Raw qPCR scores for hsf-1 were normalized to ama-1. Graphs represent cumulative data from 3 independent trials with 3 technical replicates per trial. Error bars represent ±S.E.M for RAW transcript gene values normalized to RAW ama-1 levels. In Day 4 animals normalized hsf-1 transcript levels were similar in WT and mir-80(Δ) (p = 0.90; 2-tailed Student's T-test). We noted a trend toward lower hsf-1 transcript levels at day 7 (not shown), which we think reflects modulation of the dynamic transcriptional network altered by miR-80. C. Transcriptional expression of HSF-1 target gene hsp-16.2 is elevated in mir-80(Δ). We grew age-synchronized WT (black bars) and mir-80(Δ) (red bars) under standard conditions of abundant food (20°C, OP50-1) and harvested animals at Day 4 for total RNA isolation. We normalized RAW qPCR scores for hsp-16.2 to act-1. Graphs represent cumulative data from 3 independent trials with 3 technical replicates per trial. Error bars represent ±S.E.M for RAW transcript gene values normalized to RAW act-1 levels. Data were compared using 2-tailed Student's T-test. Normalized transcript levels of hsp-16.2 are elevated in mir-80(Δ), * p<0.07. D. hsf-1 transcript levels are not regulated by the presence of miR-80 at day 4. We grew age-synchronized WT (black bars) and mir-80(Δ) (red bars) under standard conditions of abundant food (20°C, OP50-1) and harvested animals at Day 4 for RNA isolation. RAW qPCR scores for hsf-1 were normalized to act-1. Graphs represent cumulative data from 3 independent trials with 3 technical replicates per trial. Error bars represent ±S.E.M. for RAW transcript gene values normalized to RAW act-1 levels. Data were compared using 2-tailed Student's T-test. In Day 4 animals normalized hsf-1 transcript levels were similar in WT and mir-80(Δ) (p = 0.14 act-1). We noted a trend toward lower hsf-1 transcript levels at day 7 (not shown) that we think reflects modulation of the dynamic transcriptional network altered by miR-80.
(TIF)
miR-80 may directly target cbp-1 mRNA in the posterior gut, is a conserved miRNA and potential binding sites for miRNAs in human CREBBP are present at sites analogous to those in C. elegans. A. Constructs used to test candidate miR-80 binding sites present in cbp-1 for roles in translational repression. We used the native cbp-1 promoter to express GFP reporters that included i) GFP lacking any candidate miR-80 binding sites (NBS); ii) the 5′ UTR candidate miR-80 binding site and the exon 8 candidate miR-80 binding site from cbp-1 (5+8BS). B. Shown are GFP intensities for extrachromosomal p cbp-1GFP constructs without candidate miR-80 binding sites (NBS, left panel) or with the 5′ and exon 8 candidate miR-80 binding sites added (5+8BS); in WT (black line) or mir-80(Δ) (red line) backgrounds, day 7, n>30, posterior gut. Animal segments imaged and fluorescence measured as in Fig. 3. The 5+8bs construct is expressed at a higher level when mir-80 is lacking, whereas the NBS construct is not. The 5+8BS high copy number construct also variably exhibited some “sponge” effects that might be attributed to titrating out endogenous miR-80 and family members (not shown). C. Exon structure of C. elegans cbp-1 is indicated by thick blue bars, introns in thin blue lines (see WormBase for details). The rna22 algorithm [62] predicts that miR-80 binds cbp-1 within the 5′ UTR and within exon 8. The potential alignments of miR-80 (red) to C. elegans cbp-1 (blue) sequences are indicated. Note that the seed match to the exon 8 region is a perfect 10 bp match for C. elegans, and that the target sequence is conserved to some degree in mouse and human CBP1. However, human miR-136 is even a better match in this region (shown here). D. The predicted miR-80 target sites are conserved in the various Caenorhabditis spp. E. Alignments of miR-80 family members from Drosophila melanogaster (bantam) and human with C. elegans miR-80. Seed region is indicated by black bar; yellow highlight, full conservation; blue highlight, conserved. Note that there are 4 close members of the miR-80 family encoded in the C. elegans genome (mir-58, mir-81, mir-82; see [12], 2008, for alignments and discussion).
