Abstract
SOCS3 has been shown to be an important and non-redundant feedback inhibitor of several cytokines including LIF, IL-6, IL-11, CNTF, leptin and G-CSF. Loss of SOCS3 in vivo has profound effects on placental development, inflammation, fat-induced weight gain and insulin sensitivity. SOCS3 expression is induced by JAK/STAT signaling and it then binds to specific cytokine receptors (including gp130, G-CSF and leptin receptors). SOCS3 then inhibits JAK/STAT signaling in two distinct ways. First, SOCS3 is able to directly inhibit the catalytic activity of JAK1, JAK2 or TYK2 whilst remaining bound to the cytokine receptor. Second, SOCS3 recruits elongins B/C and cullin 5 to generate an E3 ligase that ubiquitinates both JAK and cytokine receptor targeting them for proteasomal degradation. Detailed in vivo studies have revealed that SOCS3 action not only limits the duration of cytokine signaling to prevent over-activity but it is also important in maintaining the specificity of cytokine signaling.
Keywords: Suppressor of cytokine signaling 3, SOCS3, JAK, STAT, cytokine receptor, cytokine signaling
JAK/STAT signaling and the SOCS family
Maintenance of the hematopoietic and immune systems is largely controlled by the secretion of cytokines. Cytokine exposure initiates an intracellular signaling cascade that is driven by the activation of a family of receptor-bound tyrosine kinases known as JAKs (Janus Kinases). Once activated, JAKs phosphorylate the receptor and then a family of transcription factors normally sequestered in the cytoplasm, the Signal Transducers and Activators of Transcription (STATs)(41, 86). Activated STATs dimerise and translocate into the nucleus, where they upregulate transcription of the appropriate genes and thereby effect the correct biological response.
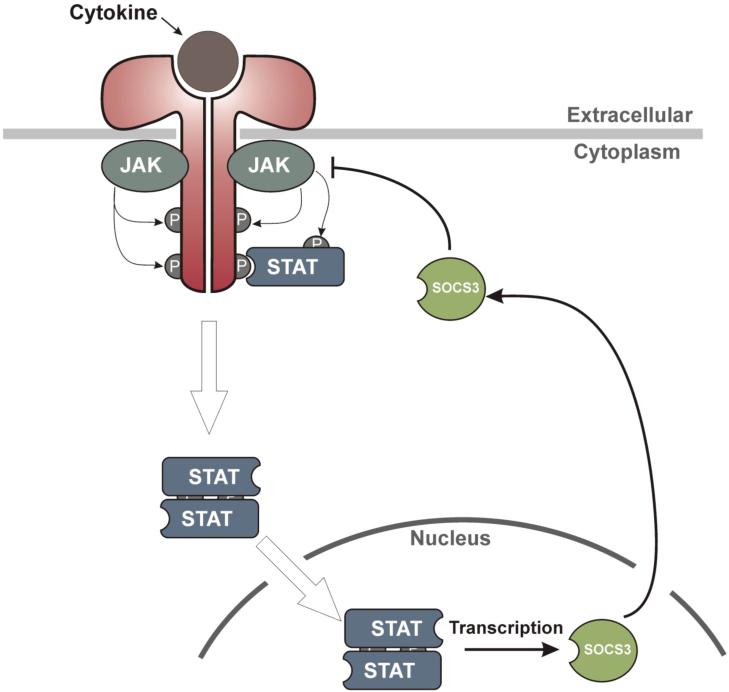
In order to avoid prolonged cytokine signaling, which could result in chronic inflammation and promote aberrant proliferation and tumorigenesis, the JAK/STAT pathway is tightly regulated at a number of levels. The primary regulators are a family of proteins known as the Suppressors of Cytokine Signaling (SOCS)(30, 36, 53, 77). The expression of these proteins is induced by STAT activation; they then inhibit the signaling cascade, creating a negative feedback loop (See Figure 1).
Figure 1.
Negative feedback regulation of JAK/STAT signaling. Cytokine binding to specific receptors activates associated Janus Kinases (JAKs) which then tyrosine phosphorylate the intracellular domain of the receptor. These newly phosphorylated tyrosine residues on the receptor recruit the STAT family of transcription factors. STATs normally reside in the cytoplasm in an inactive state however, once bound to the receptor, they are activated by JAK and then dimerise and translocate into the nucleus where they initiate transcription of cytokine-responsive genes. SOCS3 is one of the proteins whose expression is upregulated by STAT and it then feeds back into the signaling cascade thereby inhibiting signaling and allowing the cell to return to its basal state.
There are eight SOCS proteins in the human genome and all share similar overall domain architecture. In brief, they all contain: (1) an N-terminal domain of varying length and function (2) a central SH2 domain which allows them to bind to tyrosine-phosphorylated signaling molecules and (3) a C-terminal SOCS box which allows them to recruit components of an E3 ubiquitin ligase and ubiquitinate signaling molecules bound to (1) and (2). An analysis of available genome sequences suggests that the SOCS family first arose in insects where they have been implicated in the regulation of EGF signaling (16). However the advent of vertebrates saw an expansion in the SOCS family and the first appearance of SOCS1, SOCS2, SOCS3 and CIS (insect SOCS proteins are direct homologues of SOCS4-7). SOCS1-3 and CIS have much shorter N-terminal domains than their more ancient counterparts. In the case of SOCS1 and SOCS3 this short N-terminal domain enables them to interact directly with, and inhibit the catalytic activity of, JAKs. SOCS1 and SOCS3 are unique amongst the SOCS family in having this activity (71, 91).
SOCS1 and SOCS3 share the highest degree of sequence similarity to each other within the SOCS family and, because of their ability to directly inhibit JAK, are the two most potent inhibitors of cytokine signaling. In addition, they both contain sequence alterations within their SOCS box domain that renders them less able to recruit the other components of their cognate E3 ubiquitin ligases (5). These facts taken together suggest they can be considered as a separate sub-class of SOCS proteins. This review concentrates on SOCS3 regulation of cytokine signaling however it is likely that much that we know about the mechanism of SOCS3 action could be equally well applied to SOCS1. To date, the inability to produce purified, recombinant SOCS1 has hindered a detailed analysis of its mode-of-action.
The structure of SOCS3
(1) The SOCS box
The SOCS box is a small domain found in over 50 human proteins (37). This domain binds the elongin B/C heterodimer and the ternary complex can then subsequently interact with Cullin5, a protein which forms the scaffold of an E3 ubiquitin ligase (96). In this way, proteins that are bound by the upstream domains of SOCS3 (SH2 and N-terminal domains) can be ubiquitinated and subsequently degraded by the proteasome.
The SOCS box-containing proteins, such as SOCS3, interact specifically with Cullin5 (as opposed to other members of the Cullin family) whereas other proteins that contain structurally similar domains and also bind elongin B/C, such as the VHL α-domain, bind to Cullin2 (59, 60). Whilst neither the SOCS box nor elongin B/C bind Cullin5 in isolation, when in complex with each other they then interact with the N-terminal domain of Cullin5 with high affinity (4). Cullin5 binds the RING (really interesting new gene) domain-containing protein Rbx2 at its C-terminus (42, 49) and it is this protein that then recruits the remaining components of the ubiquitination cascade and catalyses the ubiquitination of SOCS-bound proteins.
The SOCS box of SOCS3 is 40 residues in length. It, like other SOCS boxes, is considered to be composed of two smaller motifs, the elongin B/C box and Cullin5 box (see Figure 2) (49). The elongin B/C box consists of approximately 12 residues at the N-terminus of the SOCS box and forms an amphipathic alpha-helix that binds an extremely hydrophobic patch on the surface of elongin C (4, 14, 15). The Cullin5 box is a shorter motif towards the C-terminus of the SOCS box of which the most conserved feature is a Leu-Pro-Leu-Pro (LPLP) motif that binds Cullin5. In SOCS3, this LPLP motif is altered to LPGP. This results in a 10-fold weaker affinity for Cullin5 than the majority of the SOCS family (in this, SOCS3 is similar to SOCS1 which has its LPLP motif altered to IPLN, resulting in a 100-fold lower affinity for Cullin5) (5). All other members of the SOCS family contain the sequence LPLP at this position. Nuclear Magnetic Resonance (NMR) studies on the SOCS box of SOCS3 have shown that the entire domain is unfolded in isolation and becomes partially folded upon elonginBC association. Even when bound to elongin B/C, the Cullin5 box remains unstructured and is presumably only completely folded when Cullin5 is present (4).
