Abstract
This article describes the methods and techniques used to produce mutagenized mice to conduct high-throughput forward genetic screens for circadian rhythm mutants in the mouse. In particular, we outline methods to safely prepare and administer the chemical mutagen N-nitrosoN-ethylurea (ENU) to mice. We also discuss the importance of selecting mouse strain and outline breeding strategies, logistics, and throughput to produce these mutant mice. Finally, we discuss the breeding strategies that we use to confirm mutation heritability.
Introduction
High-throughput forward mutagenesis screens are ongoing at a variety of institutions to identify mouse behavioral mutants using the strategy of forward genetics (from phenotype to gene) (Justice et al., 1999; Moldin et al., 2001). Unlike reverse genetic (from gene to phenotype) approaches, forward genetic approaches make no assumptions concerning the underlying genes involved to illicit a given behavior. Random point mutations are introduced into the mouse genome with the chemical mutagen N-nitroso-N-ethylurea (ENU), and mutant mice are identified based on their altered phenotype. In forward genetic screens, a deep understanding of phenotype is the essence of successful endeavors.
We use circadian wheel running activity to identify mice expressing abnormal circadian rhythms. Although activity rhythms are robust, quantitative, and automated, other factors, such as mouse strain, ENU preparation and dosage, and colony breeding strategies are integral to the success of the screen. This article describes the methods employed to produce mutagenized mice to conduct forward genetic screens for circadian rhythm mutants in mouse.
Strain Choice
The choice of mouse strain is a major consideration for any mouse mutagenesis production colony. Strain choice dictates the effectiveness of the ENU to introduce mutations, the efficiency of the breeding scheme to produce mutant mice, and ultimately the ability to detect mutants in the phenotypic screens. To avoid inconsistencies and variability in any and all of these factors, we have elected to use only inbred strains for our mutagenesis program. The two strains that we have used, BTBR/J and C57BL/ 6J, were chosen for their ENU mutation rate, their fertility, and, most importantly, their circadian behavior. Bear in mind, however, that a high mutation rate and/or great fecundity can never compensate for a weak or inconsistent phenotype.
ENU is a very potent and powerful mutagen (Russell et al., 1979). Male mice are treated with the highest dose of ENU that can be tolerated without causing infertility. The primary target of ENU in the germline of mice is the spermatogonia, which can be highly mutagenized and go on to produce mutant gametes. Any animal sired from one of these mutagenized males will harbor multiple mutations in its genome. In behavioral phenotypes such as circadian rhythms, the number of gene targets is relatively low (perhaps 20–30 genes contribute to the behavior). The success of any mutation screen, therefore, is dependent on the forward mutation rate. Some mouse strains, such as BTBR/J, can tolerate a single high dose of ENU to achieve relatively high forward mutation rates (~1/250 per locus per gamete) (McDonald et al., 1990; Shedlovsky et al., 1993). Other strains, such as C57BL/6J, required multiple weekly injections of lower ENU doses to achieve a somewhat comparable forward mutation rate (~1/800 per locus per gamete) (Justice et al., 2000).
Second, it is important to select a mouse strain that is a good breeder. Average litter size and the tendency of female mice to eat their first litters will influence the breeding strategy and logistics used to produce mutant mice. In general, we have found that our mice breed better as trios (one male and two females) as opposed to pairs. Whenever possible, we rotated males through cages of females to increase the number of litters sired by each male.
It is possible to use mixed strain F1s to increase mutation frequencies and to increase fecundity. Although these hybrid mice are better breeders overall, it is far better from a phenotyping perspective to use inbred strains for a mutagenesis production colony. Hybrid and outbred strains perform inconsistently in most behavioral assays and their mixed genetic backgrounds can make it difficult to detect true phenotypic outliers. For example, [C57BL/6J × BALB/cJ]F2 hybrid mice display a wider range of circadian behavioral measurements as compared with parental inbred strains (Shimomura et al., 2001). If used in a mutagenesis screen, it would be difficult to determine whether any altered circadian phenotypes were due to a mutation or simply a result of the mixed genetic background.