(TIF)
An RNAi screen identifies hsf-1 and cbp-1 as required for the DR-associated Exmax shift phenotype in the mir-80(Δ) background. We examined the literature for genes experimentally implicated in DR and used RNAi to knock these genes down in the mir-80(Δ) background (Day 4, 20°C, three independent trials, 50 animals/trial). Since the Exmax shift is diagnostic of the DR state, we reasoned that genetic interventions that reversed the shift would define genes needed for the mir-80(Δ) DR pathway. P-values were calculated using two-tailed unpaired Students T-test; gene knockdowns that confer statistically significant p-values are in bold.
(PDF)
RNAi directed against known DR genes in the mir-80(Δ) background identify genes required for the low age pigment level phenotype. We used RNAi to knockdown genes previously implicated in DR in the mir-80(Δ) background (Day 4, 20°C, three independent trials, 50 animals/trial) and quantitated age pigment levels relative to endogenous Trp levels. Since low age pigment/Trp ratios (AGE/TRP) are associated with the DR state [19], we reasoned that genetic interventions that elevated age pigment levels would help identify genes needed for the mir-80(Δ) DR pathway and/or for enhanced healthspan. P-values were calculated using the Unpaired two-tailed Student's T-test, values within the 90% confidence level are bold, increase or decrease indicated by arrows.
(PDF)
Strains used in this study.
(PDF)
Summary of lifespan experiments. Median lifespan is calculated from the GraphPad Prism Software while Maximum lifespan was calculated by the mean lifespan of the oldest 10% cohort in each experiment. P-values are calculated from Log-Rank Tests for the entire population for the complete lifespan.
(PDF)
Supplementary methods.
(PDF)
References used exclusively in the supplemental information.
(PDF)
Funding Statement
The work here was supported by NIH R01 AG033858, R21 AG033369 and Arra supplement. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kenyon CJ (2010) The genetics of ageing. Nature 464: 504–512. [DOI] [PubMed] [Google Scholar]
- 2. Tissenbaum HA (2012) Genetics, life span, health span, and the aging process in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci 67: 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fontana L, Partridge L, Longo VD (2010) Extending healthy life span–from yeast to humans. Science 328: 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greer EL, Brunet A (2009) Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell 8: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee GD, Wilson MA, Zhu M, Wolkow CA, de Cabo R, et al. (2006) Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell 5: 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaeberlein TL (2006) Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell 5: 487–494. [DOI] [PubMed] [Google Scholar]
- 7. Bishop NA, Guarente L (2007) Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447: 545–549. [DOI] [PubMed] [Google Scholar]
- 8. Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR (2003) Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp Gerontol 38: 947–954. [DOI] [PubMed] [Google Scholar]
- 9. Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, et al. (2007) An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans . Curr Biol 17: 1646–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rottiers V, Naar AM (2012) MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 13: 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pasquinelli AE (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13: 271–282. [DOI] [PubMed] [Google Scholar]
- 12. Ibanez-Ventoso C, Vora M, Driscoll M (2008) Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS One 3: e2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fabian MR, Sonenberg N (2012) The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 19: 586–593. [DOI] [PubMed] [Google Scholar]
- 14. Berezikov E (2011) Evolution of microRNA diversity and regulation in animals. Nat Rev Genet 12: 846–860. [DOI] [PubMed] [Google Scholar]
- 15. Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, et al. (2005) RAS is regulated by the let-7 microRNA family. Cell 120: 635–647. [DOI] [PubMed] [Google Scholar]
- 16. Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, et al. (2010) Regression of murine lung tumors by the let-7 microRNA. Oncogene 29: 1580–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, et al. (2008) Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res 18: 2005–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alvarez-Saavedra E, Horvitz HR (2010) Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol 20: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gerstbrein B, Stamatas G, Kollias N, Driscoll M (2005) In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans . Aging Cell 4: 127–137. [DOI] [PubMed] [Google Scholar]