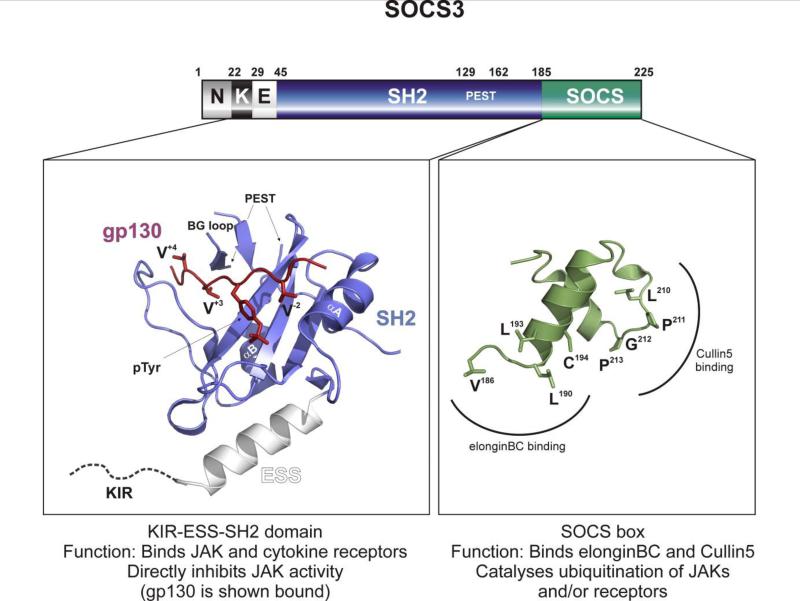
Figure 2. The structure of SOCS3.
SOCS3 can be divided into three overall domains: The N-terminal domain (Residues 1-29, including the KIR-residues 22-29), the extended SH2 domain (Residues 30-185) and the SOCS box (Residues 186-225). An atomic resolution structure exists of the extended SH2 domain (which includes the SH2 domain, blue and the ESS helix, white) of SOCS3 bound to a phospho-peptide from the gp130 receptor (red) (PDB ID: 2HMH) (8). The phosphotyrosine of gp130 is indicated (pTyr), as are three important valines: V−2, V+3 and V+4 (numbers indicate position relative to pTyr). The SOCS box of SOCS3 can be modeled on that of SOCS2 (PDB ID: 2C9W) (14) and is shown above in green. Important residues are labeled. Note that the kinase inhibitory region is unstructured and is shown as a dotted line that joins the ESS helix. The PEST motif is also unstructured.
Genetic deletion of the SOCS3 SOCS box in mice has shown that the rest of the protein is still partially active (11, 12). In this, once again, SOCS3 is similar to SOCS1 which is also partially active in the absence of its SOCS box (94). In contrast, over-expression studies have shown that the presence of the SOCS box domain is necessary for the function of the other six SOCS proteins (7, 34, 43, 55, 61, 65, 84). This suggests that, unlike most SOCS proteins, the dominant mode-of-action of SOCS3 is not to promote the ubiquitination of target proteins.
(2) The SH2 domain
SH2 domains bind phosphotyrosine residues in peptides and proteins (35, 47, 85). Studies by Nicholson et al., showed that the highest affinity ligands for the SOCS3 SH2 domain was not, as previously supposed, phosphotyrosine residues on JAKs but instead were phosphotyrosine residues on certain cytokine receptors to which JAK is bound (54). In particular pTyr 757 on gp130, a co-receptor shared by IL-6, IL-11, LIF and OSM, was shown to bind SOCS3 with >1000-fold higher affinity that pTyr1007 on the JAK activation loop (26, 54). Other high affinity SOCS binding sites were subsequently found on the EPO, Leptin and G-CSF receptors (26, 38, 39). It is noteworthy that studies involving genetic deletion of SOCS3 have identified IL-6, Leptin, LIF and G-CSF as those cytokines most susceptible to SOCS3 inhibition (22, 23, 29, 52, 81).
The pTyr-binding site on the surface of SH2 domains consists of a long shallow groove along one face of the domain which binds not only the pTyr residue itself but also accommodates several residues on either side. In this way specificity for particular pTyr containing sequences is generated. Most SH2 domains bind their target proteins with approximately micromolar affinity (47) whereas SOCS3 binds its favoured substrate with 10-fold higher affinity (6). In part this is due to making additional contacts with residues upstream of the pTyr, most SH2 domains only contact residues downstream from pTyr, resulting in a more limited binding interface (3, 8). This ability of SOCS3 allows it to specifically target sequences with a hydrophobic residue located two residues upstream of a phosphotyrosine (referred to as the pY-2 position) (10, 26, 38). Structural studies have shown that a shallow hydrophobic patch on the surface of SOCS3 (at the beginning of the αA helix, see Figure 2) accommodates the pY-2 residue. Of all other SH2 domains, only the SH2 domain of SHP-2 is similar in this regard and SHP-2 is known to target SOCS3 binding sites on several receptors (32, 54, 72).
The most unusual feature of the SOCS3 SH2 domain was highlighted when structural studies identified a large unstructured loop, consisting of 35 amino acids, inserted into the domain between the αB helix and the BG loop (3, 6). This unstructured insertion has the sequence hallmarks of being a PEST (Pro-Glu-Ser-Thr-rich) motif (70). PEST motifs were originally identified as being common to proteins that have a high turnover rate inside the cell and are believed to stimulate proteolytic degradation of the protein. In most but not all cases this is via the proteasome. Removing the PEST motif results in a modest increase in the half-life of SOCS3 without affecting its ability to bind phosphotyrosine-containing peptides.
The other unusual feature of the SOCS3 SH2 domain, a feature that is shared by all eight SOCS proteins, is the presence of a 14-residue alpha-helix immediately prior to the N-terminus of the domain. Originally labeled the N-ESS (N-terminal extended SH2 subdomain) this helix is integral to the stability (or at least solubility) of the SOCS3 SH2 domain as its removal results in the production of an unstable protein. The structure of SOCS3 shows that this helix covers a large hydrophobic surface on the under-side of the central β-sheet of the SH2 domain. This gives the helix a very fixed geometry relative to the rest of the domain and may be important for positioning the Kinase Inhibitory Region (KIR), discussed below.
(3) The Kinase inhibitory region
Seminal work by Yoshimura and colleagues (71, 91) showed that both SOCS3 and SOCS1 can directly inhibit the catalytic activity of JAK1 and JAK2 and that this ability requires a short (approximately 12 residue) motif immediately N-terminal to the ESS. This motif was termed the Kinase Inhibitory Region (KIR) (71, 91). Subsequent structural studies have shown that the first eight residues of the KIR are unstructured whilst the final four residues form the first turn of an alpha-helix (See Figure 2) (3, 8). Based on this we suggest the re-classification of the KIR as being residues 22-29 of SOCS3 and the ESS consisting of residues 30-44. The KIR has a modest degree of sequence similarity to the activation loop of the four JAKs. The JAKs, like other kinases, contain a motif called the “activation loop”. This loop blocks the catalytic sites of these kinases and must be phosphorylated in order for the kinase to become active. Phosphorylation of this loop is performed by the JAK itself, but in trans. The unphosphorylated activation loop blocks either ATP and/or substrate binding essentially by acting as a pseudosubstrate. The similarity in sequence between the activation loop and the KIR led Yoshimura and colleagues to propose that it acts as a pseudosubstrate and blocks the active site of JAK1 and JAK2, leading to kinase inhibition.
Direct inhibition of JAK signaling by SOCS3
Mechanism
Given the sequence similarity of the SOCS3 KIR and the activation loop of JAK and given the known requirement of the SH2 domain of SOCS3 for its correct function, the original model of SOCS3 inhibition proposed that the SH2 domain of SOCS3 would bind the phosphorylated activation loop of JAK and the KIR would then block the active site of JAK. In the absence of structural data, one method of investigating this is to perform Michealis-Menten kinetics in the presence of SOCS3 and a range of ATP and substrate concentrations. If SOCS3 binds the active site of JAK then the interaction will be antagonized in the presence of high ATP or substrate concentrations and this will alter the IC50 of SOCS3. However, a recent study has shown that the IC50 of SOCS3 is independent of ATP and substrate concentration, suggesting that it acts as a non-competitive inhibitor, rather than a pseudosubstrate (2).
Specificity
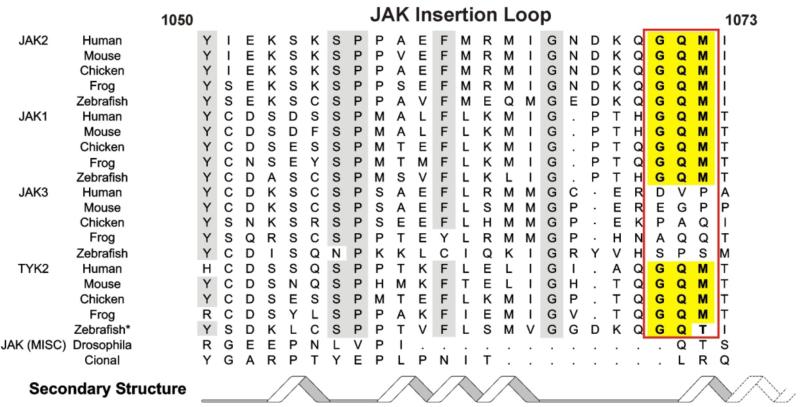
SOCS3 can inhibit JAK1, JAK2 and TYK2 but not JAK3. At the molecular level, this is due to the presence of a short, highly conserved, motif on the surface of the former three kinases that is absent in JAK3. This motif (Gly-Gln-Met, “GQM”) is located at the C-terminal end of the JAK Insertion Loop, first identified by Lucet et al., when they solved the structure of the kinase domain of JAK2 (Figure 3). The JAK insertion loop is found only in JAKs and not in other kinases and only JAK1, JAK2 and TYK2 contain the GQM motif within this loop. An evolutionary comparison of the four mammalian JAKs is telling in this regard. The GQM motif is conserved in JAK1, JAK2 and TYK2 in all vertebrates and is always absent in JAK3. The sequence in this region of JAK3 is not conserved throughout vertebrate evolution suggesting that it is not the target of another SOCS or SOCS-like protein. Lower organisms, such as insects, contain only a single JAK and no SOCS3 (or SOCS1) homologues. Therefore it appears that vertebrates evolved an expanded JAK system, consisting of four JAKs, alongside the ability to inhibit three of them.
