Abstract
After the original identification of thyroid transcription factor 1 (TTF-1 or NKX2-1) biochemical activity as a transcriptional regulator of thyroglobulin in 1989, the bulk of the ensuing research has concentrated on elucidating the roles of NKX2-1 in the development of lung and thyroid tissues. Motivated by its specific expression pattern, pathologists adopted the NKX2-1 immunoreactivity to distinguish pulmonary from nonpulmonary nonthyroid adenocarcinomas. Interestingly, the concept of NKX2-1 as an active participant in lung tumorigenesis did not take hold until 2007. This minireview contrasts the recent advancements of NKX2-1-related observations primarily in the realm of pulmonary malignancies.
Keywords: Gene Amplification, Gene Regulation, Lung Cancer, Metastasis, MicroRNA, NKX2-1, TTF-1, Lung Lineage
Introduction
In lung cancer, NK2 homeobox 1 (NKX2-1) was proposed to be an appealing candidate lineage-survival oncogene in 2006 (1). This was a logical conjecture based on the frequent NKX2-1 overexpression in lung adenocarcinomas (ADs)2 (2, 3) and the requirement of NKX2-1 for branching morphogenesis of normal lung development as well as the differentiation of lung epithelial cells (4–6). In 2007, four studies uncovered that NKX2-1 is recurrently amplified in human lung cancer (7–10), implying that NKX2-1 is likely functionally relevant to the pulmonary tumorigenic process, beyond just a marker of lung ADs. Indeed, a race was on to tease out the oncogenic mechanism of NKX2-1. In parallel, a much unexpected finding related to Nkx2-1 was reported by Winslow et al. (11): Nkx2-1 prevents primary tumors from metastasizing. This anti-metastatic activity of Nkx2-1 conceptually contradicts the functional ramification of the NKX2-1 gene amplification seen in human lung cancer. Nevertheless, the latest data from a number of investigators provide a deeper glimpse of the mechanistic intricacy to the anti-oncogenic function of NKX2-1. Evidently, NKX2-1 joins an expanding list of cancer genes with both pro- and anti-oncogenic activities. These genes include MYC (12), AKT1 (13), MDM2 (14), WT1 (15), REST (16), and others. In this minireview, I succinctly retrace and contrast the studies from the original discovery of NKX2-1 gene amplification to the multifunctionalities of NKX2-1 in cancers.
Genetic Alterations of NKX2-1 in Lung Cancer
NKX2-1 is located at the 14q13.3 region in the human genome. Earlier studies using lower resolution genomic tools had identified 14q13 amplification (17). In 2006, this region was again reported to undergo focal and wide DNA copy number increases in a panel of human lung adenocarcinoma cell lines using array-based comparative genomic hybridization (aCGH) (18). The minimal amplified area across all of the cancer cell lines with the 14q13.3 amplicon contains nine known genes: PSMA6, NFKBIA, INSM2, BRMS1, MBIP, NKX2-1, NKX2-8, PAX9, and SLC25A21. However, the target gene of amplification in this amplicon was not pursued by functional analyses at that time. Within 2 years of this 2006 study, four independent reports documented the gene amplification of NKX2-1 from different angles. Prompted by the fact that NKX2-1 is a reliable marker of the terminal respiratory unit type of lung ADs (19), Tanaka et al. (9) investigated the potential involvement of NKX2-1 in the pathogenesis of lung ADs. After obtaining the evidence that RNAi-induced knockdown of NKX2-1 in NKX2-1+ lung adenocarcinoma cell lines retarded cell growth, they searched for genetic alterations of NKX2-1 in AD specimens. No somatic point mutation was uncovered; nevertheless, 2.3% of lung ADs contained an NKX2-1 gene copy number increase in their analysis. Curiously, they also detected a higher frequency of increased NKX2-1 gene copies at metastatic sites compared with primary sites. However, the 14q13.3 minimal amplified area, i.e. the core amplicon, was not studied, and thus, it was not known how many other genes were coamplified with NKX2-1 (9).
In the next two studies, Kendall et al. (7) and Weir et al. (10) employed different types of aCGH-based tools to profile the genomic DNA copy number landscapes of human lung cancer. Although both studies identified the 14q13.3 amplicon as the most recurrent focal amplicon not containing an apparent lung oncogene, the sizes and boundaries of the core amplicon varied from 413 kb (covering three genes: NKX2-1, NKX2-8, and PAX9) (7) to 480 kb (covering two genes: MBIP and NKX2-1) (10). It is not clear why the core amplicon boundaries varied. Presumably, it is a reflection of the intrinsic sample differences. Alternatively, it may be related to the actual methodology in scoring/determining the amplified region. Kendall et al. used quantitative PCR to refine the amplicon boundaries, whereas Weir et al. used a statistical method (genomic identification of significant targets in cancer, or GISTIC (20)) to score amplicons. Nevertheless, NKX2-1 was found to be amplified in both studies. More importantly, the NKX2-1 amplicon occurs only in lung cancer, in line with the lung cell lineage link of NKX2-1. Defining the core amplicon is important because each gene within the core amplicon may be a driver oncogene targeted by the amplification. Thus, every gene inside a core amplicon is a candidate driver gene and must be analyzed for oncogenic characteristics. In the study by Kendall et al. (7), the 14q13.3 core amplicon seems to contain functionally cooperating driver genes, as coexpression of the three individual coamplified genes in a pairwise manner enhanced the growth potential of the immortalized premalignant lung epithelial cells (BEAS-2B). A subsequent study, in which a gene expression signature-based strategy was used, suggests that the co-activation of the biological pathways of NKX2-1 and NKX2-8 is correlated with poor prognosis for patients with lung ADs (21), reminiscent of the in vitro functional cooperation between the coamplified 14q13.3 genes (7). However, transgenic mice with targeted lung overexpression of Nkx2-1/Nkx2-8 or Nkx2-1/Pax9 did not develop lung tumors, and overexpression of Nkx2-1 alone in the murine lung failed to initiate tumor formation (22). Considering that normal mammalian lung epithelia are notorious for their transformation to full malignancies (23), other genetic lesions would most likely be needed to manifest these coamplified genes. For the two-gene core amplicon detected by Weir et al. (10), only the anti-NKX2-1 RNAi decreased the colony-forming ability of NKX2-1+ lung cancer cells with or without the 14q13.3 amplicon, but not that of NKX2-1− lung cancer cells, consistent with NKX2-1 being the driver gene of the 14q13.3 amplification. Like Tanaka et al. (9), Weir et al. did not detect somatic exon mutations of NKX2-1 in 384 lung adenocarcinoma DNA samples. Clearly, gene amplification is the main mechanism activating NKX2-1 in lung cancer. However, as I will discuss below, point mutations and gene rearrangements are invoked to activate the NKX2-1 oncogene in malignancies outside lung cancer.
Kwei et al. (8) also detected the 14q13.3 cytoband amplification as the most frequent focal lung cancer genomic amplification not associated with a known lung oncogene. Following the initial cDNA-based aCGH profiling, they used a custom oligonucleotide tiling array consisting of probes covering 14q13.2∼q13.3 at 300-bp intervals to fine map the core amplicon boundaries. In this case, the core amplicon covered eight genes (NFKB1A, INSM2, GARNL1, BRMS1L, MBIP, NKX2-1, NKX2-8, and PAX9). The authors chose to concentrate on a potential functional connection between NKX2-1 amplification and lung cancer in light of the fact that NKX2-1 is a critical lung developmental factor (24–26) and a histologic marker of lung ADs (27). Transfection of siRNAs against NKX2-1 into amplified human lung adenocarcinoma cell lines diminished cell proliferation, which was attributed to both decreased cell cycle progression and increased apoptosis. Interestingly, human lung adenocarcinoma cell lines without the NKX2-1 amplicon, albeit with detectable NKX2-1 expression, did not exhibit a similar behavior toward anti-NKX2-1 siRNAs, suggesting the putative functional importance of the amplification-driven NKX2-1 up-regulation.