- 20. Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, et al. (2007) Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet 3: e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang C, Xiong C, Kornfeld K (2004) Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans . PNAS 101: 8084–8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, et al. (2002) Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans . Nature 419: 808–814. [DOI] [PubMed] [Google Scholar]
- 23. Wolkow CA, Kimura KD, Lee M, Ruvkun G (2000) Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290: 147–150. [DOI] [PubMed] [Google Scholar]
- 24. Chow DK, Glenn CF, Johnston JL, Goldberg IG, Wolkow CA (2006) Sarcopenia in the Caenorhabditis elegans pharynx correlates with muscle contraction rate over lifespan. Exp Gerontol 41: 252–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Onken B, Driscoll M (2010) Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans Healthspan via AMPK, LKB1, and SKN-1. PLoS One 5: e8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hansen M, Chandra Al, Mitic LL, Onken B, Driscoll M, et al. (2008) A role for autophagy genes in the extension of lifespan by dietary restriction in C. elegans. PLoS Genetics 4: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lakowski B, Hekimi S (1998) The genetics of caloric restriction in Caenorhabditis elegans . Proceedings of the National Academy of Sciences of the United States of America 95: 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaeberlein TL, Smith ED, Tsuchiya M, Welton KL, Thomas JH, et al. (2006) Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell 5: 487–494. [DOI] [PubMed] [Google Scholar]
- 29. Shibata Y, Fujii T, Dent JA, Fujisawa H, Takagi S (2000) EAT-20, a novel transmembrane protein with EGF motifs, is required for efficient feeding in Caenorhabditis elegans. Genetics 154: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hansen M, Hsu AL, Dillin A, Kenyon C (2005) New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet 1: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mair W, Dillin A (2008) Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem 77: 727–754. [DOI] [PubMed] [Google Scholar]
- 32. Mair W, Panowski SH, Shaw RJ, Dillin A (2009) Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS One 4: e4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. An JH, Blackwell TK (2003) SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev 17: 1882–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, et al. (2010) MicroRNAs both promote and antagonize longevity in C. elegans. Curr Biol 20: 2159–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kato M, de Lencastre A, Pincus Z, Slack FJ (2009) Dynamic expression of small non-coding RNAs, including novel microRNAs and piRNAs/21U-RNAs, during Caenorhabditis elegans development. Genome Biol 10: R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Steinkraus KA, Smith ED, Davis C, Carr D, Pendergrass WR, et al. (2008) Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell 7: 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang M, Poplawski M, Yen K, Cheng H, Bloss E, et al. (2009) Role of CBP and SATB-1 in aging, dietary restriction, and insulin-like signaling. PLoS Biol 7: e1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mastaitis JW, Wurmbach E, Cheng H, Sealfon SC, Mobbs CV (2005) Acute induction of gene expression in brain and liver by insulin-induced hypoglycemia. Diabetes 54: 952–958. [DOI] [PubMed] [Google Scholar]
- 39. Badman MK, Flier JS (2005) The gut and energy balance: visceral allies in the obesity wars. Science 307: 1909–1914. [DOI] [PubMed] [Google Scholar]
- 40. Nasrin N, Ogg S, Cahill CM, Biggs W, Nui S, et al. (2000) DAF-16 recruits the CREB-binding protein coactivator complex to the insulin-like growth factor binding protein 1 promoter in HepG2 cells. Proc Natl Acad Sci U S A 97: 10412–10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hong S, Kim SH, Heo MA, Choi YH, Park MJ, et al. (2004) Coactivator ASC-2 mediates heat shock factor 1-mediated transactivation dependent on heat shock. FEBS Lett 559: 165–170. [DOI] [PubMed] [Google Scholar]
- 42. Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799. [DOI] [PubMed] [Google Scholar]
- 43. Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I (2008) MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128. [DOI] [PubMed] [Google Scholar]