Figure 3.
The JAK insertion loop and the GQM motif that encodes specificity for SOCS3 is not present in JAK3.
Biological Roles of SOCS3
Since over-expression or expression at the wrong time and place of SOCS3 can result in unphysiological suppression of a range of signaling molecules we will focus primarily on non-redundant roles of SOCS3 as revealed in genetic deletion studies in mice. For the most part these studies have revealed that the non-redundant roles of SOCS3 are associated with a few cytokines (Table 2). These cytokines have in common the fact that they signal via receptors that contain SOCS3 binding sites and therefore associate with SOCS3 with relatively high affinity (gp130, G-CSF and leptin receptors). Indeed mice in which the SOCS3-binding site on the gp130 shared co-receptor had been mutated showed hyper-responsiveness to cytokines that utilize gp130 and developed IL-11-dependent chronic gastric inflammation and associated tumorigenesis (31)
Table 2.
| Knockout | Tissue | Cytokines Dysregulated | Ref |
|---|---|---|---|
| SOCS3−/− | All | LIF | 69 |
| cKO (vav-cre) | Haematapoietic | G-CSF/IL-6 | 22,23 |
| cKO (LysM-cre) | Macrophage | IL-6 | 92 |
| cKO (Alb-cre) | Liver | IL-6 | 57 |
| cKO (Nes-cre) | Brain | Leptin | 44,52 |
| cKO (MBP-cre) | oligodendrocytes | LIF | 28 |
| cKO (Lck-cre) | T-cells | IL-6 | 46 |
| cKO (Ad-cre*) | Retina | CNTF | 75 |
Denotes adenoviral-cre delivery
Genetic deletion of SOCS3: Leukemia inhibitory Factor (LIF) signaling
SOCS3−/− mice died between day 11-13 of gestation due to grossly altered morphology of the placenta with expanded numbers of giant trophoblasts and altered blood vessels. The embryos were otherwise small but apparently normal (69). The embryonic lethality of SOCS3−/− mice could be rescued by tetraploid aggregation, reduction in either LIF or LIF receptor, or by transplantation of trophoblast stem cells (68, 80, 82) suggesting that the placental defects were a result of over-activity of LIF signaling in trophoblasts. Rescued mice were born normally but died in the perinatal period due to inflammatory lesions or cardiac hypertrophy (68, 80).
LIF utilises a receptor complex consisting of LIF receptor alpha and gp130 and is required for mouse embryonic stem (ES) cell maintenance in the undifferentiated state as well as for blastocyst implantation in pregnant females. Socs3−/− ES cells exhibited less self-renewal and greater differentiation than wild type cells when cultured in LIF. This was correlated with enhanced signaling through both the JAK/STAT and MAP kinase pathways. The latter effect was probably due to increased activation of the phosphatase SHP2. SHP2 is a key component of the MAP kinase pathway and is known to bind at the same site as SOCS3 on the gp130 shared co-receptor. Indeed MAP kinase inhibitors reversed the differentiation phenotype of Socs3−/− ES cells cultured in LIF although it did not fully rescue the LIF-dependent increase in cell numbers (33).
Selective deletion of SOCS3 in the vitreous body of adult mice (using adeno-associated virus cre) prior to optic nerve injury increased axon regeneration from retinal ganglion cells and this was enhanced with the application of CNTF (75). This effect was further enhanced and sustained for longer if PTEN was simultaneously deleted (79).
Deletion of SOCS3 in the hemopoietic system: IL-6, G-CSF signaling
As global SOCS3 deletion in mice leads to early embryonic lethality, tissue-specific deletion of SOCS3, using the cre recombinase system, has been applied to assess the role of SOCS3 in the mature hemopoietic system (22, 23, 46, 57).
Deletion of SOCS3 in hemopoietic and endothelial cells using a cre recombinase construct under control of the vav promoter (vav-cre) resulted in development in late adult mice of a spectrum of inflammatory pathologies in several organs and depletion of neutrophils from the bone marrow (22, 23). Assay of hemopoietic progenitor cells in vitro revealed increased colony size and numbers in response to G-CSF and IL-6 but not to a range of other hemopoietic growth factors. Indeed, these mice responded to injected G-CSF with exaggerated neutrophilia, mobilization of progenitor cells into the blood and splenomegaly demonstrating hyper-responsiveness to G-CSF. However, such mice also displayed pronounced inflammation in various organs and into the spinal cord resulting in hind leg paresis, a phenotype never seen in wild type animals Much but not all of this pathology was recapitulated in wild type mice transplanted with SOCS3-null hemopoietic cells suggesting that there are effects of SOCS3 both on hemopoietic and non-hemopoietic cells (23). In a mouse model of acute inflammatory arthritis (induced with methylated BSA and IL-1) deletion of SOCS3 in hemopoietic and endothelial cells resulted in a particularly severe pathology with enhanced neutrophil (and macrophage) infiltration into the synovium and increased bone destruction (88). Much of this pathology was ameliorated in mice also lacking IL-6 indicating that hyper-responsiveness to IL-6 as a result of SOCS3 loss is a major component of IL-1-induced pathology in this model system (21). Conversely, adenoviral expression of SOCS3 in articular joints dramatically reduced inflammatory pathologies of both acute antigen (methylated BSA)-induced or collagen-induced arthritis models in mice (74). In all the above systems there is evidence that IL-6 and G-CSF played a role in the SOCS3-dependent pathologies.
Deletion of SOCS3 in macrophages by using cre recombinase under control of the LysM promoter (LysM-cre) resulted in hyper-responsiveness to IL-6 but unaltered IL-10 responses (45) despite the fact that both cytokines require STAT3 for many of their biological effects (see later). However, several aspects of IL-6 signaling were qualitatively altered in the absence of SOCS3. Firstly the IL-6-induced transcriptional profile was altered to include genes normally induced only by interferons and STAT1 (hyper-inflammatory) and secondly IL-6 could now inhibit cytokine production induced by LPS (IL-10-like anti-inflammatory action) (22, 48, 92). In addition the differentiation of myeloid progenitors in response to IL-6 and G-CSF was skewed towards macrophages in SOCS3-null animals compared to neutrophils in wild type animals (24).
Deletion of SOCS3 in liver using cre recombinase under the control of the albumin promoter (albumin-cre) resulted in prolonged signaling in response to IL-6 but not interferon in hepatocytes in vivo (57, 58). This enhanced signaling not only included STAT3 and STAT3-induced genes as expected but surprisingly also resulted in the induction of genes normally induced by STAT1. STAT1 is associated with interferon signaling, rather than IL-6 signaling and it therefore appears that SOCS3 not only limits signaling duration in response to interleukin-6 but also maintains signaling specificity. This latter effect is via a more dramatic inhibition of STAT1 than of STAT3 (22, 48). Liver-specific SOCS3 deletion was also associated with enhanced hepatocyte proliferation and weight recovery after partial hepatectomy and enhanced incidence of chemically-induced hepatocellular carcinoma and fibrosis (57, 58, 67). Conversely adenoviral delivery of SOCS3 suppressed growth of hepatocellular tumours in vivo (25). These data are consistent with the observation that human hepatocellular carcinoma growth is associated with activation of the JAK/STAT pathway and a high incidence of gene silencing at the SOCS1 or SOCS3 genetic locus (56, 93).
Genetic deletion of SOCS3 has also been shown to increase signaling by other cytokines known to use gp130 including oncostatin M (78), IL-27 (13) and IL-11 (31) but the roles of signaling by these cytokines in the hemopoietic and inflammatory defects observed in SOCS3 −/− mice have not yet been delineated. Interestingly, SOCS3 does not appear to bind to the other signaling sub-unit (LIF receptor) present in several other cytokine receptors that also utilize gp130 (eg LIF, CNTF, CLC, CT-1) (19) so it is unlikely that additional complexity is generated in these receptor systems with respect to SOCS3 action.
Conditional knockout of SOCS3 in the brain: Leptin signaling
Elevated levels of SOCS3 in the arcuate nucleus of the hypothalamus have been associated with leptin resistance and obesity in mice (9). Deletion of SOCS3 in neurons using nestin-cre and Synapsin-cre constructs highlighted the role of SOCS3 in suppressing leptin signaling (44, 52). The Leptin receptor is highly expressed in the hypothalamus and biochemical analysis showed enhanced STAT3 activation in that organ in SOCS3-deficient animals. These mice gained significantly less weight when placed on a high fat diet compared to wild-type littermates, had a lower food intake and did not develop leptin resistance. In addition, these mice did not develop insulin resistance when placed on a high fat diet, unlike control animals. Many of these phenotypes, such as heightened sensitivity to exogenous leptin as well as protection from high fat-induced obesity and insulin resistance, were re-capitulated in mice haploinsufficient for SOCS3 (40).