Finally, the specific inbred strain used in a mutagenesis study can determine whether a mutant phenotype can even be detected. For example, C57BL/6J mice are robust runners with a somewhat invariant free running period. BALB/cJ mice, however, are sloppy runners with a more variable free running period (Schwartz and Zimmerman, 1990; Shimomura et al., 2001). It would be difficult to detect true outliers using BALB/cJ mice. Strain also influences the observed phenotype. For example, on a CF1 background the epidermal growth factor receptor (EGF-R) knockout is lethal at embryotic day 7.5 (Sibilia and Wagner, 1995; Threadgill et al., 1995). The same EGF-R knockout on a 129/Sv background is lethal at midgestation, lethal perinatally on a CD-1 background, and lethal postnatally on a 129/Sv × C57BL/6J background. Although we cannot predict a priori, which inbred strain is best suited to display a given mutant behavior or phenotype, we can select strains that have consistent and somewhat invariant phenotypic measurements to ensure that we will be able to detect a true phenotypic outlier.
ENU Safety Procedures
N-Nitroso-N-ethylurea is purchased in ISOPAC containers (Sigma # N3385 1 g/bottle) as a wetted solid and stored at −20° until needed. ISOPAC containment minimizes investigator contact with the ENU. We prepare the ENU and inject our mice within a SPF barrier facility. All investigators handling ENU wear a disposable laboratory coat, latex gloves, hair bonnet, disposable shoe covers, and safety goggles. To further minimize contact with the ENU, all procedures are performed within a biological safety cabinet lined with two layers of disposable bench paper. All solutions are introduced into and removed from the ISOPAC bottle through needles inserted through the rubber stopper of the container. All ENU-contaminated materials are placed in a solid waste container, collected, and removed from our facility by our hazardous waste staff. Per recommendation of our office of research safety, ENU-contaminated materials are not decontaminated with an alkaline sodium isothiosulfate solution. Although alkaline treatment neutralizes ENU, EPA guidelines dictate that investigators do not attempt to neutralize and dispose of hazardous materials within the laboratory. The following protocol produces approximately 100 ml of ENU stock solution. We rarely use more than 5 ml of solution per week. All excess ENU stock solution is disposed of immediately after use, and we prepare fresh ENU stocks for each round of injections.
ENU Preparation
To prepare an ENU solution stock, the powdered ENU is first dissolved in 10 ml of 95% ethanol within the ISOPAC bottle. The ethanol solution is injected into the bottle with an 18-gauge needle attached to a 10-ml syringe, the needle and syringe are removed, and the solution is agitated gently until the ENU is completely dissolved. An 18-gauge needle is inserted through the stopper of the bottle to act as a pressure vent. A second 18-gauge needle attached to a 60-ml syringe is used as an injection port to introduce 70 ml of PC buffer (0.1 M Na2HPO4, 0.05 M Na citrate pH adjusted to 5.0 and 0.2 Jm filter sterilized). The needles are removed and the solution is swirled gently. A small aliquot of ENU solution is removed ( 300 Jl) using a 1-ml tuberculin syringe, the sample is diluted 1:5 with PC buffer, and the OD398 is determined. ENU concentration is calculated where 1 OD398 ••• 0.72 mg/ml. For safety, the spectophotometer is kept in the biological safety cabinet and is used exclusively for ENU concentration determinations. Plastic disposable cuvettes are used in place of quartz cuvettes.
Enough additional PC buffer is added to the ENU solution to create a 10-mg/ml stock solution. Between injections the bottled ENU solution is vented with an 18-gauge needle. If the solution is not vented often, air bubbles accumulate within the injection syringes, preventing accurate ENU dose administration to the mice.
Injections
ENU treatments are initiated at 6 weeks of age. The mice are individually weighed and injected in the intraperitoneal cavity (ip) using 28-gauge short-needle insulin syringes (Becton-Dickson # 328438) with the proper dose of ENU. For C57BL/6J mice, we typically administer three weekly doses of 100 mg ENU/kg body weight (3 × 100). We mutagenize BTBR/J mice with a single dose of 250 mg/kg body weight (1 × 250).
To inject a mouse, we hold it by the scruff of the neck with the thumb and forefinger to splay the front legs back to prevent head movement. The tail is held between the ring and little fingers to extend the rear legs and expose the dorsal surface of the mouse. ENU is injected just anterior to the leg and 1 cm to the right or left of midline. This ensures that the ENU is not injected either into the bladder or into any major organ. If the ENU is injected into the bladder, the ENU is excreted and little, if any, ENU reaches the spermatogonia. If the ENU is injected into a major organ, the mouse does not survive more than 24 h postinjection. Because the final ENU stock solution contains 10% ethyl alcohol, all of the mice exhibit some symptoms of acute alcohol intoxication. The effect is short-lived and the mice resume their normal level of activity 2–4 h later.