Dynamic Nature of the Known Gene Content of a Core Amplicon
It is important to recall that the human genome is dynamic in that new functional elements continue to be discovered (28). Therefore, the known gene content of a core amplicon may change with time. Reanalysis of the 413-kb core amplicon of Kendall et al. (7) revealed that there is a new RefSeq gene termed surfactant-associated 3 (SFTA3), which is <3 kb away from NKX2-1 (Fig. 1A) and which was not known to be residing at 14q13.3 in 2007. Apparently, this gene is part of the 14q13.3 core amplicon of varying sizes and boundaries discovered by multiple groups. Without further expression and functional studies of SFTA3, it would not be possible to determine whether this gene constitutes a target of 14q13.3 amplification.
FIGURE 1.
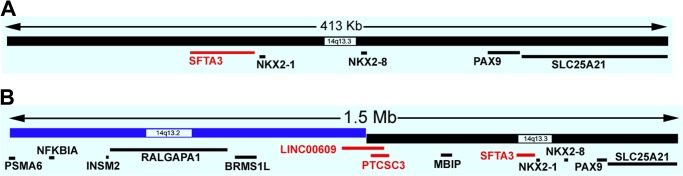
Genes in the current UCSC Genome Browser annotation of the 14q13.3 core amplicons. A, the core amplicon of 413 kb per Kendall et al. (7). The human genome coordinate shown is from GRCh37/hg19: chr14: 36,828,082–37,241,292. SLC25A21 is not fully contained in the core amplicon. B, the core amplicon of 1.5 Mb per Kwei et al. (8). SLC25A21 and PSMA6 are not fully contained in the core amplicon. The human genome coordinate shown is from GRCh37/hg19: chr14: 35,778,082–37,311,292. Only the RefSeq-annotated genes are shown (95). Genes shown in red were not known previously to be part of the core amplicons. RALGAPA1 is also known as GARNL1.
NKX2-1 Amplification in Squamous Cell Carcinomas of the Lung
Non-small cell lung cancer (NSCLC) accounts for nearly 80∼85% of all lung cancer cases (29). Within NSCLC, ADs and squamous cell carcinomas (SQCCs) are the two most frequent histologic subtypes, representing 50 and 30% of NSCLC, respectively (30). Although the histologic distinction between pulmonary ADs and SQCCs is obvious when these tumors are well differentiated, it can be difficult when they are poorly differentiated, especially in biopsies and cytology specimens. A well accepted immunostaining outcome of SQCCs is the expression of p63 and its N-terminal truncated form (ΔNp63) and the absence of NKX2-1 expression, i.e. ΔNp63+/NKX2-1− (27, 31, 32). This strategy is related to the notion that NKX2-1 is rarely expressed in SQCCs but is frequently expressed in ADs. Although there are studies reporting NKX2-1 expression in SQCCs (33–35), a thorough review of all published studies on positive NKX2-1 expression in SQCCs suggests that the NKX2-1 immunoreactivity may have been due to the type of antibody used and/or the selection of the SQCC cases containing either glandular differentiation or an AD component (36). Under this backdrop, one may anticipate that the NKX2-1-containing 14q13.3 amplicon would not occur in SQCCs as it does in ADs. Indeed, a single nucleotide polymorphism microarray-based analysis of 47 primary SQCC DNA samples did not score the 14q13.3 amplicon (37). In contrast, The Cancer Genome Atlas (TCGA) recently uncovered the 14q13.3 cytoband as one of the 32 significant amplification hubs in lung SQCCs; the core amplicon contains these eight genes: RALGAPA1, BRMS1L, MBIP, SFTA3, NKX2-1, NKX2-8, PAX9, and SLC25A21 (38). The existence of the NKX2-1 amplicon in lung SQCCs was also detected by others (8, 39, 40). In particular, Tang et al. (40) reported that NKX2-1 is amplified in 20.2% of the 99 lung SQCCs examined by fluorescence in situ hybridization, but the NKX2-1 protein is not expressed in any SQCC. Obviously, NKX2-1 is unlikely to drive the 14q13.3 amplicon in the amplified SQCCs lacking NKX2-1 expression. The possibility of NKX2-1 being a passenger amplified gene may also play out in a small fraction of lung ADs, as Barletta et al. (41) reported two NKX2-1-amplified lung ADs exhibiting NKX2-1 expression only in the cytoplasm, not in the nucleus. To reconcile this issue, I offer these hypotheses. (a) Other coamplified genes near NKX2-1 may functionally substitute for NKX2-1, as suggested by Kendall et al. (7). These coamplified genes should include the recently discovered gene SFTA3. Moreover, the larger core amplicon uncovered by Kwei et al. (8), in the current genome annotation according to the UCSC Genome Browser (42), contains two more RefSeq non-protein-coding genes than previously realized: LINC00609 and PTCSC3 (Fig. 1B). Although there is nothing known about these two genes in lung cancer, there is a study implicating PTCSC3 as a tumor suppressor in thyroid cancer (43). In view of the demonstrated cancer roles of long noncoding RNAs (44, 45), it is not impossible that these two genes also transmit biological consequences of the NKX2-1-containing amplification. (b) The 14q13.3 amplicon is a nonfunctional passenger genetic alteration in SQCCs. Evidence against this hypothesis came from the amplified SQCC cell line (NCI-H2170), which became less tumorigenic when either NKX2-8 or PAX9 was knocked down by stable RNAi (7). FOXA1 at 14q21.1, ∼1.07 Mb away from NKX2-1 at the telomeric side, has been proposed to be the driver of 14q DNA copy number gains in SQCCs (17, 46). Although this may be true for some larger scale chromosomal gains at 14q, it does not explain the focal 14q13.3 amplification over NKX2-1 in SQCCs. Future functional studies of the 14q13 amplification in SQCCs are needed to shed more light on this puzzle.
To further complicate the matter, Harris et al. (47) detected an allelic loss at 14q13 in lung tumors. The allelic loss, i.e. loss of heterozygosity, occurs in ADs by gene amplification and in SQCCs/adenosquamous tumors by deletion (47). These observations suggest that the 14q13.3 cytoband can undergo DNA copy number alterations in both directions in lung cancer. Seven genes are affected by the 1.2-Mb core-deleted region found by Harris et al.: MBIP, SFTA3, NKX2-1, NKX2-8, PAX9, SLC25A21, and MIPOL1. The main target gene(s) of this core deletion is not defined. However, the report that Nkx2-8 null mice developed spontaneous bronchial adenomas and squamous cancer suggests that Nkx2-8 may be targeted by this DNA loss (48). Nevertheless, the genomic analysis of lung SQCCs by TCGA did not score a significant deletion at 14q13 (38). Perhaps, the 14q13 deletion occurs at a frequency below the threshold of the data analysis algorithm of TCGA.
