Abstract
MicroRNAs (miRNAs) are key regulators of gene expression. They are conserved across species, expressed across cell types, and active against a large proportion of the transcriptome. The sequence-complementary mechanism of miRNA activity exploits combinatorial diversity, a property conducive to network-wide regulation of gene expression, and functional evidence supporting this hypothesized systems-level role has steadily begun to accumulate. The emerging models are exciting and will yield deep insight into the regulatory architecture of biology. However, because of the technical challenges facing the network-based study of miRNAs, many gaps remain. Here, we review mammalian miRNAs by describing recent advances in understanding their molecular activity and network-wide function.
Keywords: microRNA, miRNA, biogenesis, Drosha, DGCR8, Dicer, TRBP, gene expression, network, system
MicroRNAs (miRNAs) are ~22-nucleotide RNAs that post-transcriptionally repress gene expression by base-pairing to mRNAs. More than half of all mRNAs are estimated to be targets of miRNAs and each miRNA is predicted to regulate up to hundreds of targets. Consistent with this pervasive activity, miRNAs regulate a broad range of processes, including proliferation, differentiation, and apoptosis. These short RNAs are particularly critical during development, their total loss in the embryo leading to lethality. Although many studies focus on binary miRNA-target interactions in defining phenotypes, it is becoming increasingly evident that each miRNA exerts its influence by targeting multiple functionally-related genes that constitute gene expression networks. In this review, we provide a network-level perspective of mammalian miRNAs and describe their genomic organization, molecular activity, and biological function.
The molecular biology of miRNAs
Biogenesis and genomic organization
The biogenesis of miRNAs has been reviewed extensively elsewhere1 and is summarized briefly here. Genes encoding miRNAs are transcribed by RNA Polymerase II into long primary transcripts (pri-miRNAs) that are processed by the RNase III enzyme Drosha to yield precursor miRNAs (pre-miRNAs)2 (Figure 1). Pre-miRNAs are subsequently transported into the nucleus by Exportin 53; 4; 5 and then processed by another RNase III enzyme, Dicer, to yield a mature miRNA as well as a star strand that is degraded6; 7; 8; 9; 10. The miRNA is then loaded into an Argonaute protein within the RNA induced silencing complex (RISC), the effector complex that mediates repression of targets. Within the RISC, the miRNA acts as a guide strand conferring target specificity through a sequence termed the “seed”, which spans positions 2–7 of the miRNA11; 12; 13; 14. miRNAs with identical seed sequences are grouped into families and, since target specificity is typically dictated by the seed, members of a family generally repress the same mRNAs. Examples of targeting mediated by regions outside the seed have also been reported but are uncommon15; 16.
Figure 1.
The miRNA biogenesis pathway. MiRNA genes are transcribed by RNA Polymerase II, in combination with specific transcription factors (TF), as long primary transcripts (pri-miRNA). These transcripts are then processed in the nucleus by the RNase III enzyme Drosha, in complex with DGCR8, into precursor miRNAs (pre-miRNA), which are exported into the cytoplasm by Exportin 5. Pre-miRNAs are processed by the RNase III enzyme Dicer, in complex with TRBP, into a duplex consisting of a guide strand (miRNA) and passenger star strand (miRNA*). The mature miRNA is loaded into the RNA-induced silencing complex (RISC) and acts as a guide strand that recognizes target mRNAs based on sequence complementarity. The RISC subsequently represses targets by inhibiting translation or promoting destabilization of target mRNAs.
Genes encoding miRNAs belong to several classes of gene structures, the result of various events during the course of evolution (reviewed in detail elsewhere17). miRNAs can be transcribed from intergenic regions, where an individual miRNA gene or a cluster of miRNAs forms an independent transcriptional unit, or from introns of coding genes. Based on the exhaustive annotation described in a landmark study by Bartel and colleagues, 38% of murine miRNAs fall within introns of mRNAs18. In most cases, the miRNA is processed from the intron of the host transcript, thus the miRNA and host gene are coordinately expressed. Additionally, multiple miRNA hairpins are often encoded as clusters within a single primary transcript. In mammals, 61% of miRNAs are expressed from polycistronic clusters18. These clusters can encode multiple miRNA seed families. While expression between clustered miRNAs is strongly correlated, it is not absolute, indicating regulation at the level of processing18.
The clustered organization of miRNAs suggests shared biological function among unrelated miRNAs present in the same primary transcript. For example, let-711 and mir-125/lin-412; 19, both of which control the timing of development in worm, are clustered in most animals (though not in C. elegans), in line with a common and conserved function in developmental timing20. Similarly, the miR-1–2~133 cluster encodes two miRNAs that are not related by seed but are related by function, each regulating muscle development and myogenic gene expression networks21. Additional evidence for shared function between clustered miRNAs is provided below (“miR-17~92 and c-Myc“).
Although clustering may serve an important biological function, it nevertheless poses an obstacle when interpreting classical genetic loss-of-function studies in which deletion of a miRNA gene ablates expression of multiple clustered miRNA seed families. In such instances, the phenotype may not be attributable to any single seed family and, therefore, would require phenotypic rescue with the expression of individual, transgenic miRNAs. Examples of such rescue experiments are provided below in the characterization of miR-17~92 in mouse tumor models. Although technically challenging, selective genetic ablation of an individual miRNA within a cluster can be achieved, as reported for the deletion of miR-1–2, which is clustered with miR-13322.
A single miRNA seed family can also be expressed from multiple genomic loci, exemplified by the expression of let-7 from eight different loci in the mouse and human genomes. This organization may provide greater flexibility in modulating miRNA activity, for example by allowing promoter- and context-specific expression of different members of a single seed family. Like clustering, this genomic dispersion complicates genetic analysis of certain miRNA families. Several studies have reported the deletion of multiple loci encoding paralogous clusters of miRNAs, for example for miR-17~92 or miR-34 and their related clusters as described below. Coupled with the fact that such loci can also include clusters of unrelated miRNA seed families, genetic analysis of an isolated miRNA seed family can, in some cases, be unfeasible. The use of “sponge” constructs that titrate miRNA activity based on seed complementarity may circumvent these technical challenges23; 24; 25. In total, the functional impact of the genomic organization of miRNAs is underexplored but made feasible by the groundwork of tools and knowledge established by existing studies.
Activity of the RISC
The RISC is the key effector complex of the RNAi pathway. When loaded with a guide strand, it can inhibit mRNAs by two different mechanisms. When the guide strand and target are perfectly complementary, as observed for siRNA-mediated RNA interference, the slicer activity of Argonaute 2 (Ago2) cleaves the target between nucleotides complementary to positions 10 and 11 of the guide strand, leading to rapid decay of the resulting products26. When guide and target are imperfectly complementary, as is the case for almost all miRNA-target interactions in animals, the RISC initiates a cascade of inhibitory events that direct targets to canonical degradation pathways27. First, the mRNA is translationally inhibited through a poorly understood mechanism that likely occurs at the step of translational initiation28. Then, the GW182 proteins (TNRC6A–C), which are direct binding partners of Argonaute and well-established members of the RISC, recruit two deadenylase complexes, namely PAN2-PAN3 and CCR4-NOT, to deadenylate the targeted mRNA29; 30; 31. Finally, the deadenylated transcript is decapped by DCP2 and degraded by XRN1, the cytoplasmic 5’-to-3’ exonuclease32.
The seed-based target complementarity by which mRNAs are bound and inhibited allows combinatorial diversity in gene regulation. This property is used by several algorithms, such as TargetScan14, miRanda13, and PicTar33, to predict targets. A single miRNA seed family may be predicted to target 100–1000 mRNAs. Computational analysis has revealed enrichment for functional pathways among targets of individual seed families34. Experimentally, the pleiotropy of miRNA activity has been demonstrated by transfection or overexpression of miRNAs followed by gene expression profiling35.
In addition to the regulation of multiple mRNAs by a single miRNA seed family, a single target may itself possess multiple seed matches for a given miRNA family, thereby leading to enhanced repression. Notable examples include Hmga236 and Igf2bp137; 38, which are strongly repressed by multiple let-7 target sites in their 3’UTRs. Additionally, a single mRNA can be repressed by multiple miRNA families. Computational analysis has revealed that co-targeting of mRNAs by functionally-related miRNAs is prevalent, particularly for clustered miRNAs34. While some experimental evidence supports this observation for certain miRNA families39, a systematic experimental investigation of co-targeting relationships has not been carried out.
Structure of Argonaute
Argonaute is the catalytic engine of RISC and serves as a platform to recruit additional regulators of mRNA stability. Therefore, intense effort has been devoted to understanding its function at the structural level. Until recently, structural insights were obtained either from crystals of isolated domains of Argonaute, which provided minimal information on the spatial and functional relationships between domains, or full-length prokaryotic Argonaute, which elucidated overall architecture but did so for a homologue whose biological function in its respective organism was unknown40.
Three recent studies report the crystal structures of full length eukaryotic Argonaute from humans (HsAgo2)41; 42 and the budding yeast Kluyveromyces polysporus (KpAgo)43. In agreement with previous studies, the new structures demonstrate that Argonaute is a bilobed protein with a multidomain conformation (Figure 2). The architecture of eukaryotic Argonaute is similar to that of the prokaryotic protein, indicating high structural conservation across kingdoms. The guide RNA is anchored at each end and threads through the central cleft of the protein, interacting with every domain and loop. This extensive threading structurally stabilizes HsAgo2, as demonstrated by the resistance of the binary complex to limited proteolysis relative to free protein41.
Figure 2.
