Abstract
The top-down approach to proteomics offers compelling advantages due to the potential to provide complete characterization of protein sequence and post-translational modifications. Here we describe the implementation of 193 nm ultraviolet photodissociation (UVPD) in an Orbitrap mass spectrometer for characterization of intact proteins. Near-complete fragmentation of proteins up to 29 kDa is achieved with UVPD including the unambiguous localization of a single residue mutation and several protein modifications on Pin1 (Q13526), a protein implicated in the development of Alzheimer’s disease and in cancer pathogenesis. The 5 nanosecond, high-energy activation afforded by UVPD exhibits far less precursor ion-charge state dependence than conventional collision-based and electron-based dissociation methods.
Recent advances in instrumentation and experimental design have propelled mass spectrometry to the forefront of proteome research. Conventional bottom-up proteome analysis is based on the ability to sequence the constituent peptides of an enzymatically digested protein mixture. By this approach, the identification of many proteins and post-translation modifications (PTMs) is possible; however, complete characterization of any protein is rare.1 This limits the ability to determine sequence truncations, single nucleotide polymorphisms (SNPs) and the combinatorial nature of PTMs. The top-down approach,2–4 an alternative that involves the interrogation of intact proteins, provides both intact protein and fragment mass measurements and has the potential to overcome the analysis deficiencies noted above (splice variants, PTMs, mutations, etc.).
Despite the promise of the top-down approach, complete characterization (in terms of sequence coverage) has generally been limited to proteins less than 10 kDa due to the inability to efficiently dissociate larger proteins and thus unambiguously characterize whole proteoforms. Slow heating methods, such as collision induced dissociation (CID) and infrared multiphoton dissociation (IRMPD), often yield selective cleavage of the most labile bonds (e.g. non-covalent interactions and amide bonds N-terminal to proline residues) resulting in limited sequence coverage.5,6 The advent of electron capture dissociation (ECD)7 and electron transfer dissociation (ETD)8 has extended the utility of the top-down approach as a result of dissociation occurring prior to energy randomization. This attribute greatly decreases the degree of amino acid-specific cleavages seen with slow heating methods, yielding more random and extensive fragmentation. The major limitations of ECD and ETD stem from the strong dependency on charge density and thus the gas-phase structure of the ions selected for dissociation. The use of collisional activation or infrared photoactivation prior to or during the ECD/ETD process, termed activated-ion ECD/ETD,9–11 has been implemented to disrupt protein tertiary structure and allows effective generation and detection of ECD/ETD products of larger proteins. In addition, nozzle-skimmer dissociation12,13 has extended the mass range of proteins that can be dissociated to greater than 200 kDa; however, nozzle-skimmer dissociation is inherently nonspecific because it occurs prior to selection of a precursor. The challenge of protein separations remains a hurdle associated with top-down proteomics, but this factor primarily limits throughput rather than impeding sequence coverage.14
193 nm ultraviolet photodissociation (UVPD) has been shown to be a viable dissociation method for the characterization of peptides15–20 and small proteins cations.21–23 UVPD at 193 nm of peptide cations yields extensive fragmentation producing a, b, c, x, y, Y, z, v, w and d ions.17,20,21,23,19 Here we present the implementation of 193 nm UVPD in a hybrid linear ion trap Orbitrap mass spectrometer (Thermo Scientific Orbitrap Elite24) and the application of UVPD to top-down proteomics.
Experimental
Materials
Ubiquitin from bovine erythrocytes, carbonic anhydrase from bovine erythrocytes, and myoglobin from equine skeletal muscle were purchased from Sigma-Aldrich and used without further purification. All other chemicals were from Thermo Fisher Scientific.
Human Pin1 expression, purification and oxidation
The human Pin1 gene was subcloned in a pHIS8 vector, a derivative of pET28a vector (Novagene).25 The Pin1 R14A mutant was produced using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, CA). The purification of Pin1 R14A mutant was similar to previous reported procedure.26 Briefly, the Pin1 R14A was overexpressed in using E. coli BL21(DE3) strain at 16°C overnight induced by isopropyl-β-D-thiogalactopyranoside (IPTG). The N-terminal polyhistidine tag was removed by thrombin protease (Novegen Germany) during the overnight dialysis after eluted from Ni-NTA (Invitrogen NY) purification. The protein was further purified by gel filtration superdex75 (GE Healthcare) in 20 mM HEPES 7.5 and 50 mM NaCl.