Pro-oncogenic Activities of NKX2-1
The initial mindset following the discovery of NKX2-1 amplification was that NKX2-1 is an oncogene. Consistent with this perception, a reduction of endogenous NKX2-1 expression in NKX2-1+ lung cancer cells compromised the cellular capacity to grow without attachment (10) and decreased cell proliferation with a concomitant higher level of apoptosis (8, 9). These evidences were taken as proof that NKX2-1+ lung cancer cells are “addicted” to NKX2-1 functionally. Mechanistic studies identified receptor tyrosine kinase-like orphan receptor 1 (ROR1) as a direct transcriptional target of NKX2-1; it also mediates the NKX2-1-dependent survival signaling in lung ADs (49). Genome-wide chromatin immunoprecipitation followed by sequencing (ChIP-seq) and mRNA profiling identified another direct transcriptional target of NKX2-1 (50), LMO3, which is a member of the LMO family of oncogenes that are translocated in T-cell acute lymphoblastic leukemia (T-ALL) (51, 52). Interestingly, LMO3 suppression in NKX2-1-amplified cell lines increased apoptosis but had no effect on cell proliferation (50), suggesting multiple downstream effectors of amplified NKX2-1. Many NKX2-1-binding sites are enriched with neighboring recognition sequences of other oncogenic transcription factors such as AP1 and FOXA1 (22, 50), indicating cooperative transcriptional regulation between NKX2-1 and other oncogenic transcription factors.
NKX2-1 itself is translocated with different partner genes in T-ALL (53). In these T-ALL patients, the NKX2-1 gene product is ectopically overproduced, and cell-based assays implicate it as oncogenic (53). The NKX2-1 translocation in T-ALL harbors a route to investigate the oncogenic mechanism of NKX2-1 in reference to the recent finding that the TLX1 homeodomain oncoprotein mediates T-cell maturation arrest in T-ALL via interaction with ETS1 and suppression of T-cell receptor-α gene expression (54). The parallel may be drawn between NKX2-1 and TLX1 because some of the TLX1-translocated T-ALLs cocluster with NKX2-1-translocated cases by gene expression profiling (53). Ectopic expression of oncogenic NKX2-1 need not be driven by genetic alterations. Recently, it was documented that NKX2-1 expression is switched on in diffuse large B-cell lymphomas by epigenetic modifications (55). Through proteomic analyses of sera derived from a number of mouse models of lung cancer, Taguchi et al. (56) detected the presence of multiple proteins whose genes are known transcriptional targets of NKX2-1. These results manifest an NKX2-1-driven serum protein signature in lung cancer and suggest that NKX2-1 may be a master regulator supervising the lung secretome.
Finally, although point mutations of NKX2-1 do not seem to occur in lung cancer, a germ-line mutation (A339V, 1016C→T) of NKX2-1 was found in patients with multinodular goiter and papillary thyroid cancer (57). Overexpression of this NKX2-1 mutant in normal thyroid cells enhanced cell proliferation (57). If indeed the oncogenic activity of NKX2-1 can be activated via a point mutation, the mystery is then why such a mechanism is never utilized in lung cancer. By the same token, a cohort of 216 primary and metastatic thyroid tumors were analyzed and found to be negative for NKX2-1 gene amplification (39). In light of the importance of NKX2-1 to thyroid biology (58–60) and the identification of 14q13.3 as a risk locus for thyroid cancer by a genome-wide association study (61), the lack of NKX2-1 gene amplification in thyroid malignancies remains a mystery.
Anti-oncogenic Activities of NKX2-1
In a 2004 study by Kang et al. (62), a progressive decrease in Nkx2-1 expression was detected from the wild-type lung adenomas to ADs in a TGFβ1+/− mouse model treated with the carcinogenic ethyl carbamate. In a separate study that conditionally knocked out Nkx2-1 in the adult murine thyroid, a genotoxic carcinogen induced a higher incidence of adenomas in Nkx2-1 null mice relative to wild-type or Nkx2-1+/− mice (63). Although these data suggest an anti-cancer function of Nkx2-1, the pervasive thinking in the field was shaped by the discovery of NKX2-1 amplification and concentrated on unraveling the oncogenic mechanism of NKX2-1, largely ignoring a possible role of NKX2-1 as a tumor suppressor. However, in 2011, an unexpected finding by Winslow et al. (11) brought the anti-tumorigenic side of NKX2-1 into the spotlight. They administered lentiviral vectors expressing the Cre recombinase intratracheally into transgenic mice (KrasG12D/+;p53flox/flox), which later developed multifocal lung ADs. Some of the primary lung tumors eventually led to macroscopic metastases. The stable lentiviral integration sites allowed primary tumors to be unambiguously linked to their related metastases. Gene expression profiling of two types of primary lung tumors (non-metastatic and metastatic) indicated that Nkx2-1 was consistently and significantly depressed in the metastatic primary tumors with clonally related metastases. These observations implicate Nkx2-1 as an anti-metastatic factor in lung ADs. Winslow et al. went on to show that Nkx2-1 prevents primary tumors from metastasizing by suppressing Hmga2. Upon loss of Nkx2-1 expression, derepressed Hmga2 would then facilitate the progression to metastases. It is important to note that pathologists routinely encounter NKX2-1+ metastatic tumors derived from primary human lung ADs, testifying to the complexity of human lung cancers.
Two subsequent thorough animal studies proved that Nkx2-1 is anti-oncogenic and capable of blunting tumor initiation in specific genetic contexts (Fig. 2A) (22, 64). Maeda et al. (22) created transgenic mice harboring a constitutive loss of an Nkx2-1 allele with a conditional activation of the KrasG12D oncogene in the murine respiratory epithelium. Interestingly, the lung tumor number and volume of KrasG12D;Nkx2-1+/− mice increased compared with those of KrasG12D;Nkx2-1+/+ and control mice. The tumor histology of KrasG12D;Nkx2-1+/− mice, unlike that of KrasG12D;Nkx2-1+/+ mice, is reminiscent of human mucinous AD of the lung. To complement their loss-of-function approach, Maeda et al. made triple transgenic mice (Scgb1a1-rtTA;[tetO]-FLAG-Nkx2-1;[tetO]-Kras4bG12D) such that the expression of Nkx2-1 and KrasG12D was turned on simultaneously in the pulmonary epithelium. The result indicates that coexpression of Nkx2-1 reduces the number and volume of Kras-induced lung tumors, showcasing the anti-oncogenic activity of Nkx2-1 in retarding mutant Kras-induced tumor initiation and progression. Because EGFR is a critical lung oncogene not associated with lung mucinous ADs (32), Maeda et al. (22) created recombinant mice with a conditional EgfrL858R allele in the background of Nkx2-1 haploinsufficiency (EgfrL858R;Nkx2-1+/−). In contrast to the Kras-initiated tumors, the tumor number and volume were significantly decreased in the EgfrL858R;Nkx2-1+/− mice compared with the EgfrL858R;Nkx2-1+/+ mice, indicating that Nkx2-1 enhances Egfr-mediated tumorigenesis. The manifestation of the anti- or pro-oncogenic activities of Nkx2-1 in transgenic mice harboring different mutant oncogenes (Kras or Egfr) demonstrates the importance of the signaling network to the Nkx2-1 function. None of the Egfr-driven mouse lung ADs (EgfrL858R;Nkx2-1+/+ or EgfrL858R;Nkx2-1+/−) was of the mucinous subtype, in contrast to those seen in the KrasG12D;Nkx2-1+/− model. An interpretation of these data is that the wild-type level of Nkx2-1 expression guards against KrasG12D from steering the tumor differentiation state toward the mucinous type. It is known that raising oncogenic Ras expression in thyroid cells down-regulates Nkx2-1, which in turn inhibits thyroid differentiation (65, 66). Clearly, the differentiation state of lung tumor cells is also dictated by Nkx2-1 expression status and the coexisting oncogenic events.
FIGURE 2.