Crystal structure of human Argonaute 2 in complex with miR-20a. Ago2 is a bilobed protein with a multidomain conformation. The guide RNA is anchored at the ends by each lobe, with the MID domain binding the 5’-end, and the PAZ domain binding the 3’-end. Bases within the seed of the guide strand are solvent exposed, with a kink between nucleotides 6 and 7. The crystal structure shown (Protein Data Bank ID 4F3T) was reported by Elkayam and colleagues.41
The MID domain, which forms a lobe with the PIWI and N domains, anchors the 5’ end of the guide strand. Extensive contacts between the 5’ monophosphate, a biochemical feature of miRNAs, and multiple side chains within the MID domain define the position of the guide strand relative to the enzyme active site. As the seed sequence threads along a narrow groove adjacent to the MID domain, it is stabilized by numerous contacts between its phosphate backbone, including RNA-specific 2’ OH groups, and the protein. Nucleotides 2–6 of the guide adopt an A-form conformation that is largely sequence-independent, demonstrated by the well-defined electron density observed even when heterogeneous small RNA populations are bound by HsAgo2 or KpAgo in crystallographic preparations42; 43. Bases within the seed are solvent exposed and, therefore, accessible for base-pairing with a target. However, in HsAgo2, the stacked base-pairing within the seed is interrupted by a kink between nucleotides 6 and 7 while, in KpAgo, the bases within the seed are tilted away from an orientation optimal for base pairing. These structural features suggest a requirement for conformational changes to the protein upon nucleation of pairing with a target. In HsAgo2, a second kink is formed beyond the seed between nucleotides 9 and 10 as the guide RNA threads into the protein. The 3’ end of the guide is anchored in the PAZ domain, which forms the second lobe of Argonaute.
While the structures of HsAgo2 and KpAgo include a guide RNA, they lack the target strand. Instead, insight into ternary complexes has been obtained from crystals of a full-length catalytically inactive mutant of Thermus thermophilus Argonaute (TtAgo) bound to a 5’ phosphorylated 21-nucleotide guide DNA with or without target RNAs44. As observed for HsAgo2 and KpAgo, the guide DNA in a binary complex with TtAgo adopts an A-form conformation with the 5’ and 3’ ends anchored in the MID and PAZ domains, respectively. Upon binding a target RNA, TtAgo undergoes a conformational shift through pivot-like domain movements that release the 3’ end of the guide strand from the PAZ pocket while maintaining the DNA-RNA duplex in an A-form helix maximally spanning positions 2–16 of the guide. This conformational shift positions two Mg2+ cations and three catalytic aspartate residues within the PIWI domain, which resembles RNase H in structure, for cleavage of the target RNA.
Although the catalytic activity of Argonaute has been ascribed to a catalytic triad (“DDX”, where “X” is either aspartate or histidine) as described in TtAgo, RNase H is known to possess a “DEDD” catalytic tetrad. Indeed, the characterization of KpAgo identified a fourth conserved residue, glutamate, in the catalytic site43. Upon loading of the RNA duplex into KpAgo, the 3’end of the guide strand is released from the PAZ while the glutamate completing the catalytic tetrad is inserted into the catalytic pocket to form a “plugged-in” conformation that promotes cleavage and subsequent release of the passenger strand. KpAgo bound to guide strand retains this plugged-in conformation and is thus primed for cleavage of additional substrates. This glutamate is required for RNAi in vivo in yeast, demonstrating biological activity43. The residue is also present in HsAgo2, where it is positioned within the active site, suggesting the conservation of a catalytic tetrad in multiple species42; 43.
The new studies characterizing full length eukaryotic Argonaute integrate the previous fragmentary glimpses of Argonaute structure into a complete picture of the binary complex and, in so doing, deepen insight into the activity of this class of enzymes. These studies also complement the structural analyses, reviewed in detail elsewhere45, of other conserved RNAi components such as Dicer. The remarkable structural conservation of Argonaute proteins across three kingdoms of life indicates an ancient function for short regulatory nucleic acids. Importantly, the new structures also raise additional questions, particularly about the unexpectedly kinked trajectory of the guide strand and the nature of the second state of eukaryotic Argonaute upon formation of a ternary complex with target RNA.
The phenotypes of miRNAs
The biological function of miRNAs has been characterized at both the cellular and organismal levels. Sequencing of miRNAs from cell lines or whole tissues under a variety of treatment conditions, such as stress, has identified specific cell types or pathways associated with each miRNA seed family46. Furthermore, miRNA targets have been characterized through in vitro cell culture studies by at least two different approaches: (1) The identification and functional validation of targets through the use of prediction algorithms such as TargetScan or miRanda, and (2) unbiased identification of miRNA-responsive genes by combining miRNA overexpression or inhibition experiments with genome-wide assays such as microarrays and, more recently, mRNA sequencing. Organismal functional studies have typically been carried out with loss of function studies using either germline or conditional deletion of miRNA genes. In Table 1, we summarize the cell type- and tissue-specificity of well-studied miRNAs, as well as their archetypal targets and relevance to disease. For the remainder of the review, we will focus on the four best-characterized of these miRNAs.
Table 1.
Tissue/cell type-specific miRNAs.
| miRNA family/ cluster |
Tissue/cell type | Target mRNAs |
Target processes |
Oncogene/ tumor suppressor |
References |
|---|---|---|---|---|---|
| let-7 | Ubiquitous | Lin-28, Hmga2, Igf2bp1 | Development, proliferation, organismal growth | Tumor suppressor | 36, 38, 75, 79 |
| miR- 1~133 | Cardiac and skeletal muscle | Hand2, Irx5, Ptbp1, Ptbp2 | Cell cycle, cardiac differentiation, splicing | Tumor suppressor | 22, 161 |
| miR-15/16 | Ubiquitous | Bcl2, Cyclin D, Cyclin E | Apoptosis, cell cycle | Tumor suppressor | 162–164 |
| miR- 17~92 | Ubiquitous/enriched in B cells | c-Myc, E2F, Bim | Proliferation, apoptosis | Oncogene | 109, 115–117 |
| miR-22 | Ubiquitous/enriched in cardiac and skeletal muscle | Purb | Calcium homeostasis, stress response | Tumor suppressor | 165, 166 |
| miR-34 | Ubiquitous/enriched in testis, brain, and lung | Sirt1, Snail, PNUTS | p53 pathway, epithelial-to- mesenchymal transition | Tumor suppressor | 134–140, 144 150, 151, 157 |
| miR-122 | Liver | AldoA, Hfe2 | Cholesterol biosynthesis, lipid metabolism | Tumor suppressor | 167, 168 |
| miR-124 | Neurons | Ptbp1 | Differentiation | Tumor suppressor | 169 |
| miR- 143~145 | Ubiquitous/enriched in smooth muscle | Klf4, Elk-1, myocard in | Differentiation | Tumor suppressor | 158–160, 170 |
| miR-181 | Immune cells | Shp1, Shp2, Dusp6, Tcl1 | Differentiation, TCR signaling | Unknown | 171–173 |
| miR- 193b~365 | Brown adipocytes | Runx1t1 | Differentiation | Unknown | 174 |
| miR-200 | Epithelial tissue; olfactory bulb | Zeb1, Zeb2 | Epithelial to mesenchymal transition | Context-specific oncogene or tumor suppressor | 175–177 |
| miR-203 | Epidermis | p63 | Cell cycle, proliferation, differentiation | Tumor suppressor | 178 |
| miR-223 | Myeloid cells | Mef2c | Differentiation | Tumor suppressor | 179 |
| miR-290~295 | Embryonic stem cells | Lats2, Rbl2, p21, Casp2 | Cell cycle, proliferation, apoptosis, differentiation | Oncogene | 57–60 |
| miR-451 | Erythrocytes | 14-3-3z | Oxidant stress | Tumor suppressor | 180 |
MiRNAs participate in various circuit motifs with other regulators of gene expression, such as transcription factors47; 48; 49. At least two different network motifs have been identified within these circuits. In the first motif, termed coherent feedforward, miRNAs and the transcription factors that regulate them carry out the same activity on targets, namely coordinated repression. In so doing, each factor reinforces the activity of the other. In the second motif, termed incoherent feedforward, the miRNA and transcription factor carry out opposing functions, allowing precise modulation of the temporal dynamics of gene expression to reduce noise and confer stability. Both motifs enable biological properties critical for phenotypic robustness (resistance to fluctuations in environment)49; 50. Generally, regulators of miRNAs are poorly understood and the annotation of miRNA promoters is incomplete. However, for several examples, the relationship between a miRNA and its regulator has been characterized at both the cellular and organismal level. Below, we trace the activity of four well-studied miRNAs.
miR-290~295 and the core pluripotency transcription factors
Pluripotent embryonic stem cells (ESCs) progress from a naïve state, in the inner cell mass of a pre-implantation embryo, to a primed state in the epiblast of a post-implantation embryo51. Subsequently, the cells of the epiblast undergo gastrulation to form the three germ layers, namely mesoderm, ectoderm, and endoderm. Total loss of miRNAs, for example through loss of Dicer, results in early embryonic lethality prior to gastrulation. Dicer- or DGCR8-null ESCs, which have been characterized in vitro, are unable to inactivate self-renewal programs or initiate differentiation into the three germ layers, further demonstrating that miRNAs are critical to early development52.