We selected the 3 × 100-mg treatment as the highest dose of ENU that the C57BL/6J mice could tolerate, regain fertility, and live long enough to produce 50–100 offspring. There is some risk, however, that all of the mice injected with this high dose never regain fertility, develop tumors, and/or die. To ensure against this possibility, we also inject a second set of mice with a slightly lower dose of ENU (3 × 90 mg).
After injection, the mice are placed in a clean cage (five per cage) with fresh bedding, food, and water and are kept overnight within a biological safety cabinet. During this time, the mice excrete nearly all of the injected ENU. The following morning, the mice are placed in clean cages with fresh bedding and are returned to cage racks in the production facility. The ENU-contaminated bedding is disposed of as hazardous waste. The contaminated cages are cleaned by rinsing with soap and hot water and then washed and autoclaved along with the other cages in our animal care facility.
Breeding Strategies
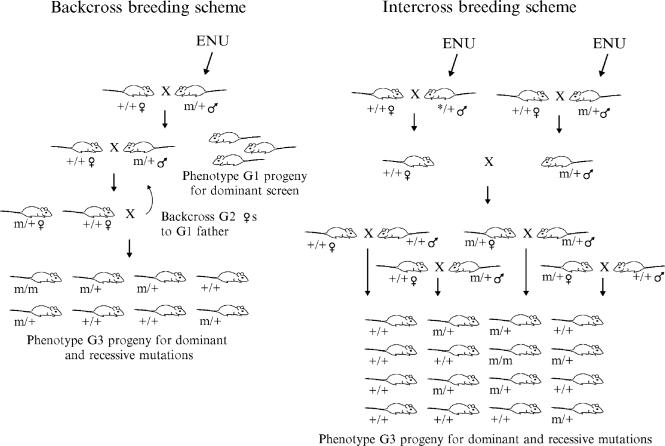
We have used two different breeding schemes to produce mutant mice— the backcross scheme and the intercross scheme (Fig. 1). Each production scheme, however, starts the same. Male mice (G0) are treated with ENU, pass through a sterile period, and eventually recover their fertility (but are now producing mutant sperm). These G0 mice are mated with wild-type females to produce G1 mice. Each G1 mouse represents one mutagenized gamete and can be phenotyped to conduct a first-generation screen for semi-dominant or dominant circadian mutants. The Clock mutant was identified in such a first-generation dominant screen (Vitaterna et al., 1994).
Figure. 1.
Backcross and intercross breeding schemes for mutant production. See the text for a full description. Note that the intercross breeding scheme only depicts the inheritance pattern of a mutation contributed by the male G1 mouse. The female G1 mouse, in fact, also contributes mutations to the pedigree, but has not been depicted for clarity.
To conduct a recessive screen for circadian rhythm mutants, G1 mice are mated to found three-generation kindreds. The resulting G3 mice are phenotyped to determine whether the G1 founder harbors any circadian mutations. These G3 mice can be produced using either a backcross scheme or an intercross scheme.
In the backcross scheme, G1 males are mated with wild-type females to produce G2 daughters. These G2 daughters are selected at random to backcross to their G1 fathers. The resulting G3 mice are phenotyped to identify mutants. One-half of all of the G2 females in the kindred will harbor the same mutation as the G1 kindred founder. In G3 litters produced by these G2 carriers, one-fourth of the pups will be homozygous for the mutation carried by the G1 founder, one-half of the pups in these litters will be heterozygous carriers, and the remaining one-fourth will be homozygous wild type. One-half of the G3 pups produced from the noncarrier G2 daughters will be heterozygous for the G1 founder mutation. Because G3 mutant carriers can be either heterozygous or homozygous for the mutation, a recessive screen is, in fact, both a dominant and a recessive screen. We conducted our BTBR recessive screen using this backcross breeding scheme.