Purified Pin1 was treated with mild oxidizing condition similarly to previous published protocol.27 2 mg/ml purified Pin1 R14A protein was incubated with 30 µ M FeSO4/1 mM H2O2 for 3hours. The oxidation was stopped by removing hydroxyl free radicals using dialysis in 20 mM HEPES, 50 mM NaCl overnight. Samples were spun down for 20 min at 20,000 × g to remove possible aggregation particles then concentrated and desalted using a 10 kDa centrifugal filter (Millipore Corporation) prior to mass spectrometric analysis.
Implementation of UVPD
UVPD was implemented in a manner similar to what we have previously described for infrared photodissociation.28 Briefly, Experiments were performed using a modified Thermo Scientific Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) equipped with ETD (Figure S1). The portion of the vacuum manifold containing the higher energy collision dissociation (HCD) cell and a portion of the ETD reagent transfer ion optics was modified to incorporate an optical window (quartz with antireflective coating) concentric with the HCD cell. An ETD reagent transfer bent quadrupole with rods rotated 45 degrees was used, so the laser beam could be transmitted between the rods. An optical periscope (Thorlabs) consisting of two mirrors (25 mm, Edmund Optics) was used to elevate and align the laser beam coaxial to the HCD cell. The excimer laser was mounted with the beam output concentric with the bottom mirror of the optical periscope. The instrument firmware was modified to allow trapping of ions in the HCD cell for a user specified amount of time and to output a TTL trigger during the HCD scan events. The TTL trigger was used gate a pulse/delay generator (Berkeley Nucleonics model 505) which triggered each pulse of an ArF excimer laser (Coherent Excistar XS). Direct measurements of the collision gas pressure were made via a flange-mounted pirani gauge connected to the HCD cell housing by 6 mm tubing.
Mass Spectrometry
Protein solutions were prepared at 10 µM in 49.5:49.5:1 (v/v/v) water:acetonitrile:formic acid, and were analyzed by direct infusion at a flow rate of 3 µL/min. Spectra for all dissociation methods were acquired using a mass range of 200–2000 m/z and resolving power of 240,000 at m/z 400. The AGC target for MS2 was set to one million, and isolation widths of 8 m/z and 25 m/z were used for model proteins and Pin1, respectively. Normalized collision energies for CID and HCD were 15–30%, and ETD reaction times were 5–15 ms with a reagent AGC target of three hundred thousand. For UVPD experiments, precursor ions were transferred to HCD cell with normalized collision energy of 1% (no collisional induced dissociation occurs at this setting), and spectra were acquired using one laser pulse at 1–4 mJ/pulse. The HCD collision gas pressure was reduced to a pressure measured as a delta of 0.1E-10 Torr in the UHV portion of the vacuum chamber containing the Orbitrap analyzer (5 mTorr collision gas pressure). To afford the most meaningful comparisons, the same amount of spectral averaging was performed for each dissociation method; however, the extent of averaging varied with protein molecular weight. 50, 200, and 500 scans were averaged for ubiquitin, myoglobin and carbonic anhydrase, respectively. This amount of averaging corresponded to acquisition times of approximately 45 sec, 2.5 min. and 6 min. for ubiquitin, myoglobin and carbonic anhydrase, respectively.
Data Processing
MS2 spectra were deconvolved using the Xtract algorithm (Thermo Fisher Scientific) with S/N threshold of 3. CID, HCD, ETD and UVPD spectra were converted to either neutral (ProSightPC) or protonated (manual interpretation) monoisotopic masses. The monoisotopic spectra for all dissociation methods were interpreted using a custom version of ProSightPC 3.0 (Thermo Fisher Scientific) which included the ability to search a/x, b/y and c/z ion pairs in a single search in order to accommodate the complete array of ions observed in UVPD spectra. Single protein mode with fragment mass tolerance set to 10 ppm was used for all fragmentation methods. Manual interpretation of UVPD data was performed using the combination of unprocessed raw spectra and deconvolved spectra by comparison to theoretical fragment masses generated by ProteinProspector v5.10.3 (http://prospector.ucsf.edu). Sequence coverage was calculated as a percentage value based on the total number of observed inter-residue cleavages (considering all ion series) divided by the number of total number of inter-residue bonds in the protein.
Results
Precursor ions were mass selected in the linear ion trap then transferred to and stored in the HCD cell where UVPD was performed. After a single 5 ns laser pulse, product ions were transferred to the Orbitrap analyzer for high resolution and mass accuracy analysis. Reduction of the collision gas pressure in the HCD cell was not required as collisional cooling is not a major competing pathway to dissociation by UVPD at 193 nm; however, reduction of the collision gas pressure does yield lower pressure in the Orbitrap analyzer and better data quality. Reduction of the collision gas pressure allowed detection of low abundance and larger product ions not observed at the standard collision gas pressure (Figure S2).