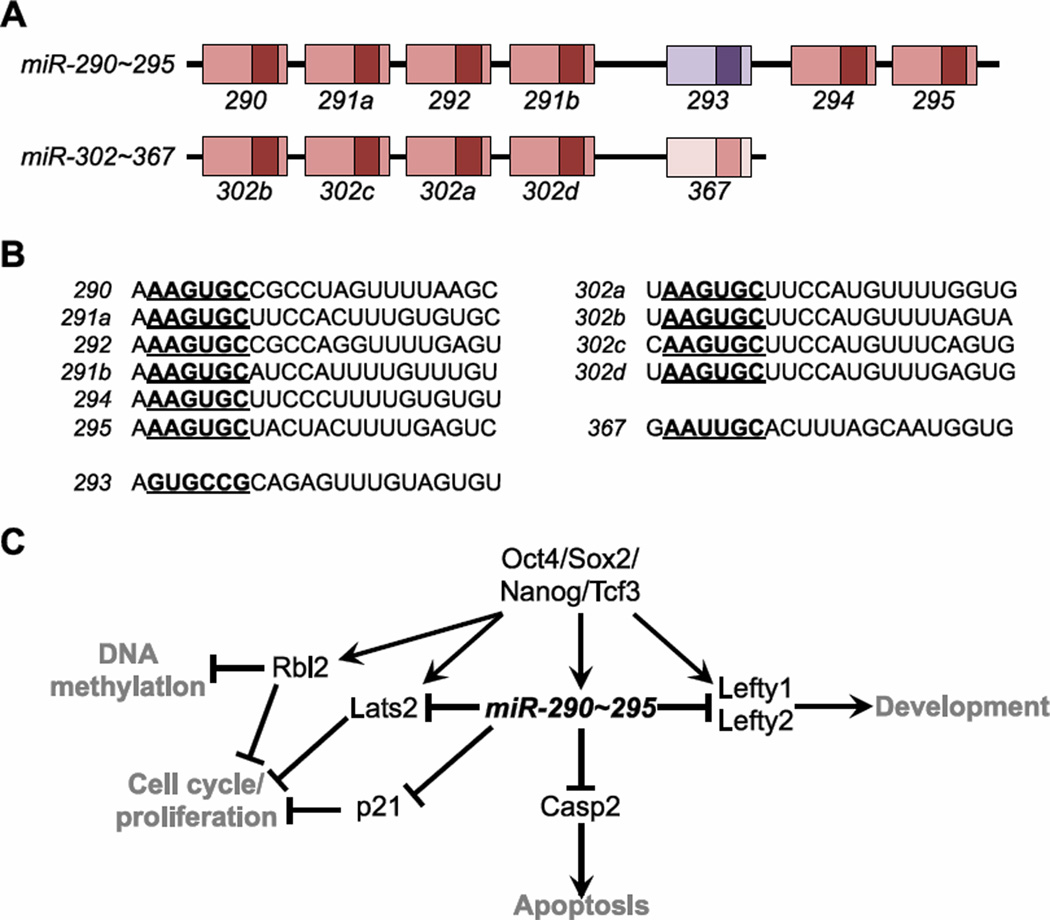
In naïve ESCs, the miR-290~295 family of miRNAs, schematized in Figure 3A, comprises ~70% of all seed families53; 54. This family is homologous to human miR-371~373, a cluster expressed in human ESCs. Although members of miR-290~295 are largely specific to ESCs, a notable exception is miR-293, which exhibits a distinct expression pattern and possesses a different, but related, seed sequence55 (Figure 3B). As naïve ESCs progress to the primed state, they downregulate miR-290~295 and activate miR-302~36756, a cluster conserved in both mouse and humans and whose members are related to miR-290 family through a 6mer seed sequence (Figure 3B). Subsequently, as the embryo develops further and differentiation progresses, expression of the miR-302~367 cluster turns off56.
Figure 3.
Embryonic stem cell-specific miRNAs. (A) Gene structure of the murine miR-290~295 and miR-302~367 clusters. The pre-miRNA sequences are indicated as boxes, with mature miRNA sequences denoted in darker shades. Related family members are indicated by color. (B) Sequences of miRNAs. Each miRNA is grouped based on seed relationship. Seeds are bold and underlined. (C) Summary of the miR-290~295 network.
miR-290~295 and miR-302~367 repress genes central to the self-renewal properties of ESCs (Figure 3C). Specifically, miR-290~295 targets regulators of cell cycle and proliferation, such as p21 and Lats257; apoptosis, such as caspase-258; and DNA methylation, such as Rbl2, a transcriptional regulator of DNA methyltransferases59; 60. The epiblast-expressed miR-302~367 represses Lefty1 and Lefty2, subtypes of TGFβ ligands that regulate specification of body axes and specification of embryonic germ layers56. Accordingly, expression or inhibition of miR-302 respectively promotes or hinders the formation of mesendodermal lineages56. Supporting a role for these ESC-specific clusters in regulating pluripotency, several studies have reported that miR-290~295, its human homologue miR-371~373, and miR-302~367, enhance reprogramming of somatic cells into an induced pluripotent state61; 62; 63. In total, these miRNA clusters are centrally positioned in pluripotency networks and regulate gene expression programs that control the proliferation, survival, and self-renewal of ESCs.
Promoters of miRNA genes in ESCs have been identified by ChIP-sequencing (ChIP-seq) of trimethylated histone H3 lysine 4 (H3K4me3), a histone mark associated with the transcriptional start sites of most genes64. The promoters of miR-290~295 and miR-302~367 are occupied by Oct4, Sox2, Nanog, and Tcf3, which are core transcriptional regulators in ESCs64. An overlap of the transcriptional circuitry of ESCs with a list of mRNAs repressed by ESC-specific miRNAs suggests a role for these clusters in fine-tuning expression of pluripotency and differentiation programs65. For example, the genes Lefty1 and Lefty2, described above, are induced by the core embryonic transcription factors, such as Oct4, an illustrative example of an incoherent feedforward loop in the embryo64. Additionally, both the core pluripotency factors and the transcriptionally repressive Polycomb complex co-occupy promoters of miRNAs that are off in ESCs but will become activated in differentiated lineages, suggesting that these promoters are poised for activation65.
To examine the importance of ESC-specific miRNAs in development, germline knockouts (KO) of miR-290~295 have been characterized and provide an interesting comparison to the Dicer KO animal as well as to in vitro studies investigating this cluster66. miR-290~295 KO animals exhibit defective migration and development of germ cells in the embryo, resulting in depletion of these cells in the adult. Males eventually recover from this early loss of germ cell count and are therefore fertile66. In contrast, adult females remain sterile. Loss of miR-290~295 also results in a partially penetrant embryonic lethal phenotype due to two distinct, abnormal phenotypes66. A subset of miR-290~295 KO embryos localize outside of the yolk sac, a phenotype that may reflect the expression and importance of this cluster in the trophectoderm, which develops into the placenta. Additionally, some miR-290~295 KOs exhibit developmental delays, including reduced somite numbers and defects in neural tube closure. Therefore, while miR-290~295 is important for normal embryonic development, it is not absolutely required for ESC differentiation or development into adulthood.
The complexity of the phenotype of miR-290~295 KO animals highlights several key properties of miRNAs. The incomplete penetrance of the phenotype suggests a role for this cluster in maintaining robustness, with stochastic variations in local environment possibly leading to defects in a subset of embryos. This model is in line with observations indicating that miR-290~295 participates with embryonic transcription factors in both coherent and incoherent feedforward loops, possibly to regulate the kinetics of gene expression during differentiation64. Alternatively, the incomplete penetrance may be explained by functional compensation by other miRNAs, derived from miR-302~367 or, alternatively, from the Sfmbt2 cluster, which possesses related seed sequences but is located in a different locus67; 68. The expression of miR-293, whose seed sequence and expression pattern differs from the other members of its cluster, also raises the possibility that the phenotypes observed in miR-290~295 KO embryos versus adults are driven by distinct miRNA seed families55. These possibilities highlight many of the challenges, and opportunities, in the study of miRNA function.
Let-7 and growth
Let-7 is one of the first miRNAs to be discovered and, as reviewed extensively elsewhere69; 70, was identified in a genetic screen in nematode for regulators of developmental timing. Caenorhabditis elegans (C. elegans) encodes several let-7 family members, namely let-7, miR-48, miR-84, and miR-241. Loss of the let-7 family in nematode results in the reiteration of developmental larval stage events. This phenotype is a result of the upregulation of multiple let-7 targets, including lin-28, an RNA-binding protein, and daf-12, a member of a nuclear hormone receptor superfamily. These two genes also feed back to regulate let-7: lin-28 binds pre-let-7 and inhibits its maturation, while daf-12 transcriptionally activates or represses let-7 expression in the presence or absence, respectively, of its ligand, dafachronic acid71; 72.