Alternatively, G3 mice can be produced by the intercross scheme. Here each kindred is founded by one G1 male and one nonsibling G1 female. In other words, two mutagenized gametes are used to found the kindred. The G2 mice produced are then intercrossed to produce G3 mice for phenotyping. This breeding strategy is similar to that used in the zebrafish where two G1 kindred founders are used to compensate for a low forward mutagenesis rate (Amsterdam et al., 1999). Because the C57BL/6J strain has a somewhat lower forward mutagenesis rate than that of BTBR/J, but is a well-characterized strain for many behavioral phenotypes (including circadian rhythm behavior), we explored the feasibility of using the intercross breeding scheme to produce mutant mice to conduct a recessive mutant screen.
The Efficiency of Scanning the Genome to Detect a Recessive Mutant
The efficiency of scanning the genome to detect a recessive mutation in a given kindred using the backcross breeding scheme is determined using the following formula (Shedlovsky et al., 1986):
or 1 minus the probability of not detecting a recessive mutation, where k is the number of G2 daughters backcrossed to the G1 founder and n is the number of progeny/G2 female. If we phenotype five offspring from each of four G2 daughters (or 20 G3 mice), we have a probability of 0.85 of detecting a recessive mutant in any given kindred.
The efficiency of scanning the genome to detect a recessive mutation in a given kindred using the intercross breeding scheme is determined using the following formula:
or 1 minus the probability of not detecting a recessive mutation, where k is the number of G2 intercross pairs per kindred and n is the number of progeny phenotyped/intercross pair. If we phenotype six offspring from each of six G2 intercrosses (or 36 G3 mice), we have a probability of 0.75 of detecting a recessive mutant in any given kindred if the kindred is founded with one G1 parent (or mutagenized gamete). Remember, however, that we initiate each kindred with two G1 mice (or two mutagenized gametes), so we are scanning 1.5 (0.75 ×2) genomes for every set of 36 G3 mice.
The intercross breeding scheme appears to be more attractive from both theoretical and practical perspectives. Theoretically, we can scan more of the genome (1.5) per 36 G3 mice as compared with 0.85 genome per 20 G3 mice in the backcross scheme. From a practical perspective, the reproductive burden placed on the G1 male mouse is less in the intercross scheme than in the backcross scheme. In the intercross scheme the G1 mice need only to produce two to three litters of G2 mice. In the backcross scheme the G1 male must first produce G2 daughters and then later sire the G3 pups once those G2 daughters are old enough to breed. In other words, the G1 males must remain viable and fertile for weeks longer than in the intercross scheme. Second, no male mice are rotated through cages of females in the intercross scheme. We simply set up mating pairs and wait for them to produce G3 litters. This reduces our work flow greatly in the production facility.
Using C57BL/6J mice, however, the intercross scheme proved to be much less effective than the backcross scheme. Fertility of the G1 and G2 females was less than expected (Table I). Matings in the intercross scheme are set up as breeding pairs. Only 45% of all G1 intercross pairs and 55% of all G2 intercross pairs were fertile. In the backcross scheme (where we set up mating trios instead of pairs), 83% of all G1 × wt matings were fertile. Second, the fraction of G2 litters that survived to weaning was greater in the backcross scheme (67%) than in the intercross scheme (44%). Finally, the establishment of G2 breeding pairs was dependent on the production of an equal number of G2 males and G2 females. Because the average litter size for the G2 litters produced by the G1 intercross pairs was five, at best we could only set up two G2 breeding pairs from each litter 33% of the time. One-third of the time we could only set up one breeding pair and the other third of the time we were unable to set up any breeding pairs. In order to compensate for the low number of G2 mating pairs, we kept the G1 breeding pairs in the colony longer to produce more G2s. The net result was a drastic increase in the number of mating cages in our breeding colony with a net decrease in the efficiency of scanning of the genome. This scheme could possibly succeed in another mouse strain with a much higher average litter size, but this theoretical strain must also breed well using mating pairs and not trios. We now use the backcross scheme exclusively to produce our G3 mutant mice.
TABLE I.