- 44. Tay YM, Tam WL, Ang YS, Gaughwin PM, Yang H, et al. (2008) MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells 26: 17–29. [DOI] [PubMed] [Google Scholar]
- 45. Forman JJ, Legesse-Miller A, Coller HA (2008) A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A 105: 14879–14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marin RM, Sulc M, Vanicek J (2013) Searching the coding region for microRNA targets. RNA 19: 467–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shi Y, Mello C (1998) A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev 12: 943–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Eastburn DJ, Han M (2005) A gain-of-function allele of cbp-1, the Caenorhabditis elegans ortholog of the mammalian CBP/p300 gene, causes an increase in histone acetyltransferase activity and antagonism of activated Ras. Mol Cell Biol 25: 9427–9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xu P, Guo M, Hay BA (2004) MicroRNAs and the regulation of cell death. Trends Genet 20: 617–624. [DOI] [PubMed] [Google Scholar]
- 50. Oh H, Irvine KD (2011) Cooperative regulation of growth by Yorkie and Mad through bantam. Dev Cell 20: 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Parrish JZ, Xu P, Kim CC, Jan LY, Jan YN (2009) The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in drosophila sensory neurons. Neuron 63: 788–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thompson BJ (2010) Developmental control of cell growth and division in Drosophila. Curr Opin Cell Biol 22: 788–794. [DOI] [PubMed] [Google Scholar]
- 53. Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, et al. (2009) A role for microRNAs in the Drosophila circadian clock. Genes Dev 23: 2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boulan L, Martin D, Milan M (2013) bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Curr Biol 23: 473–478. [DOI] [PubMed] [Google Scholar]
- 55. Brenner S (1974) The genetics of Caenorhabditis elegans . Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pinan-Lucarre B, Gabel CV, Reina CP, Hulme SE, Shevkoplyas SS, et al. (2012) The core apoptotic executioner proteins CED-3 and CED-4 promote initiation of neuronal regeneration in Caenorhabditis elegans. PLoS Biol 10: e1001331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaham S, editor(2006) Worm Book - Methods in Cell Biology (January 02, 2006).
- 58. Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Meth 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M (2010) Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods 7: 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rabindran SK, Haroun RI, Clos J, Wisniewski J, Wu C (1993) Regulation of heat shock factor trimer formation: role of a conserved leucine zipper. Science 259: 230–234. [DOI] [PubMed] [Google Scholar]
- 61. Morley JF, Morimoto RI (2004) Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell 15: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, et al. (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Individual lifespan data for lifespan analysis of mir-80(Δ). We grew age-synchronized WT (black), mir-80(Δ) (red) or the mir-80(+) (grey) under standard plate conditions (200C, OP50-1). At day 9, we placed 10 healthy animals per plate, and we scored viability as movement away from pick touch on the indicated days. Statistics were calculated using the OASIS software. Details are presented in Table format. Three additional trials that did not include the rescued strain, as well as other trials featured in the text, showed similar trends, on the order of 10–30% lifespan extension. Note the some trials with mir-80 transgene rescue suggest that overexpression of mir-80 may be deleterious, and that in a small minority of trials, we did not see life extension although the culture always trended in that direction.
(TIF)
The mir-80(Δ) mutant exhibits hypersensitivity to the DR-mimetic drug metformin, similar to DR mutant eat-2. We grew age-synchronized WT (black), mir-80(Δ) (red) or the eat-2 mutant (blue) under standard plate conditions supplemented with 50 mM metformin (20°C, OP50-1). At day 9, we placed 10 healthy animals per plate, ≥40 per strain per trial, and we scored viability as movement away from pick touch on the indicated days. The upper left graph (A) represents data combined from 3 independent trials, which are presented individually in the other panels (B-D). Statistics are calculated using the Log-rank Test. Error bars indicate ± S.E.M. Metformin reduces lifespan for mir-80(Δ) and eat-2 as compared to WT.