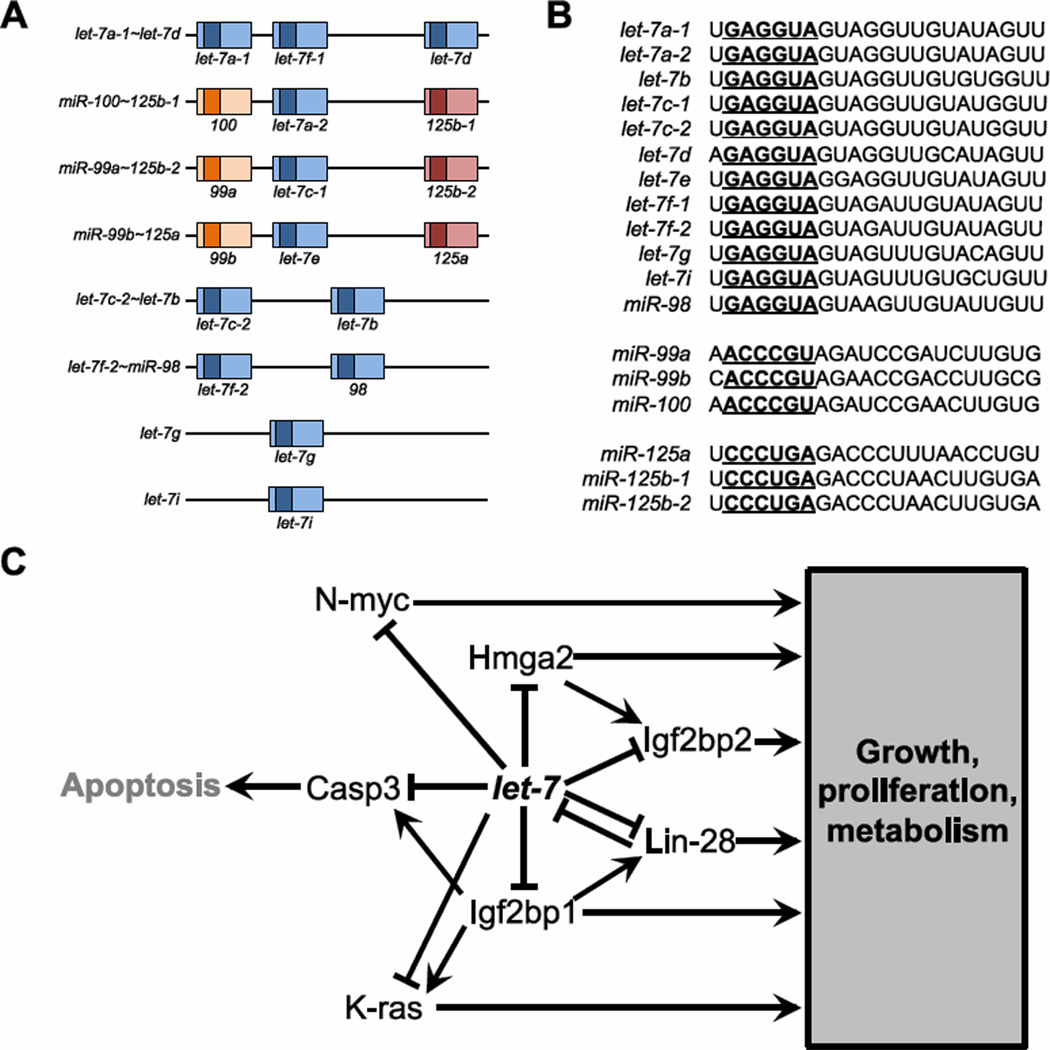
Let-7 is highly conserved across species, including mammals, and is expressed broadly across tissue types70. The number of loci encoding let-7 has expanded to eight in mouse and humans (Figure 4A). At three loci, let-7 is clustered with the miR-99/100 and miR-125 families, which possess distinct seed sequences (Figure 4B). In mammals, as in worm, expression of mature let-7 is activated during development and maintained in the adult73. Although primary transcripts of let-7 are expressed in ESCs, precursor and mature let-7 are undetectable, indicating a block at the level of processing74. During early development, maturation of pre-let-7 is inhibited by Lin-28a and Lin-28b75; 76; 77, homologues of nematode lin-28 whose functions are described in greater detail below. As in worm, the mammalian Lin-28 genes are targets of let-7, thus constituting a conserved negative feedback loop77. In many adult tissues, let-7 is expressed abundantly and functions as a tumor suppressor.
Figure 4.
The let-7 genes. (A) Gene structure of the let-7 genes. At three loci, let-7 is clustered with the miR-99/100 and miR-125 families. The pre-miRNA sequences are indicated as boxes, with mature miRNA sequences denoted in darker shades. Related family members are indicated by color. (B) Sequences of miRNAs. Each miRNA is grouped based on seed relationship. Seeds are bold and underlined. (C) Summary of the let-7 network. Let-7 genes are characterized by a shared role in regulating proliferative and metabolic pathways activated in the embryo. Let-7 targets are densely interconnected and regulate one another.
Many mammalian let-7 targets have been identified and are strongly characterized by their roles in growth, metabolism, and development (Figure 4C). The best characterized of these targets are Lin-28, Hmga2, and the Igf2bp1–3 family. These genes constitute in its entirety a class of genes termed “oncofetal” because of their expression in the embryo, inactivation in most adult tissue, and re-activation in tumors. Of these, the mammalian paralogues Lin-28a and Lin-28b are best understood and represent quintessential oncofetal genes. Lin-28a/b regulate organismal growth and metabolism78. Transgenic mice overexpressing Lin-28a exhibit increased body size and delayed onset of puberty79. Additionally, transgenic mice overexpressing either Lin-28a or LIN28B exhibit an altered metabolism, manifested as increased insulin sensitivity through activation of the insulin-PI3K–mTOR pathway80. In humans, polymorphisms in LIN28B have been associated with variations in height and the timing of menarche81; 82; 83; 84; 85. Demonstrating a role for mammalian Lin-28 in maintaining “stemness”, overexpression of Lin-28, in combination with Oct4, Sox2, and Nanog, promotes induction of pluripotency in somatic fibroblasts86. Inhibition of let-7 by overexpression of Lin-28b in adult hematopoietic stem cells (HSCs) results in the reprogramming of these HSCs into a fetal state87. Consistent with a role in proliferation and growth, mammalian Lin-28 genes are commonly activated in tumors88. Lin-28b induces neuroblastoma in patients by suppressing let-7 and enhancing expression of MYCN, another let-7 target89. In total, the Lin-28 genes regulate proliferative and metabolic pathways at least in part through their modulation of let-7.
Another well characterized oncofetal let-7 target is Hmga2, a non-histone chromatin factor. Hmga2 is normally expressed in the embryo and is off in most adult tissues. Knockout of Hmga2 in mouse leads to a dwarf phenotype in which mutant animals are smaller than wild-type littermates90. Constitutive overexpression of transgenic Hmga2 in mouse leads to increased organismal size and changes in composition of body fat91; 92, a phenotype very similar to that observed for Lin-28 transgenic mice. In humans, genome-wide association studies (GWAS) have linked polymorphisms in HMGA2 to variations in human height and predisposition to diabetes93; 94. Hmga2 is also oncogenic95. It is often translocated in benign lipomas and salivary gland tumors, leading to fusion of its AT-hook DNA-binding domains to a translocation partner96. Expression of Hmga2 is observed in high-grade tumors in various cancers, such as ovarian cancer, and associated with poor patient prognosis97. Experimental evidence supports a causal role for Hmga2 in tumorigenesis. Transgenic mice overexpressing Hmga2 develop benign lipomas and other mesenchymal tumors, as well as pituitary adenomas91; 92. Finally, changes in the 3’UTR of Hmga2 mRNA, for example by mutation, promotes resistance to let-7-mediated repression and subsequent cellular growth36.
The Igf2bp1–3 family of RNA binding proteins are yet another set of oncofetal let-7 targets that share many of the properties of Lin-28 and Hmga2. Knockout of Igf2bp1 results in a dwarf phenotype98, while its transgenic overexpression in mice leads to tumor development99. Overexpression of Igf2bp1 in vitro promotes anchorage-independent growth37. As with Hmga2, alterations in the 3’UTR of Igf2bp1, for example through the use of alternative polyadenylation sites, alters its sensitivity to repression by let-737. Igf2bp2 and Igf2bp3 are less well characterized but are likely integrated into this let-7-regulated network of growth and metabolism. Members of the Igf2bp family are associated with tumors, and are strongly correlated with each other as well as with Hmga238; 100. Demonstrating the dense interconnections in this pathway, Igf2bp1 induces expression of Lin-28b and the oncogene K-ras, another let-7 target101; 102. Additionally, Hmga2 directly induces transcription of Igf2bp2 during regeneration of muscle103. Paradoxically, let-7 also represses Caspase-3104, an activator of apoptosis that is also induced by Igf2bp1102. This pro-survival activity of let-7 is poorly understood but may reflect a role for this miRNA in balancing two related but opposing pathways, consistent with published models proposing that miRNAs regulate the dynamics of state transitions.
There are also intriguing links between targets of let-7, germ cell development, and lifespan. In fruitfly, an axis between let-7 and Imp (the fruitfly ortholog of the Igf2bp family) regulates aging of the testis stem cell niche105. In the testis, Imp stabilizes the RNA-binding protein Upd, which promotes germ line stem cell self-renewal. As the organism ages, let-7 downregulates Imp, resulting in a reduction in Upd levels, loss of self-renewal, and depletion of germ line stem cells, thus demonstrating a negative correlation between let-7 levels and germ cell proliferation. In nematode, let-7, its transcriptional activator daf-12, and its target lin-14, integrate signals from the gonad to regulate lifespan106. When the C. elegans germ line is removed, there is an increase in expression of the let-7-related miRNAs, miR-84 and miR-241, resulting in the suppression of lin-14 and akt and the subsequent stimulation of FOXO signaling. This cascade ultimately leads to an increase in lifespan. Finally, in mammals, Lin-28 is required for primordial germ cell development107. Similarly, Hmga2-deficient males mice are infertile due to a lack of spermatozoa108. These studies demonstrate a conserved role for let-7 targets in germ cell development. In total, the deeply conserved let-7 network appears to regulate the intimately linked processes of proliferation, growth, development, metabolism and longevity.