Mating Efficiencies—Intercross vs Backcross Breeding Schemes
| Mating type | G1 × G1 | G1 × 2 wt | G2 × G2 | G1 × G2 |
|---|---|---|---|---|
| Breeding scheme | Intercross | Backcross | Intercross | Backcross |
| Total matings | 1048 | 918 | 1067 | 1978 |
| Fertile matings | 475 (45%) | 760 (83%) | 590 (55%) | 886 (45%) |
| Total litters | 2030 | 2560 | 2587 | 3154 |
| Litters that survive | 898 (44%) | 1724 (67%) | 1411 (55%) | 1979 (63%) |
| Litter size at birth | 5.8 | 6.1 | 4.5 | 5.4 |
| Litter size at weaning | 5.1 | 5.4 | 4 | 4.6 |
Mouse Production Logistics
After the final ENU injection, the G0 mice go through a sterile period. During the first 4–6 weeks postinjection, the ENU treatment kills most of the spermatogonia of each mouse and mature unmutagenized sperm levels fall to zero. Mutagenized spermatogonia begin to replicate, and by the end of the sterile period sperm levels return to normal levels but the sperm produced are now highly mutagenized.
After 6 weeks, at 14-day intervals we begin to rotate the G0 mice through three to four cages of 6-to 8-week-old C57BL/6J female pairs. Because each G1 mouse produced is the product of one mutagenized gamete and the ENU treatment greatly reduces the number of different spermatogonia per G0 mouse, we limit G1 production to 25 G1 males per G0 father to prevent mutant line duplication in our colony.
Approximately 20–30% of our ENU treatment C57BL/6J mice never regain their fertility after ENU treatment. By 14 weeks postinjection, all of the G0 mice that will regain fertility have already done so. Only 7% of the fertile G0 mice reach G1 production limits. Many G0 mice die or develop tumors before they reach production goals. On average, each fertile G0 male sires 10 G1 males. Half of all G1 litters produced by the G0 males do not survive to weaning, and the average litter size at birth is six pups. Nearly all of the viable pups, however, survive to weaning. This is no different than that observed for a typical C57BL/6J wild-type colony. C57B6/6J females are not the best mothers and usually eat their first litters. We have found that production is improved when our mice are mated as trios rather than as pairs. Two mothers per cage increase the chances that at least one of the females is capable and willing to nurse and care for any litter born into that cage.
At 24 days of age, the G1 litters are weaned, G1 female mice are retired, and G1 male mice are kept as kindred founders. At 6 weeks of age, each G1 mouse is set up in a single mating with two C57BL/6J female mice. These mating trios are kept until the G1 father sires four to six G2 females. If the G1 male does not sire any viable litters within 10 weeks, he is removed from the production facility. By this time, he is 16 weeks old and unlikely to ever sire any litters. If he does sire any G2 litters after this time, he will be even older by the time his G2 daughters are old enough to backcross to produce G3 mice.
Once the first G2 daughter is weaned, the G1 male is separated from his wild-type mates and housed alone until his daughters are 6 weeks old and ready to backcross. The G1 male is then rotated at 14-day intervals through two to three cages of G2 daughter pairs. We call this set of mating cages a backcross group. Once each G2 daughter has each produced five pups, the backcross group is retired.
Production Throughput
The goal of our mutagenesis production facility is to produce 10,000 G3 mice each year for phenotyping. This target is dictated by our phenotyping capacity, in which we can initiate approximately 200 circadian assays per week. We estimate that we can produce 200 G3 mice per week from 10 G1 backcross kindreds. This means that we only need to produce 500 G1 males per year. If every G0 male survived the ENU treatment and produced 25 G1 males, we would only need to inject 20 G0 males per year. In practice, however, 50% of all G0 males do not survive the ENU treatment, many of the G1 males are infertile, G1 males sometimes produce fewer than four G2 females, and several of the G2 females are infertile. To prevent production bottlenecks, therefore, we inject 150–200 G0 males per year (50 males every 3–4 months) to overproduce G1 males. We also establish twice as many (20) G1 backcross groups than theoretically necessary to produce 10,000 G3 mice per year. We phenotype the G1 male mice not used to found kindreds to screen for dominant circadian rhythm mutants.
Mutant Heritability Tests
All putative mutants (or putants) are mated to determine trait heritability. If any offspring from the putant express the same deviant phenotype (in the expected Mendelian pattern), the putant is considered a true mutant. As a first step in this heritability test, putants are mated to wild-type mice. If the putant originates from a G1 dominant screen, we simply produce and phenotype G2 offspring. In the simplest case we would expect one-half of the G2 offspring to have the same phenotype as the putant parent. If the putant originates from a G3 recessive screen, we intercross its G4 offspring and phenotype the G5 offspring. Here, the expected phenotype of the G5 mice can vary depending on the nature of the G3 mutation.