(TIF)
Published mir-80::GFP reporters are regulated by food availability. Fig. S3A. Example of expression in the Ex[Pmir-80::GFP] line VL211 ([17], wwEx18) grown in the presence of unlimited E. coli. Animals are at day 6 from the hatch for all images in this figure. + indicates anterior. Fig. S3B. Excretory duct cell in the Is[Pmir-80::GFP] in line VT1492 ([17], maIs196) grown in the presence of unlimited E. coli. White arrow indicates the fluorescent cell that is well labeled in this integrated line, tentatively identified as the excretory duct cell. . Example of expression of Ex[Pmir-80::GFP] in line VL211 grown in the presence of unlimited E. coli to young adulthood and then switched to no food for 48 hours. Fig. S3D. Example of Is[Pmir-80::GFP] line VT1492 excretory duct cell (white arrow) grown in the presence of unlimited E. coli to young adulthood and then switched to no food for 48 hours. Fig. S3 E,F. Quantitation of fluorescence signals for mir-80 promoter fusion reporter lines in food vs. food limitation. (E) Fluorescence of overall Ex[Pmir-80::GFP] line VL211, (F) excretory duct cell Is[Pmir-80::GFP] line VT1492; after 48 hrs on no-food plates. Food limitation in these studies was by dietary deprivation [5], but food dilution on solid NGM media [8] and food dilution in liquid media [37] induced similar changes in these lines (see Fig. S5). Graphs represent measured fluorescence levels (whole body for E; cell region for F) for at least 50 animals per DR regimen. Error bars represent mean intensity ± S.E.M. Pairwise comparisons were made using Two-tailed Students' T-test. *** - p<0.0005, ** - p<0.005. Same exposure times were used for complementary panels.
(TIF)
A,B. Two different transcriptional reporters for mir-80 fail to co-localize with ASI sensory neurons, so up-regulation of skn-1::GFP expression in those neurons in DR may be a non-autonomous consequence of miR-80 activity. The expression of transcription factor skn-1 in the two head ASI sensory neurons can be necessary for DR lifespan extension benefits [7], and we have shown that mir-80(Δ) increases the expression of a skn-1::GFP reporter in the ASI neurons (Fig. 2E). Thus, an important mechanistic question is whether miR-80 is present in the ASI neurons where it might cell-autonomously affect skn-1 expression. To test for mir-80 expression in ASI neurons, we took advantage of the fact that ASI neuron endings are open to the environment and can uptake fluorescent dye from their surroundings. We used a dye-filling assay to label the amphid sensory neurons in the Pmir-80 fluorescent reporter lines to test for co-expression (Pmir-80::GFP with red DiO, and Pmir-80L::mCherry with yellow DiI). We reared animals under standard conditions (ad lib OP50-1, 20°C). We labelled {A} VL211 expressing Pmir-80::GFP with red DiO, and reciprocally, {B} strain ZB3042 containing bzEx207[Pmir-80L::mCherry] with yellow fluorescent DiI, using a standard protocol that enables the amphid neurons that are open to the environment (ASI, ADL, ASK, AWB, ASH and, ASJ) to dye-fill. For both approaches, we never observed co-label of the mir-80 reporter with any amphid neurons (n = 30 per reporter); white arrows indicate ASI in representative images. Thus, although mir-80(Δ) influences skn-1::GFP expression in ASI neurons in older animals (Fig. 2E), expression data suggest miR-80 does not act cell autonomously in ASI neurons to exert this regulation. C. Absence of fluorescent bleed-through in through the GFP/DAPI filter sets in the Pmir-80L::mCherry lines Pmec-4::GFP (top row) and Pmir-80::mCherry (bottom row) lines were age-synchronized via alkaline bleaching and plated on standard NGM containing OP50-1 as food. Day 4 animals were imaged using 3 filter sets – DIC (left), FITC (to image GFP, middle) and Texas Red (to image mCherry, right). Touch neurons labeled with GFP in the Pmec-4::GFP strain are clearly and distinctly visible using the FITC channel (white arrows) and are absent in the Texas Red channel. Conversely, mCherry signal from a transcriptional reporter of mir-80 is clearly visible in the TexasRed channel (white arrows) but is absent in the FITC channel. Thus, our images of mCherry in adult intestine is unlikely to inadvertently score age pigment fluorescence.