Specific deletion of SOCS3 within pro-opiomelanocortin (POMC)-expressing neurons resulted in more modest weight reduction on high fat diets due to increased energy expenditure rather than decreased food intake (44) while specific deletion of SOCS3 in steroidogenic factor-1(SF1)-expressing neurons resulted in decreased enegy expenditure and food intake and increased insulin sensitivity (95) suggesting potentially different roles for these neuron subsets within the hypothalamus. Finally, deletion of SOCS3 in oligodendrocytes (using the myelin basic protein promoter-cre) led to increased LIF signaling in these cells and protected mice against cuprizone-induced oliogodendrocyte loss and demyelination in the central nervous system, an effect which was enhanced by application of exogenous LIF (28). This is consistent with the known role of LIF in oligodendrocyte survival (26).
Conditional knockout of SOCS3 in the eye: CNTF signaling
Selective deletion of SOCS3 in the vitreous body of the eyes of adult mice (using adenovirus-associated cre) prior to optic nerve injury increased axon regeneration from retinal ganglion cells and this was enhanced with the application of CNTF (75). This effect was further enhanced and sustained for longer if the phosphatase PTEN was simultaneously deleted (79). This suggests that both SOCS3 and PTEN are independent physiological inhibitors of CNTF signaling in the eye.
The roles of SOCS3 in the immune system
T lymphocyte polarisation is an important mechanism to deal with different types of infections. The first described polarisation of naïve T cells was either to the TH1 or TH2 phenotypes associated with viral or helminth infections, respectively. The former immune response is co-ordinated by IL12/STAT4 signaling molecules and is associated with cellular inflammation and damage while the latter is coordinated by IL4/STAT6 and is associated with allergic reactions and over-production of IgE. Since then additional polarised states including TH17 (autoimmunity and inflammation), TH3, TH9, TH22, TFH and Treg (anti-inflammatory suppressor) have been described (87).
Several (but not all) reports have demonstrated the selective expression of SOCS3 in allergic type TH2 cells (1, 27, 50) and there is evidence that the levels of SOCS3 in peripheral T-cells correlated with disease severity and IgE levels in asthmatic patients (73). Moreover transgenic over-expression of SOCS3 in T cells increased TH2 development in mice whilst reduced expression of SOCS3 led to decreased TH2 development (73). It has been suggested that the ability of SOCS3 to skew T-cell differentiation to the TH2 phenotype may relate to an ability to compete for the STAT4-binding site on the IL-12 receptor β2 chain thus inhibiting IL-12/STAT4-driven polarization to the alternative TH1 phenotype (90) or to its inhibition of interferon-induced STAT1 activation(22) that is also associated with TH1 polarization.
There are several reports that SOCS3 expression inversely correlates with TH17 T cell polarisation. Th17 cells are polarised T cells that secrete IL-17 and are induced by IL6, TGFβ and IL21. STAT3 activation is considered to be essential for TH17 development and T-cell-specific loss of SOCS3 results in increased activation of STAT3 and IL-17 secretion in response to IL-23 (18) or CD3 stimulation in atherosclerotic mice (83), Moreover inducers of SOCS3 such as LIF inhibited TH17 differentiation (17) while inhibitors of SOCS3 such as TGFβ promoted TH17 differentiation (64).
SOCS3 expression is normally low in suppressive Tregs themselves (51) but selective deficiency of SOCS3 in dendritic cells was shown to lead to expansion of the Treg population (51). Conversely over-expression of SOCS3 in Tregs themselves suppressed proliferation (63). It may be that physiologically the effects of SOCS3 on Tregs are mediated through cytokine actions on dendritic cells although the cytokines involved have not yet been identified.
Recently it was shown that injection of the cytokine IL-7 (that plays a critical role in the early expansion of B- and T-lymphocytes) was able to induce clearance of otherwise persistent viruses in mice (60). and that the effector mechanism required both the induction of IL-6 and inhibition of the expression of SOCS3. It was suggested that the previously reported conversion of an IL6 transcriptional response to one mimicking that of interferons when SOCS3 is absent (22) may be responsible for the viral clearance (62).
Overall these studies suggest that SOCS3 has a primarily immunosuppressive role by dampening cytokine-induced STAT3 and STAT1 activation. Induction of SOCS3 appears most pronounced by cytokines that strongly activate STAT3 (gp130 cytokines, G-CSF, IL-10) and inhibition specificity is then determined by cytokine receptors that have high affinity SOCS3-binding sites (gp130, G-CSF receptor but not IL-10 or interferon receptors). Thus SOCS3 can suppress both STAT3 and STAT1 signaling by IL-6 but does not affect STAT3 signaling by IL-10 or STAT1 signaling by interferons. Consequently the inhibitory effect of SOCS3 induced by IL-10 is entirely mediated in trans on IL-6 and G-CSF receptors whereas the inhibitory effect of SOCS3 induced by IL-6 and G-CSF is more akin to classical negative feedback inhibition.
A new model of JAK/STAT inhibition by SOCS3
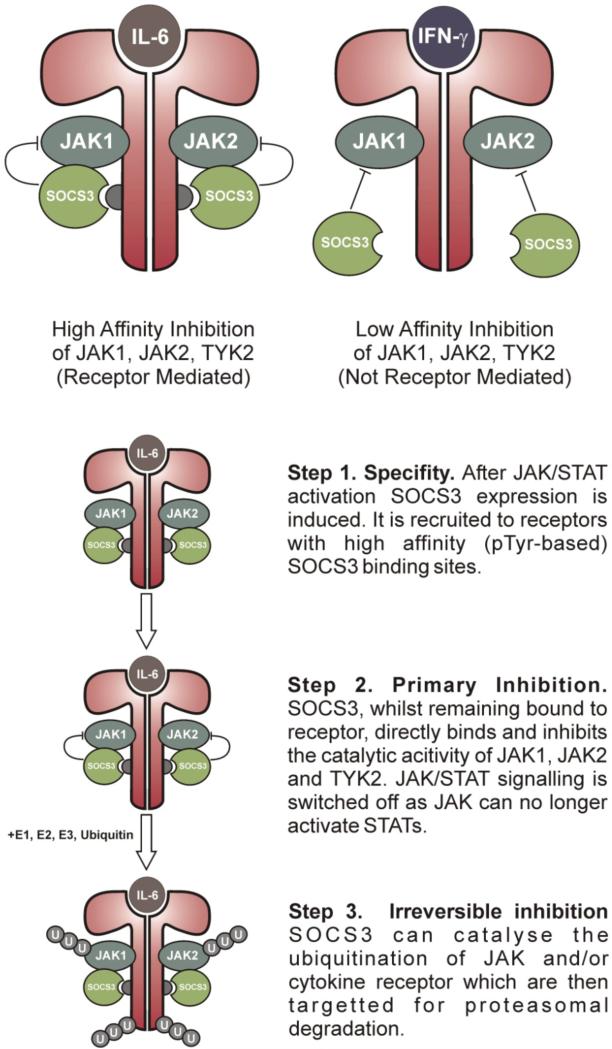
Recently, in vitro studies have shown that SOCS3 binds JAK2 and gp130 simultaneously (2). In addition, it is now clear that the SH2 domain of SOCS3 does not interact with phosphotyrosines in the activation loop of JAK (2). Taken as a whole, this suggests a different model for SOCS3 action. In this model, SOCS3 is recruited to cytokine receptors that contain high-affinity SOCS3 binding sites (such as gp130, LeptinR, G-CSFR etc). Once attached to these receptors, SOCS3 can then bind JAK1, JAK2 and TYK2 (but not JAK3) via an adjacent surface and directly inhibit the catalytic activity of those kinases. An important aspect to this model is that SOCS-JAK-Receptor forms a ternary complex in which each moiety is directly bound to the other two. JAK binds receptor through its FERM domain and SOCS3 through its kinase domain. Receptor binds JAK via its Box1 motif and SOCS3 via pY 757. Finally, SOCS3 binds gp130 via its phosphotyrosine binding groove and JAK via a surface adjacent to this. Although SOCS3 can bind JAK in the absence of receptor its affinity is relatively low (1 μM). Its affinity for pY757 of gp130 is higher (100 nM) and its affinity for a JAK2/gp130 complex would be expected to be higher still via an avidity-like effect (SOCS3 would have to separate from both JAK and receptor in order for the complex to dissociate).
This suggests that the recruitment, or scaffolding, of SOCS3 to particular receptors is its primary mode of specificity. In addition, it explains why SOCS3 is most active against cytokines that utilize receptors with SOCS binding sites rather than other cytokines, which may signal via the same JAKs and STATs but do so through receptors that lack a SOCS3 binding site (for example IFN-γ). A secondary level of specificity is present in that SOCS3 can only inhibit JAK1, JAK2 and TYK2 but not JAK3 (See Figure 4).