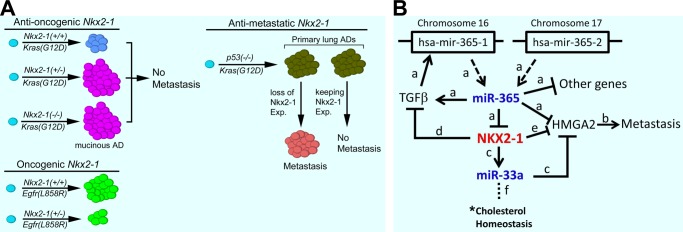
Schematics of NKX2-1 functions and its interactions with miRNAs. A, the graphical presentation summarizes the opposing functional roles of Nkx2-1 in mouse models (11, 22, 64). Exp, expression. B, NKX2-1 is imbedded in a miRNA-based signaling network, with miRNAs acting both up- and downstream. The relevant studies establishing individual interactions are indicated as follows: a, Ref. 82; b, Refs. 11 and 96; c, Ref. 84; d, Ref. 68; e, Ref. 11; and f, Refs. 87–90. *, a direct impact on cholesterol homeostasis by NKX2-1 remains to be demonstrated. This figure was modified from Qi et al. (82) with permission.
Independently, Snyder et al. (64) modeled the Nkx2-1 deletion in a variety of transgenic mice. They first generated a strain of mice that relied on the Cre recombinase to simultaneously activate a mutant KrasG12D allele and become Nkx2-1−/− (64). In doing so, the tumor burden increased by 2∼3-fold in the KrasLSL-G12D;Nkx2-1F/F mice in comparison with the KrasLSL-G12D;Nkx2-1F/+ mice, and the tumors of the KrasLSL-G12D;Nkx2-1F/F mice resembled mucinous ADs, in keeping with the findings of Maeda et al. (22). Lung-specific conditional knock-out of Nkx2-1 never gave rise to macroscopic tumors, despite alteration of the differentiation state in the adult lung (64). The investigators then temporally separated the activation of the KrasG12D allele from Nkx2-1 deletion by placing the two alleles under the control of two different recombinational events. Mice with an activated KrasG12D allele were allowed to go for 2∼7 months, followed by Nkx2-1 deletion using tamoxifen/Flp recombinase. The Nkx2-1 deletion conferred a significant increase in tumor cell proliferation without an effect on apoptosis. Six weeks after Nkx2-1 deletion, the tumor burden was 4-fold higher in the Nkx2-1-deleted mice relative to control mice, documenting that the absence of Nkx2-1 enhances the initiation and progress of Kras-driven tumorigenesis in the lung. Snyder et al. (64) also noted that Nkx2-1 appears to be involved in repressing a gastric differentiation state. The loss of Nkx2-1 may unleash this phenotype in a specific and conducive lung cell type, inducing the mucinous ADs, which are positive for gastric markers (GKN1 and CTSE) repressed by Nkx2-1. The investigators further characterized the genome-wide binding events of Nkx2-1 and analyzed the data in the context of other transcription factors, revealing functionally important links of Nkx2-1 to Foxa1, Foxa2, and Hnf4α.
Other molecules that may mediate the anti-oncogenic/anti-metastatic activities of NKX2-1 have been identified by either profiling or candidate gene approaches. Hosono et al. (67) found myosin-binding protein H (MYBPH) to be under the positive and direct transcriptional regulation of NKX2-1 through gene expression profiling of immortalized lung epithelial cells stably expressing an NKX2-1 transgene. MYBPH reduces single cell motility via negative regulation of actomyosin organization, forming a basis of the anti-metastatic function of NKX2-1. Saito et al. (68) identified that NKX2-1 transactivates E-cadherin and counters the epithelial-mesenchymal transition of lung cancer cells conferred by TGFβ. In fact, TGFβ is known to be antagonistic to NKX2-1-directed gene transcription (69). In view of the significant roles of epithelial-mesenchymal transition in cancer metastasis (70, 71), this finding could also explain the anti-metastatic role of NKX2-1. Finally, Runkle et al. (72) reported that NKX2-1 directly transactivates two molecules, occludin (OCLN) and claudin-1 (CLDN1), at the lung epithelial tight junction. Niimi et al. (73) found that claudin-18 is a transcriptional target of Nkx2-1. NKX2-1 knockdown conferred human lung cancer cell resistance to anoikis, and expression of occludin restored cellular sensitivity to anoikis; overexpression of occludin impeded migration and induced anoikis in lung carcinoma cells. These data point to a putative involvement of tight junction proteins in helping NKX2-1 suppress lung cancer progression (74). A working model derived from these data is that the anti-oncogenic/anti-metastatic activities of NKX2-1 are mediated by a variety of molecules across multiple cellular pathways.
Data Placing NKX2-1 Firmly in a MicroRNA-based Signaling Network
In the lung, microRNAs (miRNAs) play critical roles in both development and tumorigenesis (75–79). Over 100 miRNAs are dynamically regulated during organogenesis of a normal murine lung (80). Considering that NKX2-1 is a vital controller of lung development and cancer, it was surprising that a direct interaction between NKX2-1 and miRNAs was not discovered until 2012. Qi et al. (82) discovered the first miRNA (i.e. miR-365) that directly targets NKX2-1 by screening the top 10 miRNAs predicted by the TargetScan algorithm (81) to bind directly to the 3′-UTR of NKX2-1. Expression profiling identified other putative target genes of miR-365 and miR-365*. Exploration of human lung cancer genomics data uncovered that NKX2-1 gene amplification was significantly associated with DNA copy number loss at one of the two genomic loci encoding the precursor RNA of mature miR-365 (i.e. mir-365-1). This implies the putative existence of genetic selection pressure to lose the repressive miR-365 that would otherwise suppress amplified NKX2-1. Intriguingly, a signaling loop exists among TGFβ, miR-365, and NKX2-1, with TGFβ up-regulating miR-365 via the mir-365-1 precursor gene, which in turn represses NKX2-1 (82). Moreover, miR-365 feeds back to the TGF signaling pathway by specifically up-regulating TGFβ2 (Fig. 2B) (82). The observation of miR-365 repressing NKX2-1 was later reproduced in a separate study (83).
The first miRNAs that are directly regulated by NKX2-1 were uncovered by Rice et al. (84). Motivated by the finding of Nkx2-1 repressing Hmga2 by Winslow et al. (11), Rice et al. speculated that NKX2-1 directly activates miRNAs to repress HMGA2. Using two complementary strategies, gain- and loss-of-expression of NKX2-1/Nkx2-1, they found a selection of miRNAs that are putatively directly controlled by NKX2-1. One such miRNA, miR-33a, was further characterized because there are three predicted binding sites for miR-33a in the 3-kb-long 3′-UTR of HMGA2. The experimental data confirmed that HMGA2 is a genuine target of miR-33a and that NKX2-1 directly transactivates the host gene of miR-33a, SREBF2, to up-regulate miR-33a, which then keeps HMGA2 expression in check (84). An intriguing implication from this study relates to the fact that both SREBP2 (the gene product of SREBF2) (85, 86) and miR-33a (87–90) are critical players in cholesterol homeostasis, suggesting that NKX2-1 may influence cholesterol metabolism in the lung and that cholesterol metabolism may be mechanistically involved in the lung cancer biology of NKX2-1 (Fig. 2B).
Conclusions
In recent years, we have witnessed a rapid unfolding of lung cancer biology in connection with the fundamental lung developmental transcription factor gene NKX2-1. The dual faces of NKX2-1 as both a pro- and an anti-cancer factor are complex but not unique in the NK2 family of transcription factors. The NKX2-2 oncogene is translocated in T-ALL (53) and is an essential signaling mediator of the driver oncogene EWS-FLI in Ewing sarcoma (91). In contrast, NKX2-2 suppresses self-renewal of glioma-initiating cells and is frequently down-regulated in human gliomas (92). Intriguingly, the disparate outcome of NKX2-1 also extends to its correlation with clinical parameters of lung AD patients. Positive NKX2-1 expression is associated with good prognosis (41, 93, 94), but NKX2-1 amplification is linked with poor prognosis (40, 41). The biochemical activity of NKX2-1 as a transcription factor precludes it from direct drug discovery. However, as more signaling partners of NKX2-1 are uncovered, there will be opportunities to initiate translational research to create and develop NKX2-1-dependent strategies as tools to manage NKX2-1-positive lung cancer.