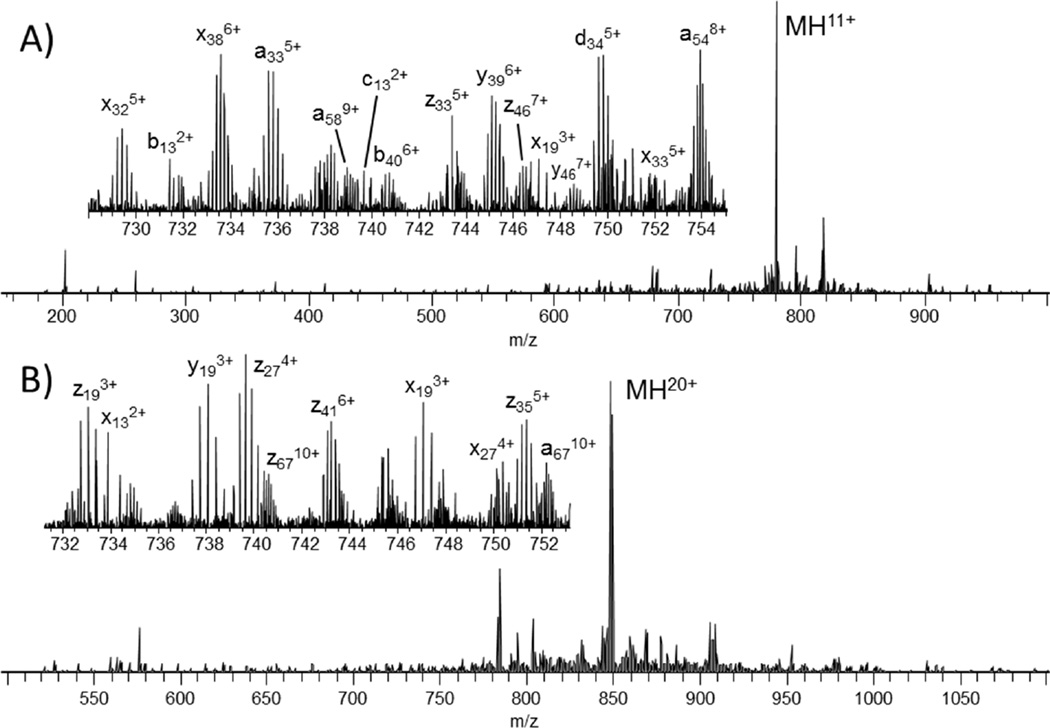
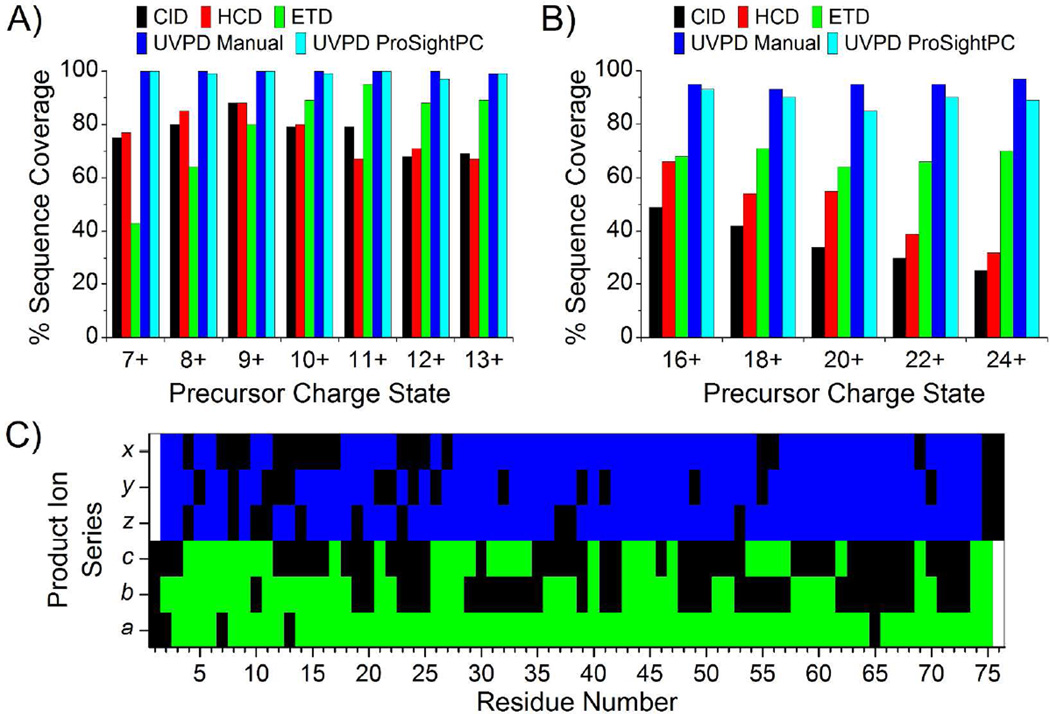
First, the small protein ubiquitin (76 residues, 8.6 kDa) was analyzed to benchmark UVPD fragmentation characteristics against other dissociation methods. A representative UVPD mass spectrum is shown in Figure 1A for the 11+ charge state of ubiquitin. The inset, which illustrates an expanded m/z region in Figure 1A, shows that even low abundance product ions are highly resolved and detected with high S/N. For comparison, UVPD, ETD, HCD and CID were performed for the 7+ to 13+ charge states of ubiquitin. The sequence coverage produced (Figure 2A) by ETD, HCD and CID varied substantially with ETD exhibiting significantly greater sequence coverage for high charge states and HCD and CID performing better for lower charge states. Unlike these well-studied activation methods, UVPD exhibited very little charge state dependence, producing 100% sequence coverage for the 7+ through 12+ charge states and only one missed cleavage (99% sequence coverage) for the 13+ charge state by manual interpretation. Manual interpretation of the UVPD spectra yielded similar results to those obtained by ProSightPC, albeit with some differences attributed to incorrect deconvolution of low mass and low charge state product ions. This discrepancy arises from low S/N of 13C isotopic peaks of low molecular weight ions and the overlap of isotopic distributions of low charge state ions with the isotopic distributions of more abundant and highly charged ions.29 Increasing the S/N threshold for deconvolution from 1.5 to 10 yielded less than a seven percent change in the sequence coverage produced by UVPD for all charge states of ubiquitin (Figure S3).
Figure 1.
UVPD spectra of the 11+ charge state of (A) ubiquitin and (B) the 20+ charge state of myoglobin.
Figure 2.
Sequence coverage produced by CID, HCD, ETD and UVPD as a function of precursor ion charge state for (A) ubiquitin and (B) myoglobin. (C) UVPD product ion array for the 10+ charge state of ubiquitin showing the distribution of cleavages throughout the protein. Green and blue areas indicate the observed cleavage for N-terminal and C-terminal product ions, respectively.