Figure 4. A model for SOCS3 inhibition of JAK/STAT signalling.
For clarity, the IL-6 receptor alpha chains are omitted and only the JAK binding (gp130) chains are shown.
Given that SOCS3 can inhibit JAK in the absence of receptor (with low, 1 μM affinity) one can predict that over-expression of SOCS3 will inhibit signaling by most cytokines that act through JAK1, JAK2 or TYK2 but that physiological levels of SOCS3 will result primarily in the inhibition of cytokines signaling through receptors with SOCS3 binding sites. This has indeed been seen in studies on the action of SOCS3 against interferon-γ (76), Growth Hormone (66), IL-2 and IL-3 (20). All of these cytokines are inhibited by SOCS3 when it is over-expressed but are relatively unaffected by physiological levels of SOCS3 as revealed in SOCS3-deleted mice.
The final part of the mechanism of SOCS3 action is its ability to catalyse the ubiquitination of signaling molecules leading to their proteolytic degradation. We have seen that in vitro SOCS3 is able to ubiquitinate both JAK and Receptor (gp130 and LIFR, JJB unpublished data). This can be viewed as long-lasting inhibition of signaling that cannot be reversed until new JAK or receptor has been synthesized by the cell and ensures that the cell is returned to its basal (unstimulated) state.
Unanswered questions
There are still many questions to answer in relation to SOCS3 biological actions. It is unclear why the highly conserved GQM motif in JAKs1,2 and Tyk is absent in JAK3 from all species. Presumably there has been evolutionary pressure to spare JAK3 from inhibition by SOCS 1 and 3 but currently we know of no receptor signaling system that depends exclusively on JAK3 and that might give a biological rationale for JAK3 sparing in this way. Similarly it is unclear why only SOCS1 and SOCS3 appear to have evolved this mechanism for direct inhibition of JAKs while other SOCS proteins rely only on SOCS-box-mediated proteolytic destruction of signaling complexes.
So far individual knockouts of each of the eight SOCS proteins have failed to demonstrate altered signaling by cytokines using the beta common receptor (GM-CSF, IL-3 and IL-5) or signaling by erythropoietin or thrombopoietin despite the fact that they are potent activators of JAK/STAT pathways. Does this reflect redundancy of different SOCS proteins acting on these receptor systems or are they immune to inhibition by SOCS proteins?
Biological specificity of SOCS1 and SOCS3, at least, appears to be highly dependent on the presence of SOCS-binding motifs in the cytoplasmic domains of particular receptors (eg in gp130 for SOCS3 and interferon receptors for SOCS1) but many cytokines induce the expression of SOCS proteins for which their receptor has no binding site (eg IL10 induces SOCS3 but its receptor has no binding site for SOCS3). Is the purpose of this induction to inhibit signaling by other cytokines (eg IL-6) in trans or does it have a different role?
Few cytokines show absolute specificity in their choices of JAKs, STATs and SOCS proteins during a signaling response yet gene deletion experiments suggest a great deal of functional specificity for particular JAKs, STATs and SOCS proteins. For example both interferon γ and IL-6 activate JAK1 and JAK2, STAT1 and STAT3 and SOCS1 and SOCS3 albeit with different kinetics and different strengths (89). Yet the transcriptional responses are quite different for these two cytokines and only SOCS1 influences interferon γ signaling while only SOCS3 influences IL6 signaling in vivo. Since deletion of SOCS proteins influences not only signal strength and duration but also transcriptional specificity it is clear that biological specificity ultimately depends on the SOCS-dependent strength and duration of activated STATs but it is unclear how this is effected in detail. Are all the effects of SOCS proteins on receptors in cis or also on non-cognate receptors in trans or even on JAKS and STATs dissociated from receptors?
Clearly there are still important aspects of the system-wide actions of SOCS proteins and their molecular mechanisms that remain to be resolved and that may provide new insights into the generation of cytokine specificity.
Table 1.
| Receptor | KD (μM) | Sequence | Ref | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EpoR | pY 425 | 69.0 | F | E | pY | T | I | L | D | P | 26 |
| EpoR | pY 453 | 1.1 | L | K | pY | L | pY | L | V | V | 38 |
| gp130 | pY 757 | 0.1 | V | E | pY | S | T | V | V | H | 54 |
| LepR | pY 985 | 8.3 | V | K | pY | A | T | L | V | S | 26 |
| LepR | pY 1077 | 26.0 | V | C | pY | L | G | V | T | S | 26 |
| G-CSFR | pY 729 | 2.8 | V | L | pY | G | Q | L | L | G | 39 |
| G-CSFR | pY 704 | 6.8 | Q | T | pY | V | L | Q | G | T | 39 |
| −2 | −1 | pY | +1 | +2 | +3 | +4 | +5 | ||||
Acknowledgements
We apologise to our colleagues that space limitations did not allow us to cite all the relevant literature. This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS. This research was supported by an NHMRC Program Grant (461219), NIH Grant (CA022556), NHMRC CDA (516777) and NHMRC Project Grant (1011804)
Footnotes
Declaration of interests
NAN is a patent holder on SOCS3 and its activities. The authors report no other conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Anderson P, Sundstedt A, Li L, O'Neill EJ, Li SL, Wraith DC, Wang P. Differential activation of signal transducer and activator of transcription (STAT)3 and STAT5 and induction of suppressors of cytokine signalling in T(h)1 and T(h)2 cells. International Immunology. 2003;15:1309–1317. doi: 10.1093/intimm/dxg130. [DOI] [PubMed] [Google Scholar]
- 2.Babon JJ, Kershaw NJ, Murphy JM, Varghese L, Laktyushin A, Lucet IS, Norton RS, Nicola NA. Suppression of Cytokine Signaling by SOCS3: Characterization of the Mode of Inhibition and the Basis of Its Specificity. Immunity. 2011;36:239–250. doi: 10.1016/j.immuni.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babon JJ, McManus EJ, Yao SG, DeSouza DP, Mielke LA, Sprigg NS, Willson TA, Hilton DJ, Nicola NA, Baca M, Nicholson SE, Norton RS. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Molecular Cell. 2006;22:205–216. doi: 10.1016/j.molcel.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 4.Babon JJ, Sabo JK, Soetopo A, Yao SG, Bailey MF, Zhang JG, Nicola NA, Norton RS. The SOCS box domain of SOCS3: Structure and interaction with the elonginBC-cullin5 ubiquitin ligase. Journal of Molecular Biology. 2008;381:928–940. doi: 10.1016/j.jmb.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babon JJ, Sabo JK, Zhang JG, Nicola NA, Norton RS. The SOCS Box Encodes a Hierarchy of Affinities for Cullin5: Implications for Ubiquitin Ligase Formation and Cytokine Signalling Suppression. Journal of Molecular Biology. 2009;387:162–174. doi: 10.1016/j.jmb.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babon JJ, Yao SG, DeSouza DP, Harrison CF, Fabri LJ, Liepinsh E, Scrofani SD, Baca M, Norton RS. Secondary structure assignment of mouse SOCS3 by NMR defines the domain boundaries and identifies an unstructured insertion in the SH2 domain. Febs Journal. 2005;272:6120–6130. doi: 10.1111/j.1742-4658.2005.05010.x. [DOI] [PubMed] [Google Scholar]
- 7.Banks AS, Li J, McKeag L, Hribal ML, Kashiwada M, Accili D, Rothman PB. Deletion of SOCS7 leads to enhanced insulin action and enlarged islets of Langerhans. J Clin Invest. 2005;115:2462–71. doi: 10.1172/JCI23853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergamin E, Wu J, Hubbard SR. Structural basis for phosphotyrosine recognition by suppressor of cytokine signaling-3. Structure. 2006;14:1285–92. doi: 10.1016/j.str.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Bjorbaek C, Elmquist JK, Frantz JD, Shoelson SE, Flier JS. Identification of SOCS-3 as a potential mediator of central leptin resistance. Molecular Cell. 1998;1:619–625. doi: 10.1016/s1097-2765(00)80062-3. [DOI] [PubMed] [Google Scholar]
- 10.Bjorbaek C, Lavery HJ, Bates SH, Olson RK, Davis SM, Flier JS, Myers MG. SOCS3 mediates feedback inhibition of the leptin receptor via Tyr(985). Journal of Biological Chemistry. 2000;275:40649–40657. doi: 10.1074/jbc.M007577200. [DOI] [PubMed] [Google Scholar]