Acknowledgments
I thank the members of my laboratory for reviewing the manuscript and K. Helsley for editorial comments.
This work was supported in part by National Institutes of Health Grant CA127547.
- AD
- adenocarcinoma
- aCGH
- array-based comparative genomic hybridization
- NSCLC
- non-small cell lung cancer
- SQCC
- squamous cell carcinoma
- T-ALL
- T-cell acute lymphoblastic leukemia
- miRNA
- microRNA.
REFERENCES
- 1. Garraway L. A., Sellers W. R. (2006) Lineage dependency and lineage-survival oncogenes in human cancer. Nat. Rev. Cancer 6, 593–602 [DOI] [PubMed] [Google Scholar]
- 2. Ordóñez N. G. (2000) Thyroid transcription factor-1 is a marker of lung and thyroid carcinomas. Adv. Anat. Pathol. 7, 123–127 [DOI] [PubMed] [Google Scholar]
- 3. Stenhouse G., Fyfe N., King G., Chapman A., Kerr K. M. (2004) Thyroid transcription factor 1 in pulmonary adenocarcinoma. J. Clin. Pathol. 57, 383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bohinski R. J., Di Lauro R., Whitsett J. A. (1994) The lung-specific surfactant protein B gene promoter is a target for thyroid transcription factor 1 and hepatocyte nuclear factor 3, indicating common factors for organ-specific gene expression along the foregut axis. Mol. Cell. Biol. 14, 5671–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kimura S., Hara Y., Pineau T., Fernandez-Salguero P., Fox C. H., Ward J. M., Gonzalez F. J. (1996) The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 10, 60–69 [DOI] [PubMed] [Google Scholar]
- 6. Minoo P., Su G., Drum H., Bringas P., Kimura S. (1999) Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev. Biol. 209, 60–71 [DOI] [PubMed] [Google Scholar]
- 7. Kendall J., Liu Q., Bakleh A., Krasnitz A., Nguyen K. C., Lakshmi B., Gerald W. L., Powers S., Mu D. (2007) Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 16663–16668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwei K. A., Kim Y. H., Girard L., Kao J., Pacyna-Gengelbach M., Salari K., Lee J., Choi Y. L., Sato M., Wang P., Hernandez-Boussard T., Gazdar A. F., Petersen I., Minna J. D., Pollack J. R. (2008) Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene 27, 3635–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tanaka H., Yanagisawa K., Shinjo K., Taguchi A., Maeno K., Tomida S., Shimada Y., Osada H., Kosaka T., Matsubara H., Mitsudomi T., Sekido Y., Tanimoto M., Yatabe Y., Takahashi T. (2007) Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 67, 6007–6011 [DOI] [PubMed] [Google Scholar]
- 10. Weir B. A., Woo M. S., Getz G., Perner S., Ding L., Beroukhim R., Lin W. M., Province M. A., Kraja A., Johnson L. A., Shah K., Sato M., Thomas R. K., Barletta J. A., Borecki I. B., Broderick S., Chang A. C., Chiang D. Y., Chirieac L. R., Cho J., Fujii Y., Gazdar A. F., Giordano T., Greulich H., Hanna M., Johnson B. E., Kris M. G., Lash A., Lin L., Lindeman N., Mardis E. R., McPherson J. D., Minna J. D., Morgan M. B., Nadel M., Orringer M. B., Osborne J. R., Ozenberger B., Ramos A. H., Robinson J., Roth J. A., Rusch V., Sasaki H., Shepherd F., Sougnez C., Spitz M. R., Tsao M. S., Twomey D., Verhaak R. G., Weinstock G. M., Wheeler D. A., Winckler W., Yoshizawa A., Yu S., Zakowski M. F., Zhang Q., Beer D. G., Wistuba I. I., Watson M. A., Garraway L. A., Ladanyi M., Travis W. D., Pao W., Rubin M. A., Gabriel S. B., Gibbs R. A., Varmus H. E., Wilson R. K., Lander E. S., Meyerson M. (2007) Characterizing the cancer genome in lung adenocarcinoma. Nature 450, 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winslow M. M., Dayton T. L., Verhaak R. G., Kim-Kiselak C., Snyder E. L., Feldser D. M., Hubbard D. D., DuPage M. J., Whittaker C. A., Hoersch S., Yoon S., Crowley D., Bronson R. T., Chiang D. Y., Meyerson M., Jacks T. (2011) Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu H., Radisky D. C., Yang D., Xu R., Radisky E. S., Bissell M. J., Bishop J. M. (2012) MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat. Cell Biol. 14, 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu H., Radisky D. C., Nelson C. M., Zhang H., Fata J. E., Roth R. A., Bissell M. J. (2006) Mechanism of Akt1 inhibition of breast cancer cell invasion reveals a protumorigenic role for TSC2. Proc. Natl. Acad. Sci. U.S.A. 103, 4134–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Manfredi J. J. (2010) The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 24, 1580–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang L., Han Y., Saurez Saiz F., Minden M. D. (2007) A tumor suppressor and oncogene: the WT1 story. Leukemia 21, 868–876 [DOI] [PubMed] [Google Scholar]
- 16. Negrini S., Prada I., D'Alessandro R., Meldolesi J. (2013) REST: an oncogene or a tumor suppressor? Trends Cell Biol. 23, 289–295 [DOI] [PubMed] [Google Scholar]
- 17. Lin L., Miller C. T., Contreras J. I., Prescott M. S., Dagenais S. L., Wu R., Yee J., Orringer M. B., Misek D. E., Hanash S. M., Glover T. W., Beer D. G. (2002) The hepatocyte nuclear factor 3α gene, HNF3α (FOXA1), on chromosome band 14q13 is amplified and overexpressed in esophageal and lung adenocarcinomas. Cancer Res. 62, 5273–5279 [PubMed] [Google Scholar]
- 18. Garnis C., Lockwood W. W., Vucic E., Ge Y., Girard L., Minna J. D., Gazdar A. F., Lam S., MacAulay C., Lam W. L. (2006) High resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int. J. Cancer 118, 1556–1564 [DOI] [PubMed] [Google Scholar]
- 19. Yatabe Y., Mitsudomi T., Takahashi T. (2002) TTF-1 expression in pulmonary adenocarcinomas. Am. J. Surg. Pathol. 26, 767–773 [DOI] [PubMed] [Google Scholar]