(TIF)
mir-80 is regulated by food availability under multiple food restriction conditions and as assayed with multiple reporters. We grew strains either under abundant food conditions or using the dietary limitation protocol indicated. Graphs represent measured fluorescence levels from areas indicated for 2 trials of at least 50 animals per DR regimen (except liquid DR for VL211, 17 worms as the strain bagged frequently), measured 48 hours after dietary limitation. Error bars represent Mean Intensity ± S.E.M. Pairwise comparisons were made using Two-tailed Students' T-test, ** - p<0.0005, ** - p<0.005. a) overall pmir-80GFP line VL211 expression, food dilution on solid NGM media. b) overall pmir-80GFP line VL211 expression, food dilution in liquid media. c) excretory duct cell pmir-80GFP line VT1492 expression, food dilution on solid NGM media. d) excretory duct cell pmir-80GFP line VT1492 expression, food dilution in liquid media. e) overall fluorescence pmir-80LmCherry expression for 4 independently derived lines, in abundant food or under dietary deprivation. In individual trials, 4/5 tested lines exhibited significant differences; one line was not regulated in this direction (not shown).
(TIF)
Relative HSF-1 target gene hsp-16.2 transcripts are elevated in mir-80(Δ), but HSF-1 transcript levels are maintained. Temperature-induced HSF-1 activation occurs post-translationally, via formation of an active trimer from inactive monomers that persist under basal conditions [60]. To address the question of whether mir-80(Δ) mutants exhibit increased HSF-1 transcriptional activity compared to WT, we used qRT-PCR to measure transcript levels of HSF-1 target gene hsp-16.2 [61], +/− mir-80. Our data suggest that loss of mir-80 indirectly upregulates HSF-1 activity to increase expression of HSF-1-dependent target genes. A. Transcriptional expression of HSF-1 target gene hsp-16.2 is elevated in mir-80(Δ). We grew age-synchronized WT and mir-80(Δ) under standard conditions of abundant food (20°C, OP50-1) and harvested animals at Day 4 for total RNA isolation. We normalized raw qPCR scores for hsp-16.2 (an hsf-1 target) to ama-1. Graphs represent cumulative data from 3 independent trials with 3 technical replicates per trial. Error bars represent ±S.E.M. for RAW transcript gene values normalized to RAW ama-1 levels. Normalized transcript levels of hsp-16.2 are elevated in mir-80(Δ), * p<0.1 (2-tailed Student's T-test). B. hsf-1 transcript levels (day 4) are not changed by the absence of miR-80. We grew age-synchronized WT animals in abundant food and harvested animals at Day 4 for RNA isolation. Raw qPCR scores for hsf-1 were normalized to ama-1. Graphs represent cumulative data from 3 independent trials with 3 technical replicates per trial. Error bars represent ±S.E.M for RAW transcript gene values normalized to RAW ama-1 levels. In Day 4 animals normalized hsf-1 transcript levels were similar in WT and mir-80(Δ) (p = 0.90; 2-tailed Student's T-test). We noted a trend toward lower hsf-1 transcript levels at day 7 (not shown), which we think reflects modulation of the dynamic transcriptional network altered by miR-80. C. Transcriptional expression of HSF-1 target gene hsp-16.2 is elevated in mir-80(Δ). We grew age-synchronized WT (black bars) and mir-80(Δ) (red bars) under standard conditions of abundant food (20°C, OP50-1) and harvested animals at Day 4 for total RNA isolation. We normalized RAW qPCR scores for hsp-16.2 to act-1. Graphs represent cumulative data from 3 independent trials with 3 technical replicates per trial. Error bars represent ±S.E.M for RAW transcript gene values normalized to RAW act-1 levels. Data were compared using 2-tailed Student's T-test. Normalized transcript levels of hsp-16.2 are elevated in mir-80(Δ), * p<0.07. D. hsf-1 transcript levels are not regulated by the presence of miR-80 at day 4. We grew age-synchronized WT (black bars) and mir-80(Δ) (red bars) under standard conditions of abundant food (20°C, OP50-1) and harvested animals at Day 4 for RNA isolation. RAW qPCR scores for hsf-1 were normalized to act-1. Graphs represent cumulative data from 3 independent trials with 3 technical replicates per trial. Error bars represent ±S.E.M. for RAW transcript gene values normalized to RAW act-1 levels. Data were compared using 2-tailed Student's T-test. In Day 4 animals normalized hsf-1 transcript levels were similar in WT and mir-80(Δ) (p = 0.14 act-1). We noted a trend toward lower hsf-1 transcript levels at day 7 (not shown) that we think reflects modulation of the dynamic transcriptional network altered by miR-80.