The extensive fragmentation and array of product ion types produced by UVPD enabled de novo sequencing of every residue of ubiquitin based on the product ions observed; however, the development of necessary informatics tools and practical implementation of this methodology is not trivial and would require significant effort for larger proteins. UVPD of the 10+ charge state yielded nearly complete sequence coverage from both N-terminal and C-terminal product ions (only 4 and 3 missed cleavages from N-terminal and C-terminal product ions, respectively). Product ions from two or more N-terminal or C-terminal ion series (i.e. a, b, c or x, y, z ions) were observed for 46 and 63 of the 75 inter-residue bonds sequenced from the N-terminus and C-terminus, respectively (Figure 2C).
The same experiments were also performed for selected charge states of myoglobin (152 residues, 17 kDa) (Figures 1B and 2B), and the advantages of UVPD are even more pronounced. As with ubiquitin, HCD and CID yielded greater sequence coverage for the lower charge states of myoglobin with HCD producing the greatest coverage (66%) for the 16+ charge state. ETD did not exhibit the same preference for higher charge states as noted for ubiquitin, yielding approximately equal sequence coverage (64–71%) for 16+ to 24+ charge states. The lower than expected performance of ETD compared to the results for ubiquitin may be due to the greater number of non-covalent interactions in myoglobin which maintain secondary and tertiary structure and prevent release and detection of many of the product ions. UVPD clearly out-performs HCD, CID and ETD for myoglobin, yielding greater than 93% sequence coverage (by manual interpretation) for each individual charge state interrogated here. There is slightly greater discrepancy in sequence coverage between the manual interpretation method and the use of ProSightPC for myoglobin compared to ubiquitin due to increasing spectral complexity and the reasons discussed above.