- 20. Beroukhim R., Getz G., Nghiemphu L., Barretina J., Hsueh T., Linhart D., Vivanco I., Lee J. C., Huang J. H., Alexander S., Du J., Kau T., Thomas R. K., Shah K., Soto H., Perner S., Prensner J., Debiasi R. M., Demichelis F., Hatton C., Rubin M. A., Garraway L. A., Nelson S. F., Liau L., Mischel P. S., Cloughesy T. F., Meyerson M., Golub T. A., Lander E. S., Mellinghoff I. K., Sellers W. R. (2007) Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. U.S.A. 104, 20007–20012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hsu D. S., Acharya C. R., Balakumaran B. S., Riedel R. F., Kim M. K., Stevenson M., Tuchman S., Mukherjee S., Barry W., Dressman H. K., Nevins J. R., Powers S., Mu D., Potti A. (2009) Characterizing the developmental pathways TTF-1, NKX2-8, and PAX9 in lung cancer. Proc. Natl. Acad. Sci. U.S.A. 106, 5312–5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maeda Y., Tsuchiya T., Hao H., Tompkins D. H., Xu Y., Mucenski M. L., Du L., Keiser A. R., Fukazawa T., Naomoto Y., Nagayasu T., Whitsett J. A. (2012) KrasG12D and Nkx2-1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J. Clin. Invest. 122, 4388–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sato M., Vaughan M. B., Girard L., Peyton M., Lee W., Shames D. S., Ramirez R. D., Sunaga N., Gazdar A. F., Shay J. W., Minna J. D. (2006) Multiple oncogenic changes (K-RASV12, p53 knockdown, mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer a full malignant phenotype on human bronchial epithelial cells. Cancer Res. 66, 2116–2128 [DOI] [PubMed] [Google Scholar]
- 24. Costa R. H., Kalinichenko V. V., Lim L. (2001) Transcription factors in mouse lung development and function. Am. J. Physiol. Lung Cell. Mol. Physiol. 280, L823–L838 [DOI] [PubMed] [Google Scholar]
- 25. Maeda Y., Davé V., Whitsett J. A. (2007) Transcriptional control of lung morphogenesis. Physiol. Rev. 87, 219–244 [DOI] [PubMed] [Google Scholar]
- 26. Minoo P. (2000) Transcriptional regulation of lung development: emergence of specificity. Respir. Res. 1, 109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Travis W. D., Brambilla E., Muller-Hermelink H. K., Harris C. C. (2004) Pathology and Genetics: Tumours of the Lung, Pleura, Thymus and Heart, IARC Press, Lyon, France [Google Scholar]
- 28. ENCODE Project Consortium, Dunham I., Kundaje A., Aldred S. F., Collins P. J., Davis C. A., Doyle F., Epstein C. B., Frietze S., Harrow J., Kaul R., Khatun J., Lajoie B. R., Landt S. G., Lee B. K., Pauli F., Rosenbloom K. R., Sabo P., Safi A., Sanyal A., Shoresh N., Simon J. M., Song L., Trinklein N. D., Altshuler R. C., Birney E., Brown J. B., Cheng C., Djebali S., Dong X., Dunham I., Ernst J., Furey T. S., Gerstein M., Giardine B., Greven M., Hardison R. C., Harris R. S., Herrero J., Hoffman M. M., Iyer S., Kelllis M., Khatun J., Kheradpour P., Kundaje A., Lassmann T., Li Q., Lin X., Marinov G. K., Merkel A., Mortazavi A., Parker S. C., Reddy T. E., Rozowsky J., Schlesinger F., Thurman R. E., Wang J., Ward L. D., Whitfield T. W., Wilder S. P., Wu W., Xi H. S., Yip K. Y., Zhuang J., Bernstein B. E., Birney E., Dunham I., Green E. D., Gunter C., Snyder M., Pazin M. J., Lowdon R. F., Dillon L. A., Adams L. B., Kelly C. J., Zhang J., Wexler J. R., Green E. D., Good P. J., Feingold E. A., Bernstein B. E., Birney E., Crawford G. E., Dekker J., Elinitski L., Farnham P. J., Gerstein M., Giddings M. C., Gingeras T. R., Green E. D., Guigo R., Hardison R. C., Hubbard T. J., Kellis M., Kent W. J., Lieb J. D., Margulies E. H., Myers R. M., Snyder M., Starnatoyannopoulos J. A., Tennebaum S. A., Weng Z., White K. P., Wold B., Khatun J., Yu Y., Wrobel J., Risk B. A., Gunawardena H. P., Kuiper H. C., Maier C. W., Xie L., Chen X., Giddings M. C., Bernstein B. E., Epstein C. B., Shoresh N., Ernst J., Kheradpour P., Mikkelsen T. S., Gillespie S., Goren A., Ram O., Zhang X., Wang L., Issner R., Coyne M. J., Durham T., Ku M., Truong T., Ward L. D., Altshuler R. C., Eaton M. L., Kellis M., Djebali S., Davis C. A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., Xue C., Marinov G. K., Khatun J., Williams B. A., Zaleski C., Rozowsky J., Roder M., Kokocinski F., Abdelhamid R. F., Alioto T., Antoshechkin I., Baer M. T., Batut P., Bell I., Bell K., Chakrabortty S., Chen X., Chrast J., Curado J., Derrien T., Drenkow J., Dumais E., Dumais J., Duttagupta R., Fastuca M., Fejes-Toth K., Ferreira P., Foissac S., Fullwood M. J., Gao H., Gonzalez D., Gordon A., Gunawardena H. P., Howald C., Jha S., Johnson R., Kapranov P., King B., Kingswood C., Li G., Luo O. J., Park E., Preall J. B., Presaud K., Ribeca P., Risk B. A., Robyr D., Ruan X., Sammeth M., Sandu K. S., Schaeffer L., See L. H., Shahab A., Skancke J., Suzuki A. M., Takahashi H., Tilgner H., Trout D., Walters N., Wang H., Wrobel J., Yu Y., Hayashizaki Y., Harrow J., Gerstein M., Hubbard T. J., Reymond A., Antonarakis S. E., Hannon G. J., Giddings M. C., Ruan Y., Wold B., Carninci P., Guigo R., Gingeras T. R., Rosenbloom K. R., Sloan C. A., Learned K., Malladi V. S., Wong M. C., Barber G. P., Cline M. S., Dreszer T. R., Heitner S. G., Karolchik D., Kent W. J., Kirkup V. M., Meyer L. R., Long J. C., Maddren M., Raney B. J., Furey T. S., Song L., Grasfeder L. L., Giresi P. G., Lee B. K., Battenhouse A., Sheffield N. C., Simon J. M., Showers K. A., Safi A., London D., Bhinge A. A., Shestak C., Schaner M. R., Kim S. K., Zhang Z. Z., Mieczkowski P. A., Mieczkowska J. O., Liu Z., McDaniell R. M., Ni Y., Rashid N. U., Kim M. J., Adar S., Zhang Z., Wang T., Winter D., Keefe D., Birney E., Iyer V. R., Lieb J. D., Crawford G. E., Li G., Sandhu K. S., Zheng M., Wang P., Luo O. J., Shahab A., Fullwood M. J., Ruan X., Ruan Y., Myers R. M., Pauli F., Williams B. A., Gertz J., Marinov G. K., Reddy T. E., Vielmetter J., Partridge E. C., Trout D., Varley K. E., Gasper C., Bansal A., Pepke S., Jain P., Amrhein H., Bowling K. M., Anaya M., Cross M. K., King B., Muratet M. A., Antoshechkin I., Newberry K. M., McCue K., Nesmith A. S., Fisher-Aylor K. I., Pusey B., DeSalvo G., Parker S. L., Balasubramanian S., Davis N. S., Meadows S. K., Eggleston T., Gunter C., Newberry J. S., Levy S. E., Absher D. M., Mortazavi A., Wong W. H., Wold B., Blow M. J., Visel A., Pennachio L. A., Elnitski L., Margulies E. H., Parker S. C., Petrykowska H. M., Abyzov A., Aken B., Barrell D., Barson G., Berry A., Bignell A., Boychenko V., Bussotti G., Chrast J., Davidson C., Derrien T., Despacio-Reyes G., Diekhans M., Ezkurdia I., Frankish A., Gilbert J., Gonzalez J. M., Griffiths E., Harte R., Hendrix D. A., Howald C., Hunt T., Jungreis I., Kay M., Khurana E., Kokocinski F., Leng J., Lin M. F., Loveland J., Lu Z., Manthravadi D., Mariotti M., Mudge J., Mukherjee G., Notredame C., Pei B., Rodriguez J. M., Saunders G., Sboner A., Searle S., Sisu C., Snow C., Steward C., Tanzer A., Tapanari E., Tress M. L., van Baren M. J., Walters N., Washieti S., Wilming L., Zadissa A., Zhengdong Z., Brent M., Haussler D., Kellis M., Valencia A., Gerstein M., Raymond A., Guigo R., Harrow J., Hubbard T. J., Landt S. G., Frietze S., Abyzov A., Addleman N., Alexander