(TIF)
miR-80 may directly target cbp-1 mRNA in the posterior gut, is a conserved miRNA and potential binding sites for miRNAs in human CREBBP are present at sites analogous to those in C. elegans. A. Constructs used to test candidate miR-80 binding sites present in cbp-1 for roles in translational repression. We used the native cbp-1 promoter to express GFP reporters that included i) GFP lacking any candidate miR-80 binding sites (NBS); ii) the 5′ UTR candidate miR-80 binding site and the exon 8 candidate miR-80 binding site from cbp-1 (5+8BS). B. Shown are GFP intensities for extrachromosomal p cbp-1GFP constructs without candidate miR-80 binding sites (NBS, left panel) or with the 5′ and exon 8 candidate miR-80 binding sites added (5+8BS); in WT (black line) or mir-80(Δ) (red line) backgrounds, day 7, n>30, posterior gut. Animal segments imaged and fluorescence measured as in Fig. 3. The 5+8bs construct is expressed at a higher level when mir-80 is lacking, whereas the NBS construct is not. The 5+8BS high copy number construct also variably exhibited some “sponge” effects that might be attributed to titrating out endogenous miR-80 and family members (not shown). C. Exon structure of C. elegans cbp-1 is indicated by thick blue bars, introns in thin blue lines (see WormBase for details). The rna22 algorithm [62] predicts that miR-80 binds cbp-1 within the 5′ UTR and within exon 8. The potential alignments of miR-80 (red) to C. elegans cbp-1 (blue) sequences are indicated. Note that the seed match to the exon 8 region is a perfect 10 bp match for C. elegans, and that the target sequence is conserved to some degree in mouse and human CBP1. However, human miR-136 is even a better match in this region (shown here). D. The predicted miR-80 target sites are conserved in the various Caenorhabditis spp. E. Alignments of miR-80 family members from Drosophila melanogaster (bantam) and human with C. elegans miR-80. Seed region is indicated by black bar; yellow highlight, full conservation; blue highlight, conserved. Note that there are 4 close members of the miR-80 family encoded in the C. elegans genome (mir-58, mir-81, mir-82; see [12], 2008, for alignments and discussion).
(TIF)
An RNAi screen identifies hsf-1 and cbp-1 as required for the DR-associated Exmax shift phenotype in the mir-80(Δ) background. We examined the literature for genes experimentally implicated in DR and used RNAi to knock these genes down in the mir-80(Δ) background (Day 4, 20°C, three independent trials, 50 animals/trial). Since the Exmax shift is diagnostic of the DR state, we reasoned that genetic interventions that reversed the shift would define genes needed for the mir-80(Δ) DR pathway. P-values were calculated using two-tailed unpaired Students T-test; gene knockdowns that confer statistically significant p-values are in bold.
(PDF)
RNAi directed against known DR genes in the mir-80(Δ) background identify genes required for the low age pigment level phenotype. We used RNAi to knockdown genes previously implicated in DR in the mir-80(Δ) background (Day 4, 20°C, three independent trials, 50 animals/trial) and quantitated age pigment levels relative to endogenous Trp levels. Since low age pigment/Trp ratios (AGE/TRP) are associated with the DR state [19], we reasoned that genetic interventions that elevated age pigment levels would help identify genes needed for the mir-80(Δ) DR pathway and/or for enhanced healthspan. P-values were calculated using the Unpaired two-tailed Student's T-test, values within the 90% confidence level are bold, increase or decrease indicated by arrows.
(PDF)
Strains used in this study.
(PDF)
Summary of lifespan experiments. Median lifespan is calculated from the GraphPad Prism Software while Maximum lifespan was calculated by the mean lifespan of the oldest 10% cohort in each experiment. P-values are calculated from Log-Rank Tests for the entire population for the complete lifespan.
(PDF)
Supplementary methods.
(PDF)
References used exclusively in the supplemental information.
(PDF)