- 11.Boyle K, Egan P, Rakar S, Willson TA, Wicks IP, Metcalf D, Hilton DJ, Nicola NA, Alexander WS, Roberts AW, Robb L. The SOCS box of suppressor of cytokine signaling-3 contributes to the control of G-CSF responsiveness in vivo. Blood. 2007;110:1466–1474. doi: 10.1182/blood-2007-03-079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyle K, Zhang JG, Nicholson SE, Trounson E, Babon JJ, McManus EJ, Nicola NA, Robb L. Deletion of the SOCS box of suppressor of cytokine signaling 3 (SOCS3) in embryonic stem cells reveals SOCS box-dependent regulation of JAK but not STAT phosphorylation. Cellular Signalling. 2009;21:394–404. doi: 10.1016/j.cellsig.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brender C, Tannahill GM, Jenkins BJ, Fletcher J, Columbus R, Saris CJ, Ernst M, Nicola NA, Hilton DJ, Alexander WS, Starr R. Suppressor of cytokine signaling 3 regulates CD8 T-cell proliferation by inhibition of interleukins 6 and 27. Blood. 2007;110:2528–36. doi: 10.1182/blood-2006-08-041541. [DOI] [PubMed] [Google Scholar]
- 14.Bullock AN, Debreczeni JE, Edwards AM, Sundstrom M, Knapp S. Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase. Proc Natl Acad Sci U S A. 2006;103:7637–42. doi: 10.1073/pnas.0601638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bullock AN, Rodriguez MC, Debreczeni JE, Songyang Z, Knapp S. Structure of the SOCS4-ElonginB/C Complex Reveals a Distinct SOCS Box Interface and the Molecular Basis for SOCS-Dependent EGFR Degradation. Structure. 2007;15:1493–504. doi: 10.1016/j.str.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–21. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- 17.Cao W, Yang YQ, Wang ZY, Liu AL, Fang L, Wu FL, Hong J, Shi YF, Leung S, Dong C, Zhang JWZ. Leukemia Inhibitory Factor Inhibits T Helper 17 Cell Differentiation and Confers Treatment Effects of Neural Progenitor Cell Therapy in Autoimmune Disease. Immunity. 2011;35:273–284. doi: 10.1016/j.immuni.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Laurence A, Kanno Y, Pacher-Zavisin M, Zhu BM, Tato C, Yoshimura A, Hennighausen L, O'Shea JJ. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8137–8142. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clahsen T, Lehmann U, Stross C, Hermanns HM, Volkmer-Engert R, Schneider-Mergener J, Heinrich PC, Schaper F. The tyrosine 974 within the LIF-R-chain of the gp130/LIF-R heteromeric receptor complex mediates negative regulation of LIF signalling. Cellular Signalling. 2005;17:559–569. doi: 10.1016/j.cellsig.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Cohney SJ, Sanden D, Cacalano NA, Yoshimura A, Mui A, Migone TS, Johnston JA. SOCS-3 is tryrosine phosphorylated in response to interleukin-2 and suppresses STAT5 phosphorylation and lymphocyte proliferation. Molecular and Cellular Biology. 1999;19:4980–4988. doi: 10.1128/mcb.19.7.4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croker BA, Kiu H, Pellegrini M, Toe J, Preston S, Metcalf D, O'Donnell JA, Cengia LH, McArthur K, Nicola NA, Alexander WS, Roberts AW. IL-6 promotes acute and chronic inflammatory disease in the absence of SOCS3. Immunol Cell Biol. 2011 doi: 10.1038/icb.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Croker BA, Krebs DL, Zhang JG, Wormald S, Willson TA, Stanley EG, Robb L, Greenhalgh CJ, Forster I, Clausen BE, Nicola NA, Metcalf D, Hilton DJ, Roberts AW, Alexander WS. SOCS3 negatively regulates IL-6 signaling in vivo. Nature Immunology. 2003;4:540–545. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- 23.Croker BA, Metcalf D, Robb L, Wei W, Mifsud S, DiRago L, Cluse LA, Sutherland KD, Hartley L, Williams E, Zhang JG, Hilton DJ, Nicola NA, Alexander WS, Roberts AW. SOCS3 is a critical physiological negative regulator of G-CSF signaling and emergency granulopoiesis. Immunity. 2004;20:153–165. doi: 10.1016/s1074-7613(04)00022-6. [DOI] [PubMed] [Google Scholar]
- 24.Croker BA, Mielke LA, Wormald S, Metcalf D, Kiu H, Alexander WS, Hilton DJ, Roberts AW. Socs3 maintains the specificity of biological responses to cytokine signals during granulocyte and macrophage differentiation. Exp Hematol. 2008;36:786–98. doi: 10.1016/j.exphem.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Q, Jiang W, Wang YX, Lv C, Luo JJ, Zhang W, Lin F, Yin YX, Cai R, Wei P, Qian C. Transfer of suppressor of cytokine signaling 3 by an oncolytic adenovirus induces potential antitumor activities in hepatocellular carcinoma. Hepatology. 2008;47:105–112. doi: 10.1002/hep.21951. [DOI] [PubMed] [Google Scholar]
- 26.De Souza D, Fabri LJ, Nash A, Hilton DJ, Nicola NA, Baca M. SH2 domains from suppressor of cytokine signaling-3 and protein tyrosine phosphatase SHP-2 have similar binding specificities. Biochemistry. 2002;41:9229–9236. doi: 10.1021/bi0259507. [DOI] [PubMed] [Google Scholar]
- 27.Egwuagu CE, Yu CR, Zhang MF, Mahdi RM, Kim SJ, Gery I. Suppressors of cytokine signaling proteins are differentially expressed in Th1 and Th2 cells: Implications for Th cell lineage commitment and maintenance. Journal of Immunology. 2002;168:3181–3187. doi: 10.4049/jimmunol.168.7.3181. [DOI] [PubMed] [Google Scholar]
- 28.Emery B, Cate HS, Marriottt M, Merson T, Binder MD, Snell C, Soo PY, Murray S, Croker B, Zhang JG, Alexander WS, Cooper H, Butzkueven H, Kilpatrick TJ. Suppressor of cytokine signaling 3 limits protection of leukemia inhibitory factor receptor signaling against central demyelination. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7859–7864. doi: 10.1073/pnas.0602574103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emery B, Merson TD, Snell C, Young KM, Ernst M, Kilpatrick TJ. SOCS3 negatively regulates LIF signaling in neural precursor cells. Molecular and Cellular Neuroscience. 2006;31:739–747. doi: 10.1016/j.mcn.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Endo TA, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, Miyazaki T, Leonor N, Taniguchi T, Fujita T, Kanakura Y, Komiya S, Yoshimura A. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–4. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 31.Ernst M, Najdovska M, Grail D, Lundgren-May T, Buchert M, Tye H, Matthews VB, Armes J, Bhathal PS, Hughes NR, Marcusson EG, Karras JG, Na SQ, Sedgwick JD, Hertzog PJ, Jenkins BJ. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. Journal of Clinical Investigation. 2008;118:1727–1738. doi: 10.1172/JCI34944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fairlie WD, De Souza D, Nicola NA, Baca M. Negative regulation of gp130 signalling mediated through tyrosine-757 is not dependent on the recruitment of SHP2. Biochemical Journal. 2003;372:495–502. doi: 10.1042/BJ20030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forrai A, Boyle K, Hart AH, Hartley L, Rakar S, Willson TA, Simpson KM, Roberts AW, Alexander WS, Voss AK, Robb L. Absence of suppressor of cytokine signalling 3 reduces self-renewal and promotes differentiation in murine embryonic stem cells. Stem Cells. 2006;24:604–614. doi: 10.1634/stemcells.2005-0323. [DOI] [PubMed] [Google Scholar]
- 34.Greenhalgh CJ, Metcalf D, Thaus AL, Corbin JE, Uren R, Morgan PO, Fabri LJ, Zhang JG, Martin HM, Willson TA, Billestrup N, Nicola NA, Baca M, Alexander WS, Hilton DJ. Biological evidence that SOCS-2 can act either as an enhancer or suppressor of growth hormone signaling. J Biol Chem. 2002;277:40181–4. doi: 10.1074/jbc.C200450200. [DOI] [PubMed] [Google Scholar]
- 35.Grucza RA, Bradshaw JM, Futterer K, Waksman G. SH2 domains: from structure to energetics, a dual approach to the study of structure-function relationships. Med Res Rev. 1999;19:273–93. doi: 10.1002/(sici)1098-1128(199907)19:4<273::aid-med2>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 36.Hilton DJ, Richardson RT, Alexander WS, Viney EM, Willson TA, Sprigg NS, Starr R, Nicholson SE, Metcalf D, Nicola NA. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proc Natl Acad Sci U S A. 1998;95:114–9. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hilton DJ, Richardson RT, Alexander WS, Viney EM, Willson TA, Sprigg NS, Starr R, Nicholson SE, Metcalf D, Nicola NA. Twenty proteins containing a C-terminal SOCS box form five structural classes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:114–119. doi: 10.1073/pnas.95.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hortner M, Nielsch U, Mayr LM, Heinrich PC, Haan S. A new high affinity binding site for suppressor of cytokine signaling-3 on the erythropoietin receptor. European Journal of Biochemistry. 2002;269:2516–2526. doi: 10.1046/j.1432-1033.2002.02916.x. [DOI] [PubMed] [Google Scholar]