R. P., Auerbach R. K., Balasubramanian S., Bettinger K., Bhardwaj N., Boyle A. P., Cao A. R., Cayting P., Charos A., Cheng Y., Cheng C., Eastman C., Euskirchen G., Fleming J. D., Grubert F., Habegger L., Hariharan M., Harmanci A., Iyenger S., Jin V. X., Karczewski K. J., Kasowski M., Lacroute P., Lam H., Larnarre-Vincent N., Leng J., Lian J., Lindahl-Allen M., Min R., Miotto B., Monahan H., Moqtaderi Z., Mu X. J., O'Geen H., Ouyang Z., Patacsil D., Pei B., Raha D., Ramirez L., Reed B., Rozowsky J., Sboner A., Shi M., Sisu C., Slifer T., Witt H., Wu L., Xu X., Yan K. K., Yang X., Yip K. Y., Zhang Z., Struhl K., Weissman S. M., Gerstein M., Farnham P. J., Snyder M., Tenebaum S. A., Penalva L. O., Doyle F., Karmakar S., Landt S. G., Bhanvadia R. R., Choudhury A., Domanus M., Ma L., Moran J., Patacsil D., Slifer T., Victorsen A., Yang X., Snyder M., White K. P., Auer T., Centarin L., Eichenlaub M., Gruhl F., Heerman S., Hoeckendorf B., Inoue D., Kellner T., Kirchmaier S., Mueller C., Reinhardt R., Schertel L., Schneider S., Sinn R., Wittbrodt B., Wittbrodt J., Weng Z., Whitfield T. W., Wang J., Collins P. J., Aldred S. F., Trinklein N. D., Partridge E. C., Myers R. M., Dekker J., Jain G., Lajoie B. R., Sanyal A., Balasundaram G., Bates D. L., Byron R., Canfield T. K., Diegel M. J., Dunn D., Ebersol A. K., Ebersol A. K., Frum T., Garg K., Gist E., Hansen R. S., Boatman L., Haugen E., Humbert R., Jain G., Johnson A. K., Johnson E. M., Kutyavin T. M., Lajoie B. R., Lee K., Lotakis D., Maurano M. T., Neph S. J., Neri F. V., Nguyen E. D., Qu H., Reynolds A. P., Roach V., Rynes E., Sabo P., Sanchez M. E., Sandstrom R. S., Sanyal A., Shafer A. O., Stergachis A. B., Thomas S., Thurman R. E., Vernot B., Vierstra J., Vong S., Wang H., Weaver M. A., Yan Y., Zhang M., Akey J. A., Bender M., Dorschner M. O., Groudine M., MacCoss M. J., Navas P., Stamatoyannopoulos G., Kaul R., Dekker J., Stamatoyannopoulos J. A., Dunham I., Beal K., Brazma A., Flicek P., Herrero J., Johnson N., Keefe D., Lukk M., Luscombe N. M., Sobral D., Vaquerizas J. M., Wilder S. P., Batzoglou S., Sidow A., Hussami N., Kyriazopoulou-Panagiotopoulou S., Libbrecht M. W., Schaub M. A., Kundaje A., Hardison R. C., Miller W., Giardine B., Harris R. S., Wu W., Bickel P. J., Banfai B., Boley N. P., Brown J. B., Huang H., Li Q., Li J. J., Noble W. S., Bilmes J. A., Buske O. J., Hoffman M. M., Sahu A. O., Kharchenko P. V., Park P. J., Baker D., Taylor J., Weng Z., Iyer S., Dong X., Greven M., Lin X., Wang J., Xi H. S., Zhuang J., Gerstein M., Alexander R. P., Balasubramanian S., Cheng C., Harmanci A., Lochovsky L., Min R., Mu X. J., Rozowsky J., Yan K. K., Yip K. Y., Birney E. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Larsen J. E., Minna J. D. (2011) Molecular biology of lung cancer: clinical implications. Clin. Chest Med. 32, 703–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perez-Moreno P., Brambilla E., Thomas R., Soria J. C. (2012) Squamous cell carcinoma of the lung: molecular subtypes and therapeutic opportunities. Clin. Cancer Res. 18, 2443–2451 [DOI] [PubMed] [Google Scholar]
- 31. Bishop J. A., Teruya-Feldstein J., Westra W. H., Pelosi G., Travis W. D., Rekhtman N. (2012) p40 (ΔNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod. Pathol. 25, 405–415 [DOI] [PubMed] [Google Scholar]
- 32. Travis W. D., Brambilla E., Noguchi M., Nicholson A. G., Geisinger K. R., Yatabe Y., Beer D. G., Powell C. A., Riely G. J., Van Schil P. E., Garg K., Austin J. H., Asamura H., Rusch V. W., Hirsch F. R., Scagliotti G., Mitsudomi T., Huber R. M., Ishikawa Y., Jett J., Sanchez-Cespedes M., Sculier J. P., Takahashi T., Tsuboi M., Vansteenkiste J., Wistuba I., Yang P. C., Aberle D., Brambilla C., Flieder D., Franklin W., Gazdar A., Gould M., Hasleton P., Henderson D., Johnson B., Johnson D., Kerr K., Kuriyama K., Lee J. S., Miller V. A., Petersen I., Roggli V., Rosell R., Saijo N., Thunnissen E., Tsao M., Yankelewitz D. (2011) International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 6, 244–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fabbro D., Di Loreto C., Stamerra O., Beltrami C. A., Lonigro R., Damante G. (1996) TTF-1 gene expression in human lung tumours. Eur. J. Cancer 32, 512–517 [DOI] [PubMed] [Google Scholar]
- 34. Matoso A., Singh K., Jacob R., Greaves W. O., Tavares R., Noble L., Resnick M. B., Delellis R. A., Wang L. J. (2010) Comparison of thyroid transcription factor-1 expression by 2 monoclonal antibodies in pulmonary and nonpulmonary primary tumors. Appl. Immunohistochem. Mol. Morphol. 18, 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Puglisi F., Barbone F., Damante G., Bruckbauer M., Di Lauro V., Beltrami C. A., Di Loreto C. (1999) Prognostic value of thyroid transcription factor-1 in primary, resected, non-small cell lung carcinoma. Mod. Pathol. 12, 318–324 [PubMed] [Google Scholar]
- 36. Ordóñez N. G. (2012) Thyroid transcription factor-1 is not expressed in squamous cell carcinomas of the lung: an immunohistochemical study with review of the literature. Appl. Immunohistochem. Mol. Morphol. 20, 525–530 [DOI] [PubMed] [Google Scholar]
- 37. Bass A. J., Watanabe H., Mermel C. H., Yu S., Perner S., Verhaak R. G., Kim S. Y., Wardwell L., Tamayo P., Gat-Viks I., Ramos A. H., Woo M. S., Weir B. A., Getz G., Beroukhim R., O'Kelly M., Dutt A., Rozenblatt-Rosen O., Dziunycz P., Komisarof J., Chirieac L. R., Lafargue C. J., Scheble V., Wilbertz T., Ma C., Rao S., Nakagawa H., Stairs D. B., Lin L., Giordano T. J., Wagner P., Minna J. D., Gazdar A. F., Zhu C. Q., Brose M. S., Cecconello I., Jr., U. R., Marie S. K., Dahl O., Shivdasani R. A., Tsao M. S., Rubin M. A., Wong K. K., Regev A., Hahn W. C., Beer D. G., Rustgi A. K., Meyerson M. (2009) SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 41, 1238–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cancer Genome Atlas Research Network (2012) Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perner S., Wagner P. L., Soltermann A., LaFargue C., Tischler V., Weir B. A., Weder W., Meyerson M., Giordano T. J., Moch H., Rubin M. A. (2009) TTF1 expression in non-small cell lung carcinoma: association with TTF1 gene amplification and improved survival. J. Pathol. 217, 65–72 [DOI] [PubMed] [Google Scholar]