Figure S4 shows the comparative spectra for CID, HCD, ETD and UVPD of the 20+ charge state of myoglobin. For all charge states analyzed, the CID and HCD spectra were dominated by cleavages N-terminal to proline residues,30 C-terminal to acidic residues31 and neutral losses of water and ammonia. Due to the higher energy deposition of HCD compared to CID, the HCD spectra also contained a significant number of low mass internal ions produced by multiple cleavages of the protein backbone. The predominant products in the ETD spectra of myoglobin were charge reduced precursor ions. For the corresponding UVPD spectra, there were surprisingly few neutral losses from the precursor or product ions despite the high energy deposition of 193 nm UVPD (6.4 eV photons). The conversion efficiency (i.e. precursor to product ions) for both ETD and UVPD were rather low for the parameters used in these experiments, but this could be modulated by adjusting the ETD reaction time and laser pulse energy for UVPD. 100% conversion is possible for both UVPD and ETD; however, greater precursor depletion does not necessarily produce a better outcome for UVPD or ETD due to secondary dissociation of product ions.
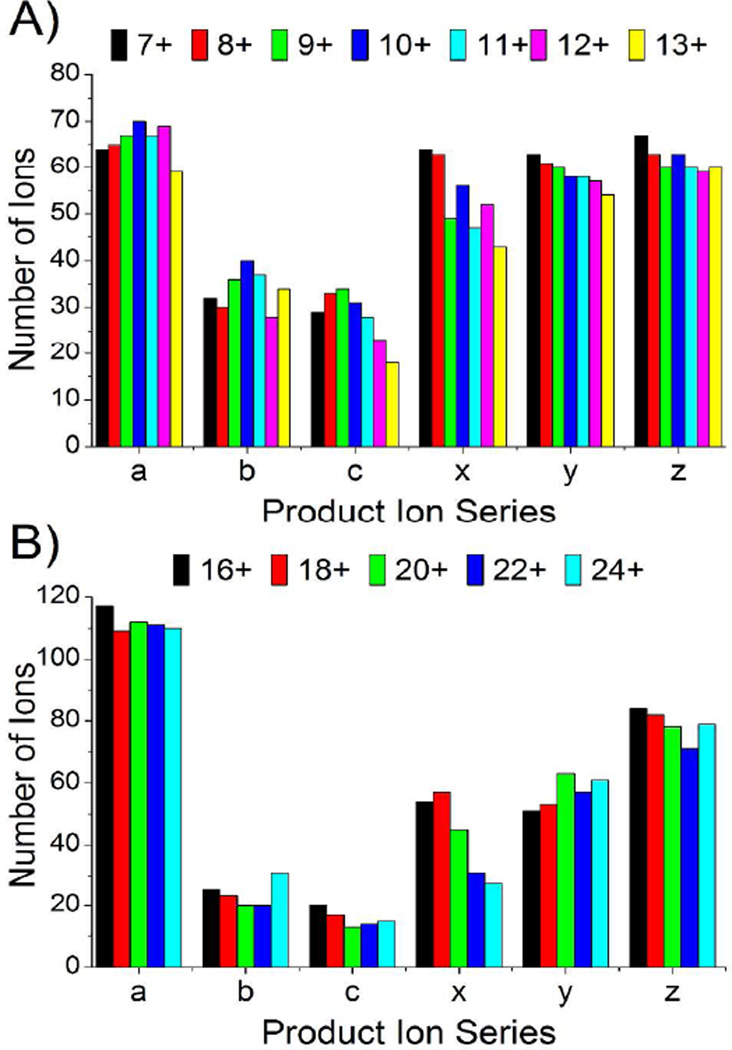
The dominant ion series produced by UVPD for all charge states of ubiquitin and myoglobin (Figure 3A, B) were a, x, y and z ions with relatively few b, c, v, w, and d ions observed. The a, x, y and z ions exhibited significant contributions from both even and odd electron forms, thus yielding mass shifts of ±1.0078 Da and favoring the a + 1, x, y − 1, and z species as summarized for myoglobin in Figure S5. In general, our results are consistent with previous peptide and small protein UVPD studies, suggesting 193 nm UVPD promotes backbone cleavage through pathways occurring prior to intramolecular vibrational re-distribution as well as others on longer time scales subsequent to intramolecular vibrational redistribution.22,32,33
Figure 3.
Number of each product ion type observed as function of precursor ion charge state for (A) ubiquitin and (B) myoglobin.