miR-17~92 and c-Myc
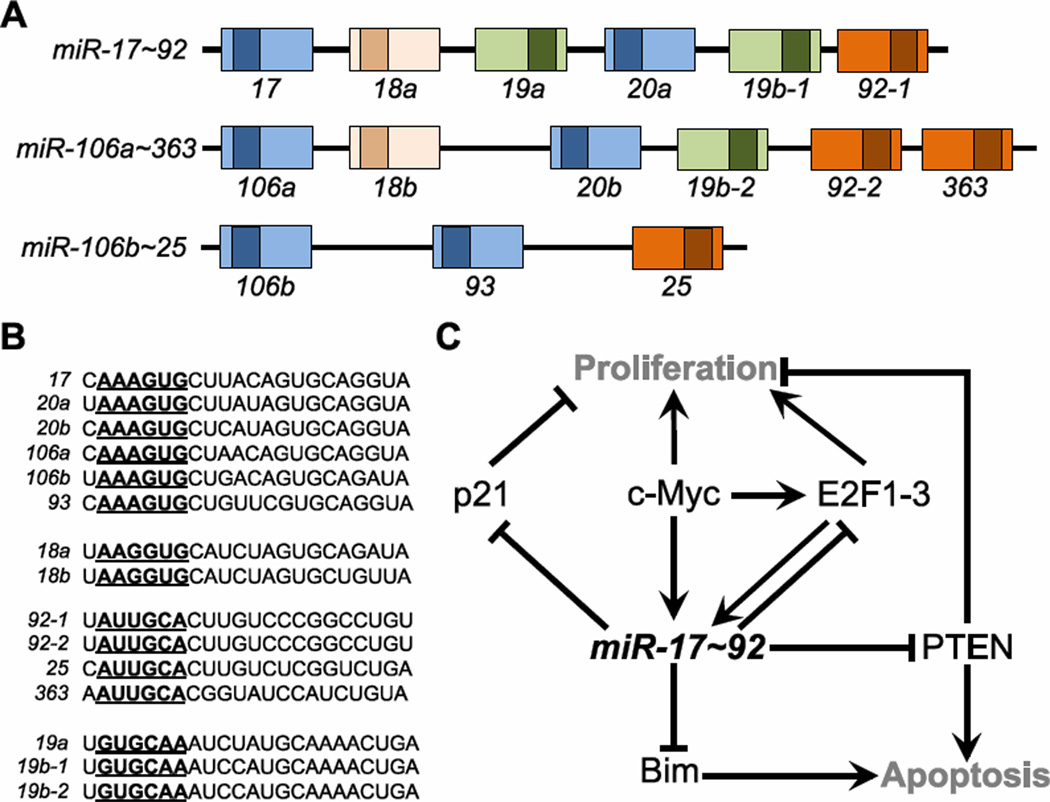
miR-17~92, also known as oncomiR-1, is a widely-studied, oncogenic cluster of miRNAs that encodes four different seed families and has two paralogues, namely miR-106b~25 and miR-106a~363 (Figure 5A,B). miRNAs derived from miR-17~92 and miR-106b~25 are broadly expressed during development, including in ESCs and midgestation embryos, and across adult tissues, including liver, heart, and brain109. miR-17~92 is critical to development. Patients with hemizygous germline deletion of miR-17~92 develop Feingold Syndrome, previously associated only with mutations in MYCN, a proliferative gene and let-7 target, as described above. This disorder is characterized by microcephaly, short stature and digital abnormalities110. miR-17~92 also regulates proliferation and is often amplified or overexpressed in various tumor types, including B cell lymphomas and small cell lung carcinoma, and drives tumorigenesis in mouse models of lymphoma and leukemia111; 112; 113; 114.
Figure 5.
The miR-17~92 genes. (A) Gene structure of paralogues miR-17~92, miR-106a~363, and miR-106b~25. The pre-miRNA sequences are indicated as boxes, with mature miRNA sequences denoted in darker shades. Related family members are indicated by color. (B) Sequences of miRNAs. Each miRNA is grouped based on seed relationship. Seeds are bold and underlined. (C) Summary of the miR-17~92 network.
The miR-17~92 cluster participates in a circuit with c-Myc and E2F115; 116; 117; 118 (Figure 5C). c-Myc transcriptionally induces miR-17~92 and the transcription factors E2F1115, E2F2 and E2F3117. E2F1–3 also induce transcription of miR-17~92115; 116; 117. miR-17 and miR-20, members of the same seed family within the miR-17~92 cluster, in turn repress translation of E2F1–3, exemplifying negative feedback in the case of E2F regulation115; 116; 117 and an incoherent feedforward loop in the case of c-Myc115. Consistent with these functional relationships, miR-17~92 cooperates with c-Myc to induce murine lymphoma111. Additionally, transcription of miR-17~92 is regulated by BMP signaling in the heart119; 120. Many additional pathways, summarized recently112, are regulated by miR-17~92: Proliferation (Cyclin D1121, p2139; 122); TGFbeta signaling (TGFbeta-R2, SMAD2, SMAD4123); survival (PTEN124; 125, BIM109; 126, Fas127); and cell type-specific processes such as differentiation (CEBPA128, GATA6128). Interestingly, the seed sequence of the miR-17 family (AAAGUGC) overlaps with the seed of the ESC-specific miR-290 family (AAGUGCU). Both families repress common targets120 that regulate proliferation, such as Lats-2, p21; and differentiation, such as Lefty1, Lefty2, TGFbeta-R2. Furthermore, miR-17~92 was recently demonstrated to enhance induction of pluripotency129. These results suggest that miR-17~92 is a somatic counterpart to the miR-290 family that contributes to the de-differentiation and plasticity of tumors.
A mouse model of miR-17~92 loss has been generated109. Germline deletion of miR-17~92 results in early postnatal lethality, while animals with germline deletion of either miR-106b~25 or miR-106a~363 are viable and fertile. Triple knockout of all paralogues, or double knockout of miR-17~92 and miR-106b~25, results in embryonic lethality by E15109, indicating at least partially redundant functions. miR-17~92 KO animals exhibit several prominent developmental abnormalities: (1) Skeletal defects, which phenocopy the symptoms observed in patients with germline mutations in this cluster110, (2) lung hypoplasia109, (3) ventricular septal defect109, and (4) a failure in fetal B cell development109. miR-17~92 is also required for adult B cell development. Transplant of miR-17~92-deficient hematopoietic cells fails to reconstitute hematopoiesis in lethally-irradiated mice109, while tissue-specific deletion of Dicer in B cell progenitors leads to increased apoptosis126. These immunological defects are partially due to de-repression, in pro-B cells, of Bim, a pro-apoptotic gene and target of miRNAs clustered in miR-17~92109; 126. In myeloid cells, the miR-17 family promotes proliferation by targeting sequestome 1, a ubiquitin-binding protein that regulates autophagy-mediated protein degradation130. In total, miR-17~92 plays an important role in various tissue types, consistent with its broad expression pattern.
In addition to elucidating the function of this cluster in normal development and physiology, mouse models of miR-17~92 function have been used to characterize the role of this cluster in promoting tumorigenesis. In an Eµ-Myc model of murine lymphoma, deletion of miR-17~92 results in reduced lymphoma burden due to increased apoptosis of tumor cells124. Overexpression of miR-19, a distinct seed family within miR-17~92, is necessary and sufficient to promote c-Myc-induced tumorigenesis and rescues the phenotype of miR-17~92 deletion124; 125. The activity of miR-19 is mediated by its downregulation of the tumor suppressor PTEN124; 125. A second mouse model of cancer, specifically retinoblastoma, also demonstrates that miR-17~92 participates in the development of tumors131; 132. While overexpression of miR-17~92 alone does not induce tumors, combining this transgene with mutations in the Rb pathway promotes the formation and metastasis of retinoblastoma131. In contrast to the Eµ-Myc model, in which miR-19 represses apoptosis, seed family-specific inhibition of miRNAs with antagomirs suggests that the activity of this cluster in retinoblastoma is independent of miR-19 as well as apoptosis, instead promoting proliferation through repression of p21 by the miR-17 family131. In complementary loss-of-function studies, deletion of miR-17~92 in retinal progenitor cells in the context of combined Rb- and p53-loss results in suppression of retinoblastoma, a result again attributable primarily to the miR-17 family132. Thus, various family members within the miR-17~92 cluster possess oncogenic activities.
miR-17~92 exemplifies many of the properties of miRNAs relevant to the regulation of gene expression networks. This cluster contains multiple miRNA seed families whose members are expressed from multiple, paralogous loci. Clustered, unrelated seed family members, such as miR-17 and miR-19, regulate distinct but related biological functions, namely proliferation and apoptosis, respectively. In normal tissues and under pathological conditions, miR-17~92 promotes growth by targeting genes that participate in common pathways, including the c-Myc/Rb/E2F axis and its targets. One of the many outstanding questions in the field concerns the tissue-specific phenotypes of the knockout animals. For example, do the lung and cardiac defects result from misregulation of the same developmental and proliferative axes observed in blood or are distinct pathways responsible for these phenotypes? What are the functions of the additional family members expressed from these clusters? The experimental tools currently available, such as miR-17~92-conditional mice, tissue-specific Cre transgenes, and miRNA expression constructs, may be sufficient to answer these questions.
miR-34 and the p53-response
p53 is a commonly mutated tumor suppressor that coordinates multiple pathways, ranging from damage repair to cell death, to counter stress133. Given its profound biological and clinical importance, the discovery that p53 induced the expression of a miRNA was met with great interest. However, follow-up studies have led to conflicting results that suggest a functional complexity not predicted by current models.