If the G3 putant is heterozygous for the mutation, then only one-half of its offspring are carriers of the mutation and only some of the G4 intercrosses will be between heterozygous carriers. The phenotypes of G5 mice, therefore, can vary from completely wild type to that of the G3 putant to some extreme and previously undetected phenotype (including homozygous lethal). If the G3 putant is a homozygous carrier, all of the G4 offspring are heterozygous carriers and one-fourth of G5 mice will be wild type, one-half will be heterozygous for the mutation, and one-fourth homozygous of the mutation.
Taken together, the patterns of behavior of all of the G3 mice from a given kindred can hint at the mutation mode of inheritance, and we adjust our heritability mating strategies accordingly. Occasionally a G3 putant will have siblings or cousins with the similar or weaker, but still similar, phenotype. This strongly suggests (but does not prove) that the putant is, in fact, a true mutant. Every one of these siblings or cousins is mated to increase the probability that the putant line is not lost to infertility. Once we have reproduced the behavior in putant line offspring, the putant is upgraded to mutant and we expand the kindred to generate enough mice for genetic mapping and to create a homozygous mutant mouse line.
Acknowledgments
This work was supported by NIH grant #U01_MH61915. J. S. Takahashi is an Investigator in the Howard Hughes Medical Institute.
References
- Amsterdam A, Burgess S, Golling G, Chen W, Sun Z, Townsend K, Farrington S, Haldi M, Hopkins N. A large-scale insertional mutagenesis screen in zebrafish. Genes Dev. 1999;13:2713–2724. doi: 10.1101/gad.13.20.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice MJ, Carpenter DA, Favor J, Neuhauser-Klaus A, Hrabe de Angelis M, Soewarto D, Moser A, Cordes S, Miller D, Chapman V, Weber JS, Rinchik EM, Hunsicker PR, Russell WL, Bode VC. Effects of ENU dosage on mouse strains. Mamm. Genome. 2000;11:484–488. doi: 10.1007/s003350010094. [DOI] [PubMed] [Google Scholar]
- Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A. Mouse ENU mutagenesis. Hum. Mol. Genet. 1999;8:1955–1963. doi: 10.1093/hmg/8.10.1955. [DOI] [PubMed] [Google Scholar]
- McDonald JD, Bode VC, Dove WF, Shedlovsky A. The use of N-ethylN-nitrosourea to produce mouse models for human phenylketonuria and hyperphenylalaninemia. Prog. Clin. Biol. Res. 1990;340C:407–413. [PubMed] [Google Scholar]
- Moldin SO, Farmer ME, Chin HR, Battey JF., Jr. Trans-NIH neuroscience initiatives on mouse phenotyping and mutagenesis. Mamm. Genome. 2001;12:575–581. doi: 10.1007/s00335-001-4005-7. [DOI] [PubMed] [Google Scholar]
- Russell WL, Kelly EM, Hunsicker PR, Bangham JW, Maddux SC, Phipps EL. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc. Natl. Acad. Sci. USA. 1979;76:5818–5819. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz WJ, Zimmerman P. Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J. Neurosci. 1990;10:3685–3694. doi: 10.1523/JNEUROSCI.10-11-03685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shedlovsky A, Guenet JL, Johnson LL, Dove WF. Induction of recessive lethal mutations in the T/t-H-2 region of the mouse genome by a point mutagen. Genet. Res. 1986;47:135–142. doi: 10.1017/s0016672300022977. [DOI] [PubMed] [Google Scholar]
- Shedlovsky A, McDonald JD, Symula D, Dove WF. Mouse models of human phenylketonuria. Genetics. 1993;134:1205–1210. doi: 10.1093/genetics/134.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura K, Low-Zeddies SS, King DP, Steeves TD, Whiteley A, Kushla J, Zemenides PD, Lin A, Vitaterna MH, Churchill GA, Takahashi JS. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 2001;11:959–980. doi: 10.1101/gr.171601. [DOI] [PubMed] [Google Scholar]
- Sibilia M, Wagner EF. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, Barnard JA, Yuspa SH, Coffey RJ, Magnuson T. Targeted disruption of mouse EGF receptor: Effect of genetic background on mutant phenotype. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]