The excess energy imparted upon absorption of a 193 nm photon (~3 eV greater than typical peptide backbone bond energies) and the combination of fast and slow fragmentation timescales are likely the key to the success of UVPD for the dissociation of larger proteins. UVPD of the 34+ charge state of carbonic anhydrase II (29 kDa, 259 residues) yielded 150 a, 38 b, 15 c, 54 x, 71 y, and 64 z ions (Figure S6). These fragments corresponded to cleavage at 225 (87%) of the 258 interresidue sites. Extensive fragmentation is observed at the termini of the protein as well as the central portion of the sequence. This is the greatest sequence coverage that has been reported for carbonic anhydrase (to the best of our knowledge). HCD and ETD of the 34+ charge state of carbonic anhydrase yielded 30% and 70% sequence coverage, respectively, and are consistent with results recently published also using a Thermo Scientific Orbitrap Elite mass spectrometer.24 Previously, plasma-ECD was shown to produce cleavages at 197 of the 258 inter-residue bonds of carbonic anhydrase II from spectra acquired under multiple experimental conditions.34
The potential to provide unambiguous determination of SNPs, sequence truncations and the combinatorial nature of PTMs are the major attractors of top-down mass spectrometry, and 193 nm UVPD provides a means for fulfilling these goals. To demonstrate, we studied the oxidation of peptidyl-prolyl cis/trans isomerase Pin1. Human Pin1 (UniProt Accession Number: O13526) regulates the signal transduction by conformational changes of phosphorylated Ser/Thr-Pro peptide bonds upon proline isomerization.35 Due to the prevalence of the Ser/Thr-Pro motif in signaling pathways, the physiological function of Pin1 has been demonstrated to be critical for the development of pathological processes including cancer and Alzheimer’s disease.36 Pin1 has been shown to be inactivated in neurons of patients with Alzheimer’s disease consistent with the accumulation of cis form of phosphorylated tau observed at this stage.37 Pin1 activity is reduced in the early stage of AD through modification by oxidation.38,39 The identification of oxidation sites of Pin1 requires considerable sequencing detail and has not been accomplished.
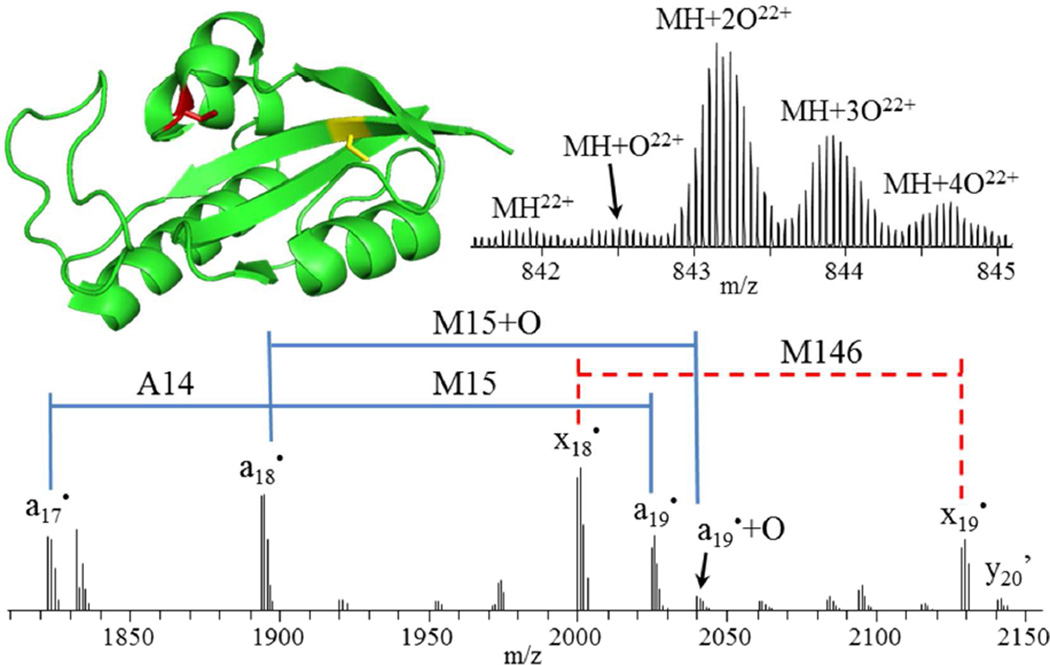
Intact mass analysis of Pin1, incubated with hydrogen peroxide for 3 hours (Figure 4 top right), reveals that the primary products are doubly, triply and quadruply oxidized species. 193 nm UVPD of all 22+ charge state species produced 96% sequence coverage for Pin1 and allowed unambiguous determination of dioxidation of C113 (to cysteine sulfinic acid) as the major oxidation product with lower abundance oxidations detected at M5 and M19 and no detectable oxidation present at C57, M130 or M146. Inspection of a crystal structure of Pin140 (Figure 4 top left) shows the catalytically-essential C113 is located in a loop at the surface of the C-terminal PPIase domain, whereas C57 is buried at the interface of two β-sheets and an α-helix. This supports the observation of C113 oxidation and no oxidation of C57. Consecutive a ions allow unambiguous localization of a single residue mutation of R14A and partial oxidation of M15, while consecutive x ions show that oxidation of M146 is not observed in detectable abundance (Figure 4 bottom). A mechanistic rationale for this particular oxidation motif with respect to the progression of Alzheimer’s disease would be premature, but this result demonstrates the ability of UVPD to provide such rich fragmentation spectra that the precise characterization of individual proteoforms associated with a certain disease type becomes possible.