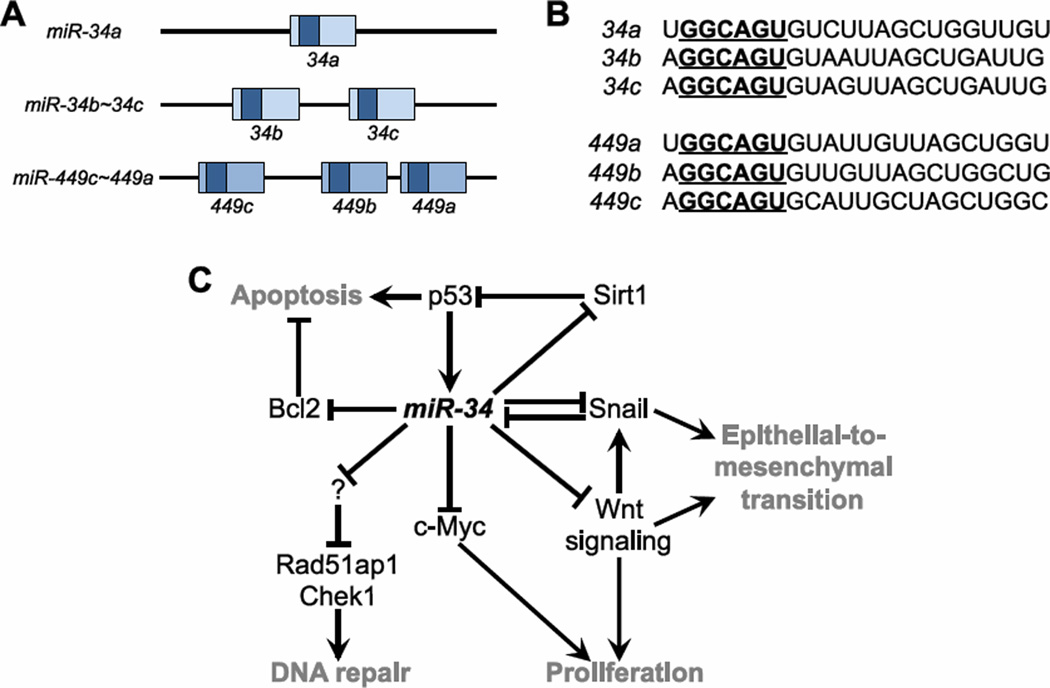
Upon exposure to stress, p53 induces the miR-34 family of miRNAs in murine embryonic fibroblasts134, a mesenchymal cell type, and HCT116 colon cancer cells135, an epithelial cell type. Three miRNAs comprise the miR-34 family (Figure 6A,B): miR-34a is expressed as a single miRNA from an intergenic locus, while miR-34b and miR-34c are expressed as an intergenic cluster. The promoters of both loci possess p53 binding sites and are bound and activated by p53. Therefore, these miRNAs are direct transcriptional targets of p53134; 135; 136; 137; 138; 139. Additionally, the miR-449 family, encoded as a cluster of three miRNAs in a single locus, shares a seed sequence with miR-34 and therefore belongs to the same seed family140. However, miR-449 and miR-34 are divergent in sequence outside of the seed region (Figure 6B). The p53-responsiveness of miR-449 has not been systematically characterized.
Figure 6.
The miR-34 and miR-449 genes. (A) Gene structure of miR-34a, miR-34b~34c, and miR-449c~449a. The pre-miRNA sequences are indicated as boxes, with mature miRNA sequences denoted in in darker shades. Related family members are indicated by color. miR-34 and miR-449 share the same seed sequence. (B) Sequences of miRNAs. Each miRNA is grouped based on seed relationship and sequence similarity. Seeds are bold and underlined. (C) Summary of the miR-34 network.
Supporting a role in the p53 pathway, exogenous expression of miR-34 mediates anti-proliferative effects, p53-mediated apoptosis137, and senescence134. Multiple strategies, including mRNA expression profiling135, proteomics141, and capture of miRNA-bound mRNAs142, have identified targets of miR-34 in these pathways143 (Figure 6C). miR-34 participates in a positive feedback loop with its transcriptional activator by repressing SIRT1, a gene responsible for deacetylation, and subsequently reduced activity, of p53144; 145. Additional targets of miR-34135; 143 include regulators of cell cycle, such as CDK4134; 146 and Cyclin E134; 139; apoptosis, such as Bcl2 and DcR3139; and proliferation, such as c-Myc147. miR-34 also regulates multiple genes in the DNA damage pathway142. This activity includes induction of DNA repair genes, such as Rad51ap1 and Chek1135, presumably through incompletely-characterized intermediate targets. Consistent with a common functional role, both p53 and miR-34 pose a barrier to the reprogramming of somatic cells into induced pluripotent stem cells, although this activity may be due solely to the regulation of proliferation148. Additionally, gene expression analyses followed up with functional experiments have demonstrated a role for miR-34 in downregulating multiple components in canonical Wnt-signaling, a pathway important in development, epithelial-to-mesenchymal transition (EMT), and tumorigenesis149. SNAIL, a transcription factor regulated by Wnt-signaling, participates in a double-negative feedback loop with miR-34 to form a bistable switch that regulates EMT150; 151. As a further illustration of the complexity and redundancy of miRNA-regulated pathways, p53 also induces the miR-200 family of miRNAs, which repress ZEB1 and ZEB2, transcriptional activators of EMT152. Clinically, multiple studies have reported reduction in miR-34 expression in a variety of tumor types, including lung and breast cancer143; 152, and in vivo functional follow-ups, such as in the case of hepatocellular carcinoma, have confirmed a tumor suppressive role for this miRNA in tumor-derived cell lines153.
While overexpression studies have demonstrated that miR-34 is sufficient to activate p53-related pathways, an elegant and exhaustive genetic loss-of-function study in mouse suggests that miR-34 is not necessary for a canonical p53 response. Deletion of both miR-34 loci, and consequently all three miR-34 family members, in mouse has revealed a surprisingly mild phenotype140. The mutant animals develop normally, an observation consistent with the normal development of p53 KO mice. However, unlike p53 KO cells, miR-34 KO cells respond normally to genotoxic stress by activating cell cycle arrest or apoptosis in murine embryonic fibroblasts and thymocytes, respectively. Additionally, both wild type and miR-34 KO animals are sensitive to gamma-irradiation while, in contrast, p53 KO animals are resistant. miR-34 KO animals also do not form the spontaneous tumors that are a hallmark of both p53 heterozygosity and loss140. Furthermore, miR-34 is expressed in testes, lung, and brain independent of p53 expression, suggesting functions additional to the p53 pathway. The observation that miR-34 is expressed in brain supports recent reports demonstrating that this miRNA regulates development of the nervous system154; 155. The miR-449 gene, a transcriptional target of E2F1 and regulator of cell cycle progression, encodes miRNAs that share a seed sequence with miR-34156. Since this gene is intact in miR-34 KO animals, it could in principle compensate for miR-34 loss. However, with the exception of testis, this miRNA is not appreciably expressed in the same tissues as miR-34. Additionally, miR-449 is not upregulated in miR-34 KO animals140. Nonetheless, a formal test of functional redundancy will require additional compound mutant mice.
Despite its strong transcriptional link to p53, miR-34 in the mouse appears to be dispensable for canonical p53 stress response, suggesting context-dependent functions for this miRNA family. Consistent with this possibility, a recent study demonstrated a role for miR-34 in promoting age-associated cardiomyocyte cell death through repression of PNUTS, a regulator of apoptosis and DNA damage response157. Additional analyses of miR-34 KO animals in various tumor models with greater numbers of mice may uncover tissue-specific roles for this miRNA in p53-mediated tumor suppression. Nonetheless, even in these initial studies, the findings contrast with reports that inhibition of miR-34 through complementary antagomirs compromises p53 response. These results may reflect a phenotypic difference between acute loss of miR-34 activity in the inhibition experiments and early developmental loss in the mouse model. The observation of a surprisingly mild mouse KO phenotype under basal, unstressed conditions is a common feature in miRNA studies. A notable example is miR-143~145, a cluster of two unrelated miRNAs implicated as tumor suppressors in leukemia and as inhibitors of pluripotency158; 159; 160. Mouse KOs of this cluster progress normally into adulthood without developing spontaneous tumors. However, these animals exhibit intestinal collapse due to defects in smooth muscle function. The discrepancies between in vivo and in vitro models raise the possibility that early loss of a miRNA family, for example by deletion in the germline, leads to functional compensation mediated by the re-wiring of miRNA-regulated networks. For many such miRNA KO animals, it will be important to determine if acute, tissue-specific deletion of conditional alleles yields phenotypes more consistent with those observed in cell culture through overexpression or, conversely, antagomir-mediated inhibition.
Concluding Remarks
Much progress has been made in understanding miRNA function. These small RNAs orchestrate the activities of functionally related genes within discrete and isolatable networks. As additional targets of miRNAs are delineated through global gene expression profiling and animal studies, the thus far binary sets of interactions that have been identified will be placed into a larger context, yielding greater insight not only into miRNA function but also into the circuitry of gene expression.
Highlights.