- 39.Hortner M, Nielsch U, Mayr LM, Johnston JA, Heinrich PC, Haan S. Suppressor of cytokine signaling-3 is recruited to the activated granulocyte-colony stimulating factor receptor and modulates its signal transduction. Journal of Immunology. 2002;169:1219–1227. doi: 10.4049/jimmunol.169.3.1219. [DOI] [PubMed] [Google Scholar]
- 40.Howard JK, Cave BJ, Oksanen LJ, Tzameli I, Bjorbaek C, Flier JS. Enhanced leptin sensitivity and attenuation of diet-induced obesity in mice with haploinsufficiency of Socs3. Nature Medicine. 2004;10:734–738. doi: 10.1038/nm1072. [DOI] [PubMed] [Google Scholar]
- 41.Ihle JN, Witthuhn BA, Quelle FW, Yamamoto K, Thierfelder WE, Kreider B, Silvennoinen O. Signaling by the cytokine receptor superfamily: JAKs and STATs. Trends Biochem Sci. 1994;19:222–7. doi: 10.1016/0968-0004(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 42.Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 2004;18:3055–65. doi: 10.1101/gad.1252404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kario E, Marmor MD, Adamsky K, Citri A, Amit I, Amariglio N, Rechavi G, Yarden Y. Suppressors of cytokine signaling 4 and 5 regulate epidermal growth factor receptor signaling. J Biol Chem. 2005;280:7038–48. doi: 10.1074/jbc.M408575200. [DOI] [PubMed] [Google Scholar]
- 44.Kievit P, Howard JK, Badman MK, Balthasar N, Coppari R, Mori H, Lee CE, Elmquist JK, Yoshimura A, Flier JS. Enhanced leptin sensitivity and improved glucose homeostasis in mice lacking suppressor of cytokine signaling-3 in POMC-expressing cells. Cell Metabolism. 2006;4:123–132. doi: 10.1016/j.cmet.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Kimura A, Kinjyo I, Matsumura Y, Mori H, Mashima R, Harada M, Chien KR, Yasukawa H, Yoshimura A. SOCS3 is a physiological negative regulator for granulopoiesis and granulocyte colony-stimulating factor receptor signaling. Journal of Biological Chemistry. 2004;279:6905–6910. doi: 10.1074/jbc.C300496200. [DOI] [PubMed] [Google Scholar]
- 46.Kinjyo I, Inoue H, Hamano S, Fukuyama S, Yoshimura T, Koga K, Takaki H, Himeno K, Takaesu G, Kobayashi T, Yoshimura A. Loss of SOCS3 in T helper cells resulted in reduced immune responses and hyperproduction of interleukin 10 and transforming growth factor-beta 1. Journal of Experimental Medicine. 2006;203:1021–1031. doi: 10.1084/jem.20052333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ladbury JE, Lemmon MA, Zhou M, Green J, Botfield MC, Schlessinger J. Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc Natl Acad Sci U S A. 1995;92:3199–203. doi: 10.1073/pnas.92.8.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lang R, Pauleau AL, Parganas E, Takahashi Y, Mages J, Ihle JN, Rutschman R, Murray PJ. SOCS3 regulates the plasticity of gp130 signaling. Nature Immunology. 2003;4:546–550. doi: 10.1038/ni932. [DOI] [PubMed] [Google Scholar]
- 49.Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, Staehling-Hampton K, Washburn MP, Conaway RC, Conaway JW. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J Biol Chem. 2008;283:8005–13. doi: 10.1074/jbc.M706987200. [DOI] [PubMed] [Google Scholar]
- 50.Matsumoto A, Seki Y, Watanabe R, Hayashi K, Johnston JA, Harada Y, Abe R, Yoshimura A, Kubo M. A role of suppressor of cytokine signaling 3 (SOCS3/CIS3/SSI3) in CD28-mediated interleukin 2 production. Journal of Experimental Medicine. 2003;197:425–436. doi: 10.1084/jem.20020939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumura Y, Kobayashi T, Ichiyama K, Yoshida R, Hashimoto M, Takimoto T, Tanaka K, Chinen T, Shichita T, Wyss-Coray T, Sato K, Yoshimura A. Selective expansion of Foxp3-positive regulatory T cells and immunosuppression by suppressors of cytokine signaling 3-deficient dendritic cells. Journal of Immunology. 2007;179:2170–2179. doi: 10.4049/jimmunol.179.4.2170. [DOI] [PubMed] [Google Scholar]
- 52.Mori H, Hanada R, Hanada T, Aki D, Mashima R, Nishinakamura H, Torisu T, Chien KR, Yasukawa H, Yoshimura A. Socs3 deficiency in the brain elevates leptin sensitivity and confers resistance to diet-induced obesity. Nature Medicine. 2004;10:739–743. doi: 10.1038/nm1071. [DOI] [PubMed] [Google Scholar]
- 53.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, Akira S, Kishimoto T. Structure and function of a new STAT-induced STAT inhibitor. Nature. 1997;387:924–9. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 54.Nicholson SE, De Souza D, Fabri LJ, Corbin J, Willson TA, Zhang JG, Silva A, Asimakis M, Farley A, Nash AD, Metcalf D, Hilton DJ, Nicola NA, Baca M. Suppressor of cytokine signaling-3 preferentially binds to the SHP-2-binding site on the shared cytokine receptor subunit gp130. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6493–6498. doi: 10.1073/pnas.100135197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicholson SE, Metcalf D, Sprigg NS, Columbus R, Walker F, Silva A, Cary D, Willson TA, Zhang JG, Hilton DJ, Alexander WS, Nicola NA. Suppressor of cytokine signaling (SOCS)-5 is a potential negative regulator of epidermal growth factor signaling. Proc Natl Acad Sci U S A. 2005;102:2328–33. doi: 10.1073/pnas.0409675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niwa Y, Kanda H, Shikauchi Y, Saiura A, Matsubara K, Kitagawa T, Yamamoto J, Kubo T, Yoshikawa H. Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene. 2005;24:6406–6417. doi: 10.1038/sj.onc.1208788. [DOI] [PubMed] [Google Scholar]
- 57.Ogata H, Chinen T, Yoshida T, Kinjyo I, Takaesu G, Shiraishi H, Iida M, Kobayashi T, Yoshimura A. Loss of SOCS3 in the liver promotes fibrosis by enhancing STAT3-mediated TGF-beta 1 production. Oncogene. 2006;25:2520–2530. doi: 10.1038/sj.onc.1209281. [DOI] [PubMed] [Google Scholar]
- 58.Ogata H, Kobayashi T, Chinen T, Takaki H, Sanada T, Minoda Y, Koga K, Takaesu G, Maehara Y, Iida M, Yoshimura A. Deletion of the SOCS3 gene in liver parenchymal cells promotes hepatitis-induced hepatocarcinogenesis. Gastroenterology. 2006;131:179–193. doi: 10.1053/j.gastro.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 59.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, Pavletich N, Chau V, Kaelin WG. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–7. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 60.Ohh M, Takagi Y, Aso T, Stebbins CE, Pavletich NP, Zbar B, Conaway RC, Conaway JW, Kaelin WG., Jr. Synthetic peptides define critical contacts between elongin C, elongin B, and the von Hippel-Lindau protein. J Clin Invest. 1999;104:1583–91. doi: 10.1172/JCI8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pazienza V, Clement S, Pugnale P, Conzelman S, Foti M, Mangia A, Negro F. The hepatitis C virus core protein of genotypes 3a and 1b downregulates insulin receptor substrate 1 through genotype-specific mechanisms. Hepatology. 2007;45:1164–71. doi: 10.1002/hep.21634. [DOI] [PubMed] [Google Scholar]
- 62.Pellegrini M, Calzascia T, Toe JG, Preston SP, Lin AE, Elford AR, Shahinian A, Lang PA, Lang KS, Morre M, Assouline B, Lahl K, Sparwasser T, Tedder TF, Paik JH, DePinho RA, Basta S, Ohashi PS, Mak TW. IL-7 Engages Multiple Mechanisms to Overcome Chronic Viral Infection and Limit Organ Pathology. Cell. 2011;144:601–613. doi: 10.1016/j.cell.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 63.Pillemer BBL, Xu H, Oriss TB, Qi ZB, Ray A. Deficient SOCS3 expression in CD4(+)CD25(+)FoxP3(+) regulatory T cells and SOCS3-mediated suppression of Treg function. European Journal of Immunology. 2007;37:2082–2089. doi: 10.1002/eji.200737193. [DOI] [PubMed] [Google Scholar]
- 64.Qin HW, Wang LF, Feng T, Elson CO, Niyongere SA, Lee SJ, Reynolds SL, Weaver CT, Roarty K, Serra R, Benveniste EN, Cong YZ. TGF-beta Promotes Th17 Cell Development through Inhibition of SOCS3. Journal of Immunology. 2009;183:97–105. doi: 10.4049/jimmunol.0801986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ram PA, Waxman DJ. Role of the cytokine-inducible SH2 protein CIS in desensitization of STAT5b signaling by continuous growth hormone. J Biol Chem. 2000;275:39487–96. doi: 10.1074/jbc.M004755200. [DOI] [PubMed] [Google Scholar]