Figure 4.
Crystal structure (PDB ID 3NTP) of Pin1 (top left) showing the locations of C57 and C113, which are observed in non-oxidized (C57, yellow) and oxidized (C113, red) forms, respectively. Mass spectrum of intact Pin1 (top right) showing doubly and triply oxidized species as the most abundant. Zoomed-in section of a 193 nm UVPD mass spectrum (bottom) of the 22+ charge state species, deconvolved to singly protonated species. Apostrophe (’) indicates the neutral loss of water.
Overall, 193 nm UVPD yields major performance gains for the characterization of intact proteins compared to any existing collision- or electron-based dissociation methods. A single 5 ns laser pulse results in extensive sequence coverage and the ability to identify and localize PTMs in exquisite detail, demonstrating the significant potential of UVPD to drive an expansion of top-down proteomics.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the following funding sources: NSF (CHE-1012622 to JSB), the Welch Foundation (F-1155 to JSB), and NIH (GM067193 to NLK).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Experimental details and additional spectra are available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Duncan MW, Aebersold R, Caprioli RM. Nat Biotech. 2010;28:659–664. doi: 10.1038/nbt0710-659. [DOI] [PubMed] [Google Scholar]
- 2.Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. J. Am. Chem. Soc. 1999;121:806–812. [Google Scholar]
- 3.McLafferty FW, Breuker K, Jin Mi, Han Xuemei, Infusini G, Jiang Honghai, Kong Xianglei, Begley TP. FEBS Journal. 2007;274:6256–6268. doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhou H, Ning Z, E. Starr A, Abu-Farha M, Figeys D. Anal. Chem. 2011;84:720–734. doi: 10.1021/ac202882y. [DOI] [PubMed] [Google Scholar]
- 5.Little DP, Speir JP, Senko MW, O’Connor PB, McLafferty FW. Anal. Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 6.Raspopov SA, El-Faramawy A, Thomson BA, Siu KWM. Anal. Chem. 2006;78:4572–4577. doi: 10.1021/ac052248i. [DOI] [PubMed] [Google Scholar]
- 7.Zubarev RA, Kelleher NL, McLafferty FW. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- 8.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. P. Natl. Acad. Sci. U.S.A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horn DM, Ge Y, McLafferty FW. Anal. Chem. 2000;72:4778–4784. doi: 10.1021/ac000494i. [DOI] [PubMed] [Google Scholar]
- 10.Håkansson K, Chalmers MJ, Quinn JP, McFarland MA, Hendrickson CL, Marshall AG. Anal. Chem. 2003;75:3256–3262. doi: 10.1021/ac030015q. [DOI] [PubMed] [Google Scholar]
- 11.Ledvina AR, Beauchene NA, McAlister GC, Syka JEP, Schwartz JC, Griep-Raming J, Westphall MS, Coon JJ. Anal. Chem. 2010;82:10068–10074. doi: 10.1021/ac1020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loo JA, Edmonds CG, Smith RD. Anal. Chem. 1991;63:2488–2499. doi: 10.1021/ac00021a018. [DOI] [PubMed] [Google Scholar]
- 13.Han X, Jin M, Breuker K, McLafferty FW. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 14.Capriotti AL, Cavaliere C, Foglia P, Samperi R, Laganà A. J. Chromatogr. A. 2011;1218:8760–8776. doi: 10.1016/j.chroma.2011.05.094. [DOI] [PubMed] [Google Scholar]
- 15.Bowers WD, Delbert SS, Hunter RL, McIver RT. J. Am. Chem. Soc. 1984;106:7288–7289. [Google Scholar]
- 16.Hunt DF, Shabanowitz J, Yates JR. J. Chem. Soc. Chem. Commun. 1987;0:548–550. [Google Scholar]
- 17.Thompson MS, Cui W, Reilly JP. J. Am. Soc. Mass Spectrom. 2007;18:1439–1452. doi: 10.1016/j.jasms.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Williams ER, Furlong JJP, McLafferty FW. Journal of the American Society for Mass Spectrometry. 1990;1:288–294. doi: 10.1016/1044-0305(90)85003-5. [DOI] [PubMed] [Google Scholar]