-
-
Network-level perspective of miRNA activity
-
-
A summary of the structure of eukaryotic Argonaute
-
-
A comparison of miRNA function in vitro versus in vivo
-
-
A description of four miRNA families: miR-290~295, let-7, miR-17~92, and miR-34
Acknowledgements
We thank Victoria Lu for preparing the schematic of the miRNA biogenesis pathway. The style of the schematics depicting miRNA genes was influenced by the publications of Andrea Ventura. We thank members of the Sharp laboratory, particularly Arvind Ravi and Paul Boutz, for productive discussions. We apologize to authors whose work we could not cite due to space limitations. This work was supported by United States Public Health Service grants RO1-CA133404 from the National Institutes of Health and PO1-CA42063 from the National Cancer Institute to P.A.S., and partially by Cancer Center Support (core) grant P30-CA14051 from the National Cancer Institute. A.M.G. acknowledges support from a Leukemia and Lymphoma Society grant 5198-09.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 3.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 8.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 9.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 10.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 13.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O’Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, Becker KG, Gorospe M, Hide W, Lieberman J. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to "seedless" 3'UTR microRNA recognition elements. Mol Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 18.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, Blelloch R, Schroth GP, Nusbaum C, Bartel DP. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 20.Ambros V. The evolution of our thinking about microRNAs. Nat Med. 2008;14:1036–1040. doi: 10.1038/nm1008-1036. [DOI] [PubMed] [Google Scholar]
- 21.Townley-Tilson WH, Callis TE, Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol. 2010;42:1252–1255. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 23.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebert MS, Sharp PA. Emerging roles for natural microRNA sponges. Curr Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 27.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 28.Bethune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, Sonenberg N. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 32.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 33.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 34.Tsang JS, Ebert MS, van Oudenaarden A. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol Cell. 2010;38:140–153. doi: 10.1016/j.molcel.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 36.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayr C, Bartel DP. Widespread shortening of 3'UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 39.Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, Kim VN. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joshua-Tor L, Hannon GJ. Ancestral roles of small RNAs: an Ago-centric perspective. Cold Spring Harb Perspect Biol. 2011;3:a003772. doi: 10.1101/cshperspect.a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150:100–110. doi: 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Structure of yeast Argonaute with guide RNA. Nature. 2012;486:368–374. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sashital DG, Doudna JA. Structural insights into RNA interference. Curr Opin Struct Biol. 2010;20:90–97. doi: 10.1016/j.sbi.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shkumatava A, Stark A, Sive H, Bartel DP. Coherent but overlapping expression of microRNAs and their targets during vertebrate development. Genes Dev. 2009;23:466–481. doi: 10.1101/gad.1745709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–859. doi: 10.1038/ng.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development. 2011;138:1653–1661. doi: 10.1242/dev.056234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 54.Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ciaudo C, Servant N, Cognat V, Sarazin A, Kieffer E, Viville S, Colot V, Barillot E, Heard E, Voinnet O. Highly dynamic and sex-specific expression of microRNAs during early ES cell differentiation. PLoS Genet. 2009;5:e1000620. doi: 10.1371/journal.pgen.1000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng GX, Ravi A, Calabrese JM, Medeiros LA, Kirak O, Dennis LM, Jaenisch R, Burge CB, Sharp PA. A latent pro-survival function for the mir-290–295 cluster in mouse embryonic stem cells. PLoS Genet. 2011;7:e1002054. doi: 10.1371/journal.pgen.1002054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benetti R, Gonzalo S, Jaco I, Munoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P, Li E, Serrano M, Millar S, Hannon G, Blasco MA. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 61.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 64.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Medeiros LA, Dennis LM, Gill ME, Houbaviy H, Markoulaki S, Fu D, White AC, Kirak O, Sharp PA, Page DC, Jaenisch R. Mir-290–295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc Natl Acad Sci U S A. 2011;108:14163–14168. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Houbaviy HB, Dennis L, Jaenisch R, Sharp PA. Characterization of a highly variable eutherian microRNA gene. RNA. 2005;11:1245–1257. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng GX, Ravi A, Gould GM, Burge CB, Sharp PA. Genome-wide impact of a recently expanded microRNA cluster in mouse. Proc Natl Acad Sci U S A. 2011;108:15804–15809. doi: 10.1073/pnas.1112772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ambros V. MicroRNAs and developmental timing. Curr Opin Genet Dev. 2011;21:511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nimmo RA, Slack FJ. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405–418. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–98. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hammell CM, Karp X, Ambros V. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18668–18673. doi: 10.1073/pnas.0908131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn. 2005;234:1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 78.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 79.Zhu H, Shah S, Shyh-Chang N, Shinoda G, Einhorn WS, Viswanathan SR, Takeuchi A, Grasemann C, Rinn JL, Lopez MF, Hirschhorn JN, Palmert MR, Daley GQ. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42:626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Altshuler D, Daley GQ. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, Sanna S, Eyheramendy S, Voight BF, Butler JL, Guiducci C, Illig T, Hackett R, Heid IM, Jacobs KB, Lyssenko V, Uda M, Diabetes Genetics I, Fusion Kora, Prostate LC, Ovarian Cancer Screening T, Nurses’ Health S, SardiNia Boehnke M., Chanock SJ, Groop LC, Hu FB, Isomaa B, Kraft P, Peltonen L, Salomaa V, Schlessinger D, Hunter DJ, Hayes RB, Abecasis GR, Wichmann HE, Mohlke KL, Hirschhorn JN. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Alexandersen P, Feenstra B, Boyd HA, Aben KK, Verbeek AL, Roeleveld N, Jonasdottir A, Styrkarsdottir U, Steinthorsdottir V, Karason A, Stacey SN, Gudmundsson J, Jakobsdottir M, Thorleifsson G, Hardarson G, Gulcher J, Kong A, Kiemeney LA, Melbye M, Christiansen C, Tryggvadottir L, Thorsteinsdottir U, Stefansson K. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat Genet. 2009;41:734–738. doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- 83.Ong KK, Elks CE, Li S, Zhao JH, Luan J, Andersen LB, Bingham SA, Brage S, Smith GD, Ekelund U, Gillson CJ, Glaser B, Golding J, Hardy R, Khaw KT, Kuh D, Luben R, Marcus M, McGeehin MA, Ness AR, Northstone K, Ring SM, Rubin C, Sims MA, Song K, Strachan DP, Vollenweider P, Waeber G, Waterworth DM, Wong A, Deloukas P, Barroso I, Mooser V, Loos RJ, Wareham NJ. Genetic variation in LIN28B is associated with the timing of puberty. Nat Genet. 2009;41:729–733. doi: 10.1038/ng.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.He C, Kraft P, Chen C, Buring JE, Pare G, Hankinson SE, Chanock SJ, Ridker PM, Hunter DJ, Chasman DI. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41:724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perry JR, Stolk L, Franceschini N, Lunetta KL, Zhai G, McArdle PF, Smith AV, Aspelund T, Bandinelli S, Boerwinkle E, Cherkas L, Eiriksdottir G, Estrada K, Ferrucci L, Folsom AR, Garcia M, Gudnason V, Hofman A, Karasik D, Kiel DP, Launer LJ, van Meurs J, Nalls MA, Rivadeneira F, Shuldiner AR, Singleton A, Soranzo N, Tanaka T, Visser JA, Weedon MN, Wilson SG, Zhuang V, Streeten EA, Harris TB, Murray A, Spector TD, Demerath EW, Uitterlinden AG, Murabito JM. Meta-analysis of genome-wide association data identifies two loci influencing age at menarche. Nat Genet. 2009;41:648–650. doi: 10.1038/ng.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 87.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335:1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, Shah SP, Tanwar PS, Mermel CH, Beroukhim R, Azam M, Teixeira J, Meyerson M, Hughes TP, Llovet JM, Radich J, Mullighan CG, Golub TR, Sorensen PH, Daley GQ. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Molenaar JJ, Domingo-Fernandez R, Ebus ME, Lindner S, Koster J, Drabek K, Mestdagh P, van Sluis P, Valentijn LJ, van Nes J, Broekmans M, Haneveld F, Volckmann R, Bray I, Heukamp L, Sprussel A, Thor T, Kieckbusch K, Klein-Hitpass L, Fischer M, Vandesompele J, Schramm A, van Noesel MM, Varesio L, Speleman F, Eggert A, Stallings RL, Caron HN, Versteeg R, Schulte JH. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012;44:1199–1206. doi: 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- 90.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 91.Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, Ciarmiello A, Pallante P, Arra C, Melillo RM, Helin K, Croce CM, Fusco A. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–471. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 92.Zaidi MR, Okada Y, Chada KK. Misexpression of full-length HMGA2 induces benign mesenchymal tumors in mice. Cancer Res. 2006;66:7453–7459. doi: 10.1158/0008-5472.CAN-06-0931. [DOI] [PubMed] [Google Scholar]
- 93.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, Elliott KS, Hackett R, Guiducci C, Shields B, Zeggini E, Lango H, Lyssenko V, Timpson NJ, Burtt NP, Rayner NW, Saxena R, Ardlie K, Tobias JH, Ness AR, Ring SM, Palmer CN, Morris AD, Peltonen L, Salomaa V, Diabetes Genetics I, Wellcome Trust Case Control C, Davey Smith G, Groop LC, Hattersley AT, McCarthy MI, Hirschhorn JN, Frayling TM. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Horikoshi M, Yaghootkar H, Mook-Kanamori DO, Sovio U, Taal HR, Hennig BJ, Bradfield JP, St Pourcain B, Evans DM, Charoen P, Kaakinen M, Cousminer DL, Lehtimaki T, Kreiner-Moller E, Warrington NM, Bustamante M, Feenstra B, Berry DJ, Thiering E, Pfab T, Barton SJ, Shields BM, Kerkhof M, van Leeuwen EM, Fulford AJ, Kutalik Z, Zhao JH, den Hoed M, Mahajan A, Lindi V, Goh LK, Hottenga JJ, Wu Y, Raitakari OT, Harder MN, Meirhaeghe A, Ntalla I, Salem RM, Jameson KA, Zhou K, Monies DM, Lagou V, Kirin M, Heikkinen J, Adair LS, Alkuraya FS, Al-Odaib A, Amouyel P, Andersson EA, Bennett AJ, Blakemore AI, Buxton JL, Dallongeville J, Das S, de Geus EJ, Estivill X, Flexeder C, Froguel P, Geller F, Godfrey KM, Gottrand F, Groves CJ, Hansen T, Hirschhorn JN, Hofman A, Hollegaard MV, Hougaard DM, Hypponen E, Inskip HM, Isaacs A, Jorgensen T, Kanaka-Gantenbein C, Kemp JP, Kiess W, Kilpelainen TO, Klopp N, Knight BA, Kuzawa CW, McMahon G, Newnham JP, Niinikoski H, Oostra BA, Pedersen L, Postma DS, Ring SM, Rivadeneira F, Robertson NR, Sebert S, Simell O, Slowinski T, Tiesler CM, Tonjes A, Vaag A, Viikari JS, Vink JM, Vissing NH, Wareham NJ, Willemsen G, Witte DR, Zhang H, et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat Genet. 2012;45:76–82. doi: 10.1038/ng.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ashar HR, Chouinard RA, Jr, Dokur M, Chada K. In vivo modulation of HMGA2 expression. Biochim Biophys Acta. 2010;1799:55–1761. doi: 10.1016/j.bbagrm.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 96.Fedele M, Battista S, Manfioletti G, Croce CM, Giancotti V, Fusco A. Role of the high mobility group A proteins in human lipomas. Carcinogenesis. 2001;22:1583–1591. doi: 10.1093/carcin/22.10.1583. [DOI] [PubMed] [Google Scholar]