- 66.Ram PA, Waxman DJ. SOCS/CIS protein inhibition of growth hormone-stimulated STAT5 signaling by multiple mechanisms. J Biol Chem. 1999;274:35553–61. doi: 10.1074/jbc.274.50.35553. [DOI] [PubMed] [Google Scholar]
- 67.Riehle KJ, Campbell JS, McMahan RS, Johnson MM, Beyer RP, Bammler TK, Fausto N. Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. Journal of Experimental Medicine. 2008;205:91–103. doi: 10.1084/jem.20070820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Robb L, Boyle K, Rakar S, Hartley L, Lochland J, Roberts AW, Alexander WS, Metcalf D. Genetic reduction of embryonic leukemia-inhibitory factor production rescues placentation in SOCS3-null embryos but does not prevent inflammatory disease. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16333–16338. doi: 10.1073/pnas.0508023102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roberts AW, Robb L, Rakar S, Hartley L, Cluse L, Nicola NA, Metcalf D, Hilton DJ, Alexander WS. Placental defects and embryonic lethality in mice lacking suppressor of cytokine signaling 3. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9324–9329. doi: 10.1073/pnas.161271798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–8. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 71.Sasaki A, Yasukawa H, Suzuki A, Kamizono S, Syoda T, Kinjyo I, Sasaki M, Johnston JA, Yoshimura A. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes to Cells. 1999;4:339–351. doi: 10.1046/j.1365-2443.1999.00263.x. [DOI] [PubMed] [Google Scholar]
- 72.Schmitz J, Weissenbach M, Haan S, Heinrich PC, Schaper F. SOCS3 exerts its inhibitory function on interleukin-6 signal transduction through the SHP2 recruitment site of gp130. Journal of Biological Chemistry. 2000;275:12848–12856. doi: 10.1074/jbc.275.17.12848. [DOI] [PubMed] [Google Scholar]
- 73.Seki Y, Inoue H, Nagata N, Hayashi K, Fukuyama S, Matsumoto K, Komine O, Hamano S, Himeno K, Inagaki-Ohara K, Cacalano N, O'Garra A, Oshida T, Saito H, Johnston JA, Yoshimura A, Kubo M. SOCS-3 regulates onset and maintenance of T(H)2-mediated allergic responses. Nature Medicine. 2003;9:1047–1054. doi: 10.1038/nm896. [DOI] [PubMed] [Google Scholar]
- 74.Shouda T, Yoshida T, Hanada T, Wakioka T, Oishi M, Miyoshi K, Komiya S, Kosai K, Hanakawa Y, Hashimoto K, Nagata K, Yoshimura A. Induction of the cytokine signal regulator SOCS3/CIS3 as a therapeutic strategy for treating inflammatory arthritis. Journal of Clinical Investigation. 2001;108:1781–1788. doi: 10.1172/JCI13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith PD, Sun F, Park KK, Cai B, Wang C, Kuwako K, Martinez-Carrasco I, Connolly L, He ZG. SOCS3 Deletion Promotes Optic Nerve Regeneration In Vivo. Neuron. 2009;64:617–623. doi: 10.1016/j.neuron.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Song MM, Shuai K. The suppressor of cytokine signaling (SOCS) 1 and SOCS3 but not SOCS2 proteins inhibit interferon-mediated antiviral and antiproliferative activities. J Biol Chem. 1998;273:35056–62. doi: 10.1074/jbc.273.52.35056. [DOI] [PubMed] [Google Scholar]
- 77.Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–21. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 78.Stross C, Radtke S, Clahsen T, Gerlach C, Volkmer-Engert R, Schaper F, Heinrich PC, Hermanns HM. Oncostatin M receptor-mediated signal transduction is negatively regulated by SOCS3 through a receptor tyrosine-independent mechanism. Journal of Biological Chemistry. 2006;281:8458–8468. doi: 10.1074/jbc.M511212200. [DOI] [PubMed] [Google Scholar]
- 79.Sun F, Park KK, Belin S, Wang D, Lu T, Chen G, Zhang K, Yeung C, Feng G, Yankner BA, He Z. Sustained axon regeneration induced by co-deletion of PTEN and SOCS3. Nature. 2011 doi: 10.1038/nature10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Takahashi Y, Carpino N, Cross JC, Torres M, Parganas E, Ihle JN. SOCCS3: an essential regulator of LIF receptor signaling in trophoblast giant cell differentiation. Embo Journal. 2003;22:372–384. doi: 10.1093/emboj/cdg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takahashi Y, Carpino N, Cross JC, Torres M, Parganas E, Ihle JN. SOCS3: an essential regulator of LIF receptor signaling in trophoblast giant cell differentiation. Embo J. 2003;22:372–84. doi: 10.1093/emboj/cdg057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takahashi Y, Dominici M, Swift J, Nagy C, Ihle JN. Trophoblast stem cells rescue placental defect in SOCS3-deficient mice. Journal of Biological Chemistry. 2006;281:11444–11445. doi: 10.1074/jbc.C600015200. [DOI] [PubMed] [Google Scholar]
- 83.Taleb S, Romain M, Ramkhelawon B, Uyttenhove C, Pasterkamp G, Herbin O, Esposito B, Perez N, Yasukawa H, Van Snick J, Yoshimura A, Tedgui A, Mallat Z. Loss of SOCS3 expression in T cells reveals a regulatory role for interleukin-17 in atherosclerosis. Journal of Experimental Medicine. 2009;206:2067–2077. doi: 10.1084/jem.20090545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Verdier F, Chretien S, Muller O, Varlet P, Yoshimura A, Gisselbrecht S, Lacombe C, Mayeux P. Proteasomes regulate erythropoietin receptor and signal transducer and activator of transcription 5 (STAT5) activation. Possible involvement of the ubiquitinated Cis protein. J Biol Chem. 1998;273:28185–90. doi: 10.1074/jbc.273.43.28185. [DOI] [PubMed] [Google Scholar]
- 85.Waksman G, Shoelson SE, Pant N, Cowburn D, Kuriyan J. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell. 1993;72:779–90. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]
- 86.Wilks AF, Oates AC. The JAK/STAT pathway. Cancer Surv. 1996;27:139–63. [PubMed] [Google Scholar]
- 87.Wisniewski JA, Borish L. Novel cytokines and cytokine-producing T cells in allergic disorders. Allergy Asthma Proc. 2011;32:83–94. doi: 10.2500/aap.2011.32.3428. [DOI] [PubMed] [Google Scholar]
- 88.Wong PKK, Egan PJ, Croker BA, O'Donnell K, Sims NA, Drake S, Kiu H, McManus EJ, Alexander WS, Roberts AW, Wicks IP. SOCS-3 negatively regulates innate and adaptive immune mechanisms in acute IL-1-dependent inflammatory arthritis. Journal of Clinical Investigation. 2006;116:1571–1581. doi: 10.1172/JCI25660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wormald S, Zhang JG, Krebes DL, Mielke LA, Silver J, Alexander WS, Speed TP, Nicola NA, Hilton DJ. The comparative roles of suppressor of cytokine signaling-1 and-3 in the inhibition and desensitization of cytokine signaling. Journal of Biological Chemistry. 2006;281:11135–11143. doi: 10.1074/jbc.M509595200. [DOI] [PubMed] [Google Scholar]
- 90.Yamamoto K, Yamaguchi M, Miyasaka N, Miura O. SOCS-3 inhibits IL-12-induced STAT4 activation by binding through its SH2 domain to the STAT4 docking site in the IL-12 receptor beta 2 subunit. Biochemical and Biophysical Research Communications. 2003;310:1188–1193. doi: 10.1016/j.bbrc.2003.09.140. [DOI] [PubMed] [Google Scholar]
- 91.Yasukawa H, Misawa H, Sakamoto H, Masuhara M, Sasaki A, Wakioka T, Ohtsuka S, Imaizumi T, Matsuda T, Ihle JN, Yoshimura A. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. Embo J. 1999;18:1309–20. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yasukawa H, Ohishi M, Mori H, Murakami M, Chinen T, Aki D, Hanada T, Takeda K, Akira S, Hoshijima M, Hirano T, Chien KR, Yoshimura A. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nature Immunology. 2003;4:551–556. doi: 10.1038/ni938. [DOI] [PubMed] [Google Scholar]
- 93.Yoshikawa H, Matsubara K, Qian GS, Jackson P, Groopman JD, Manning JE, Harris CC, Herman JG. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat Genet. 2001;28:29–35. doi: 10.1038/ng0501-29. [DOI] [PubMed] [Google Scholar]
- 94.Zhang JG, Metcalf D, Rakar S, Asimakis M, Greenhalgh CJ, Willson TA, Starr R, Nicholson SE, Carter W, Alexander WS, Hilton DJ, Nicola NA. The SOCS box of suppressor of cytokine signaling-1 is important for inhibition of cytokine action in vivo. Proc Natl Acad Sci U S A. 2001;98:13261–5. doi: 10.1073/pnas.231486498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang R, Dhillon H, Yin H, Yoshimura A, Lowell BB, Maratos-Flier E, Flier JS. Selective Inactivation of Socs3 in SF1 Neurons Improves Glucose Homeostasis without Affecting Body Weight. Endocrinology. 2008;149:5654–5661. doi: 10.1210/en.2008-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]