- 19.Reilly JP. Mass Spectrom. Rev. 2009;28:425–447. doi: 10.1002/mas.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madsen JA, Boutz DR, Brodbelt JS. J. Proteome Res. 2010;9:4205–4214. doi: 10.1021/pr100515x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan Z, Kelleher NL, O’Connor PB, Aaserud DJ, Little DP, McLafferty FW. Int. J. Mass Spectrom. 1996;157–158:357–364. [Google Scholar]
- 22.Kim TY, Thompson MS, Reilly JP. Rapid Commun. Mass Spectrom. 2005;19:1657–1665. doi: 10.1002/rcm.1969. [DOI] [PubMed] [Google Scholar]
- 23.Moon JH, Shin YS, Cha HJ, Kim MS. Rapid Commun. Mass Spectrom. 2007;21:359–368. doi: 10.1002/rcm.2855. [DOI] [PubMed] [Google Scholar]
- 24.Michalski A, Damoc E, Lange O, Denisov E, Nolting D, Müller M, Viner R, Schwartz J, Remes P, Belford M, Dunyach J-J, Cox J, Horning S, Mann M, Makarov A. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.O111.013698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jez JM, Ferrer J-L, Bowman ME, Dixon RA, Noel JP. Biochemistry. 2000;39:890–902. doi: 10.1021/bi991489f. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Daum S, Wildemann D, Zhou XZ, Verdecia MA, Bowman ME, Lücke C, Hunter T, Lu KP, Fischer G. ACS Chem. Biol. 2007;2:320–328. doi: 10.1021/cb7000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sultana R, Boyd-Kimball D, Poon HF, Cai J, Pierce WM, Klein JB, Markesbery WR, Zhou XZ, Lu KP, Butterfield DA. Neurobiol. Aging. 2006;27:918–925. doi: 10.1016/j.neurobiolaging.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Vasicek LA, Ledvina AR, Shaw JB, Griep-Raming J, Westphall MS, Coon JJ, Brodbelt JS. J. Am. Soc. Mass Spectrom. 2011;22:1105–1108. doi: 10.1007/s13361-011-0119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Senko MW, Beu SC, McLafferty FW. J. Am. Soc. Mass Spectrom. 1995;6:52–56. doi: 10.1016/1044-0305(94)00091-D. [DOI] [PubMed] [Google Scholar]
- 30.Vaisar T, Urban J. J. Mass Spectrom. 1996;31:1185–1187. doi: 10.1002/(SICI)1096-9888(199610)31:10<1185::AID-JMS396>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 31.Gu C, Tsaprailis G, Breci L, Wysocki VH. Anal. Chem. 2000;72:5804–5813. doi: 10.1021/ac000555c. [DOI] [PubMed] [Google Scholar]
- 32.Morgan JW, Russell DH. J. Am. Soc. Mass Spectrom. 2006;17:721–729. doi: 10.1016/j.jasms.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Yoon SH, Chung YJ, Kim MS. J. Am. Soc. Mass Spectrom. 2008;19:645–655. doi: 10.1016/j.jasms.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Sze SK, Ge Y, Oh, McLafferty FW. Anal. Chem. 2003;75:1599–1603. doi: 10.1021/ac020446t. [DOI] [PubMed] [Google Scholar]
- 35.Ping Lu K, Hanes SD, Hunter T. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 36.Lu KP, Zhou XZ. Nat Rev Mol Cell Biol. 2007;8:904–916. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura K, Greenwood A, Binder L, Bigio EH, Denial S, Nicholson L, Zhou XZ, Lu KP. Cell. 2012;149:232–244. doi: 10.1016/j.cell.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butterfield DA, Poon HF, St Clair D, Keller JN, Pierce WM, Klein JB, Markesbery WR. Neurobiol. Dis. 2006;22:223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Sultana R, Butterfield DA. J Alzheimers Dis. 2013;33:S243–S251. doi: 10.3233/JAD-2012-129018. [DOI] [PubMed] [Google Scholar]
- 40.Xu GG, Zhang Y, Mercedes-Camacho AY, Etzkorn FA. Biochemistry. 2011;50:9545–9550. doi: 10.1021/bi201055c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.