- 97.Wu J, Liu Z, Shao C, Gong Y, Hernando E, Lee P, Narita M, Muller W, Liu J, Wei JJ. HMGA2 overexpression-induced ovarian surface epithelial transformation is mediated through regulation of EMT genes. Cancer Res. 2011;71:349–359. doi: 10.1158/0008-5472.CAN-10-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24:4448–4464. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tessier CR, Doyle GA, Clark BA, Pitot HC, Ross J. Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res. 2004;64:209–214. doi: 10.1158/0008-5472.can-03-2927. [DOI] [PubMed] [Google Scholar]
- 100.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- 101.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 102.Mongroo PS, Noubissi FK, Cuatrecasas M, Kalabis J, King CE, Johnstone CN, Bowser MJ, Castells A, Spiegelman VS, Rustgi AK. IMP-1 displays crosstalk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2. Cancer Res. 2011;71:2172–2182. doi: 10.1158/0008-5472.CAN-10-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li Z, Gilbert JA, Zhang Y, Zhang M, Qiu Q, Ramanujan K, Shavlakadze T, Eash JK, Scaramozza A, Goddeeris MM, Kirsch DG, Campbell KP, Brack AS, Glass DJ. An HMGA2-IGF2BP2 Axis Regulates Myoblast Proliferation and Myogenesis. Dev Cell. 2012 doi: 10.1016/j.devcel.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tsang WP, Kwok TT. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis. 2008;13:1215–1222. doi: 10.1007/s10495-008-0256-z. [DOI] [PubMed] [Google Scholar]
- 105.Toledano H, D’Alterio C, Czech B, Levine E, Jones DL. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature. 2012;485:605–610. doi: 10.1038/nature11061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shen Y, Wollam J, Magner D, Karalay O, Antebi A. A steroid receptor-microRNA switch regulates life span in response to signals from the gonad. Science. 2012;338:1472–1476. doi: 10.1126/science.1228967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, Surani MA, Daley GQ. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chieffi P, Battista S, Barchi M, Di Agostino S, Pierantoni GM, Fedele M, Chiariotti L, Tramontano D, Fusco A. HMGA1 and HMGA2 protein expression in mouse spermatogenesis. Oncogene. 2002;21:3644–3650. doi: 10.1038/sj.onc.1205501. [DOI] [PubMed] [Google Scholar]
- 109.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, Van Haeringen A, Genevieve D, Goldenberg A, Oufadem M, Manouvrier S, Munnich A, Vidigal JA, Vekemans M, Lyonnet S, Henrion-Caude A, Ventura A, Amiel J. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sage J, Ventura A. miR than meets the eye. Genes Dev. 2011;25:1663–1667. doi: 10.1101/gad.17454011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 114.Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, Yamada H, Suzuki M, Nagino M, Nimura Y, Osada H, Takahashi T. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17–92. Oncogene. 2007;26:6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 115.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 116.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 117.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 118.Aguda BD, Kim Y, Piper-Hunter MG, Friedman A, Marsh CB. MicroRNA regulation of a cancer network: consequences of the feedback loops involving miR-17–92, E2F, and Myc. Proc Natl Acad Sci U S A. 2008;105:19678–19683. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, Bai Y, Huang Z, Black BL, Wang F, Martin JF. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a MicroRNA-mediated mechanism. Dev Cell. 2010;19:903–912. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vidigal JA, Ventura A. Embryonic stem cell miRNAs and their roles in development and disease. Semin Cancer Biol. 2012;22:428–436. doi: 10.1016/j.semcancer.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, Wu K, Whittle J, Ju X, Hyslop T, McCue P, Pestell RG. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182:509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Inomata M, Tagawa H, Guo YM, Kameoka Y, Takahashi N, Sawada K. MicroRNA-17–92 down-regulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2009;113:396–402. doi: 10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- 123.Mestdagh P, Bostrom AK, Impens F, Fredlund E, Van Peer G, De Antonellis P, von Stedingk K, Ghesquiere B, Schulte S, Dews M, Thomas-Tikhonenko A, Schulte JH, Zollo M, Schramm A, Gevaert K, Axelson H, Speleman F, Vandesompele J. The miR-17–92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Mol Cell. 2010;40:762–773. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D’Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17–92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 127.Huang G, Nishimoto K, Zhou Z, Hughes D, Kleinerman ES. miR-20a encoded by the miR-17–92 cluster increases the metastatic potential of osteosarcoma cells by regulating Fas expression. Cancer Res. 2012;72:908–916. doi: 10.1158/0008-5472.CAN-11-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oeztuerk-Winder F, Guinot A, Ochalek A, Ventura JJ. Regulation of human lung alveolar multipotent cells by a novel p38alpha MAPK/miR-17–92 axis. EMBO J. 2012;31:3431–3441. doi: 10.1038/emboj.2012.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li Z, Yang CS, Nakashima K, Rana TM. Small RNA-mediated regulation of iPS cell generation. EMBO J. 2011;30:823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Meenhuis A, van Veelen PA, de Looper H, van Boxtel N, van den Berge IJ, Sun SM, Taskesen E, Stern P, de Ru AH, van Adrichem AJ, Demmers J, Jongen-Lavrencic M, Lowenberg B, Touw IP, Sharp PA, Erkeland SJ. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood. 2011;118:916–925. doi: 10.1182/blood-2011-02-336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, Hammond SM, MacPherson D. miR-17~92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev. 2011;25:1734–1745. doi: 10.1101/gad.17027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nittner D, Lambertz I, Clermont F, Mestdagh P, Kohler C, Nielsen SJ, Jochemsen A, Speleman F, Vandesompele J, Dyer MA, Schramm A, Schulte JH, Marine JC. Synthetic lethality between Rb, p53 and Dicer or miR-17–92 in retinal progenitors suppresses retinoblastoma formation. Nat Cell Biol. 2012;14:958–965. doi: 10.1038/ncb2556. [DOI] [PubMed] [Google Scholar]
- 133.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 137.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 138.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 139.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 140.Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D'Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Roh S, Hoffmann R, Warscheid B, Hermeking H. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol Cell Proteomics. 2011;10:M111 010462. doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lal A, Thomas MP, Altschuler G, Navarro F, O'Day E, Li XL, Concepcion C, Han YC, Thiery J, Rajani DK, Deutsch A, Hofmann O, Ventura A, Hide W, Lieberman J. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7:e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 146.Wei J, Shi Y, Zheng L, Zhou B, Inose H, Wang J, Guo XE, Grosschedl R, Karsenty G. miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2. J Cell Biol. 2012;197:509–521. doi: 10.1083/jcb.201201057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cannell IG, Kong YW, Johnston SJ, Chen ML, Collins HM, Dobbyn HC, Elia A, Kress TR, Dickens M, Clemens MJ, Heery DM, Gaestel M, Eilers M, Willis AE, Bushell M. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc Natl Acad Sci U S A. 2010;107:5375–5380. doi: 10.1073/pnas.0910015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C, Ozturk A, Hicks GG, Hannon GJ, He L. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13:1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim NH, Kim HS, Kim NG, Lee I, Choi HS, Li XY, Kang SE, Cha SY, Ryu JK, Na JM, Park C, Kim K, Lee S, Gumbiner BM, Yook JI, Weiss SJ. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci Signal. 2011;4:ra71. doi: 10.1126/scisignal.2001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 151.Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, Rowe RG, Lee S, Maher CA, Weiss SJ, Yook JI. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol. 2011;195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 153.Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, Deng Y, Zhao J, Jiang S, Yuan Y, Wang HY, Cheng SQ, Xie D, Wang XF. TGF-beta-miR-34a–CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, Zhu Y, Wang LS, Bonini NM. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482:519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW, Ventura A, Concepcion CP, Han YC, Candi E, Knight RA, Mak TW, Melino G. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci U S A. 2011;108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yang X, Feng M, Jiang X, Wu Z, Li Z, Aau M, Yu Q. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23:2388–2393. doi: 10.1101/gad.1819009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Muller OJ, Potente M, Zeiher AM, Hermeking H, Dimmeler S. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013 doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 158.Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 2010;30:1118–1126. doi: 10.1161/ATVBAHA.109.200873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, Peterson KL, Indolfi C, Catalucci D, Chen J, Courtneidge SA, Condorelli G. The knockout of miR-143 and −145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rissland OS, Hong SJ, Bartel DP. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol Cell. 2011;43:993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gurha P, Abreu-Goodger C, Wang T, Ramirez MO, Drumond AL, van Dongen S, Chen Y, Bartonicek N, Enright AJ, Lee B, Kelm RJ, Jr, Reddy AK, Taffet GE, Bradley A, Wehrens XH, Entman ML, Rodriguez A. Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation. 2012;125:2751–2761. doi: 10.1161/CIRCULATIONAHA.111.044354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, Matsunaga J, Takahashi RU, Takata T, Shimamoto A, Ochiya T, Tahara H. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol. 2011;193:409–424. doi: 10.1083/jcb.201010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 168.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 169.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 172.Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, Calin GA, Hagan JP, Kipps T, Croce CM. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 173.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 174.Sun L, Xie H, Mori MA, Alexander R, Yuan B, Hattangadi SM, Liu Q, Kahn CR, Lodish HF. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011;13:958–965. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 176.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Choi PS, Zakhary L, Choi WY, Caron S, Alvarez-Saavedra E, Miska EA, McManus M, Harfe B, Giraldez AJ, Horvitz HR, Schier AF, Dulac C. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57:41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing 'stemness'. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 180.Yu D, dos Santos CO, Zhao G, Jiang J, Amigo JD, Khandros E, Dore LC, Yao Y, D’Souza J, Zhang Z, Ghaffari S, Choi J, Friend S, Tong W, Orange JS, Paw BH, Weiss MJ. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 2010;24:1620–1633. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]