- 40. Tang X., Kadara H., Behrens C., Liu D. D., Xiao Y., Rice D., Gazdar A. F., Fujimoto J., Moran C., Varella-Garcia M., Lee J. J., Hong W. K., Wistuba I. I. (2011) Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: implications in lung cancer pathogenesis and prognosis. Clin. Cancer Res. 17, 2434–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barletta J. A., Perner S., Iafrate A. J., Yeap B. Y., Weir B. A., Johnson L. A., Johnson B. E., Meyerson M., Rubin M. A., Travis W. D., Loda M., Chirieac L. R. (2009) Clinical significance of TTF-1 protein expression and TTF-1 gene amplification in lung adenocarcinoma. J. Cell. Mol. Med. 13, 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meyer L. R., Zweig A. S., Hinrichs A. S., Karolchik D., Kuhn R. M., Wong M., Sloan C. A., Rosenbloom K. R., Roe G., Rhead B., Raney B. J., Pohl A., Malladi V. S., Li C. H., Lee B. T., Learned K., Kirkup V., Hsu F., Heitner S., Harte R. A., Haeussler M., Guruvadoo L., Goldman M., Giardine B. M., Fujita P. A., Dreszer T. R., Diekhans M., Cline M. S., Clawson H., Barber G. P., Haussler D., Kent W. J. (2013) The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 41, D64–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fan M., Li X., Jiang W., Huang Y., Li J., Wang Z. (2013) A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp. Ther. Med. 5, 1143–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prensner J. R., Chinnaiyan A. M. (2011) The emergence of lncRNAs in cancer biology. Cancer Discov. 1, 391–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spizzo R., Almeida M. I., Colombatti A., Calin G. A. (2012) Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 31, 4577–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deutsch L., Wrage M., Koops S., Glatzel M., Uzunoglu F. G., Kutup A., Hinsch A., Sauter G., Izbicki J. R., Pantel K., Wikman H. (2012) Opposite roles of FOXA1 and NKX2-1 in lung cancer progression. Genes Chromosomes Cancer 51, 618–629 [DOI] [PubMed] [Google Scholar]
- 47. Harris T., Pan Q., Sironi J., Lutz D., Tian J., Sapkar J., Perez-Soler R., Keller S., Locker J. (2011) Both gene amplification and allelic loss occur at 14q13.3 in lung cancer. Clin. Cancer Res. 17, 690–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tian J., Mahmood R., Hnasko R., Locker J. (2006) Loss of Nkx2.8 deregulates progenitor cells in the large airways and leads to dysplasia. Cancer Res. 66, 10399–10407 [DOI] [PubMed] [Google Scholar]
- 49. Yamaguchi T., Yanagisawa K., Sugiyama R., Hosono Y., Shimada Y., Arima C., Kato S., Tomida S., Suzuki M., Osada H., Takahashi T. (2012) NKX2-1/TITF1/TTF-1-induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell 21, 348–361 [DOI] [PubMed] [Google Scholar]
- 50. Watanabe H., Francis J. M., Woo M. S., Etemad B., Lin W., Fries D. F., Peng S., Snyder E. L., Tata P. R., Izzo F., Schinzel A. C., Cho J., Hammerman P. S., Verhaak R. G., Hahn W. C., Rajagopal J., Jacks T., Meyerson M. (2013) Integrated cistromic and expression analysis of amplified NKX2-1 in lung adenocarcinoma identifies LMO3 as a functional transcriptional target. Genes Dev. 27, 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boehm T., Buluwela L., Williams D., White L., Rabbitts T. H. (1988) A cluster of chromosome 11p13 translocations found via distinct D-D and D-D-J rearrangements of the human T cell receptor δ chain gene. EMBO J. 7, 2011–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McGuire E. A., Hockett R. D., Pollock K. M., Bartholdi M. F., O'Brien S. J., Korsmeyer S. J. (1989) The t(11;14)(p15;q11) in a T-cell acute lymphoblastic leukemia cell line activates multiple transcripts, including Ttg-1, a gene encoding a potential zinc finger protein. Mol. Cell. Biol. 9, 2124–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Homminga I., Pieters R., Langerak A. W., de Rooi J. J., Stubbs A., Verstegen M., Vuerhard M., Buijs-Gladdines J., Kooi C., Klous P., van Vlierberghe P., Ferrando A. A., Cayuela J. M., Verhaaf B., Beverloo H. B., Horstmann M., de Haas V., Wiekmeijer A. S., Pike-Overzet K., Staal F. J., de Laat W., Soulier J., Sigaux F., Meijerink J. P. (2011) Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell 19, 484–497 [DOI] [PubMed] [Google Scholar]
- 54. Dadi S., Le Noir S., Payet-Bornet D., Lhermitte L., Zacarias-Cabeza J., Bergeron J., Villarèse P., Vachez E., Dik W. A., Millien C., Radford I., Verhoeyen E., Cosset F. L., Petit A., Ifrah N., Dombret H., Hermine O., Spicuglia S., Langerak A. W., Macintyre E. A., Nadel B., Ferrier P., Asnafi V. (2012) TLX homeodomain oncogenes mediate T cell maturation arrest in T-ALL via interaction with ETS1 and suppression of TCRα gene expression. Cancer Cell 21, 563–576 [DOI] [PubMed] [Google Scholar]
- 55. Nagel S., Ehrentraut S., Tomasch J., Quentmeier H., Meyer C., Kaufmann M., Drexler H. G., Macleod R. A. (2013) Ectopic expression of homeobox gene NKX2-1 in diffuse large B-cell lymphoma is mediated by aberrant chromatin modifications. PLoS ONE 8, e61447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Taguchi A., Politi K., Pitteri S. J., Lockwood W. W., Faça V. M., Kelly-Spratt K., Wong C. H., Zhang Q., Chin A., Park K. S., Goodman G., Gazdar A. F., Sage J., Dinulescu D. M., Kucherlapati R., Depinho R. A., Kemp C. J., Varmus H. E., Hanash S. M. (2011) Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer Cell 20, 289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ngan E. S. W., Lang B. H. H., Liu T., Shum C. K. Y., So M.-T., Lau D. K. C., Leon T. Y. Y., Cherny S. S., Tsai S. Y., Lo C.-Y., Khoo U.-S., Tam P. K. H., Garcia-Barcelo M.-M. (2009) A germline mutation (A339V) in thyroid transcription factor-1 (TITF-1/NKX2.1) in patients with multinodular goiter and papillary thyroid carcinoma. J. Natl. Cancer Inst. 101, 162–175 [DOI] [PubMed] [Google Scholar]
- 58. Antonica F., Kasprzyk D. F., Opitz R., Iacovino M., Liao X. H., Dumitrescu A. M., Refetoff S., Peremans K., Manto M., Kyba M., Costagliola S. (2012) Generation of functional thyroid from embryonic stem cells. Nature 491, 66–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kimura S. (2011) Thyroid-specific transcription factors and their roles in thyroid cancer. J. Thyroid Res. 2011, 710213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kusakabe T., Kawaguchi A., Hoshi N., Kawaguchi R., Hoshi S., Kimura S. (2006) Thyroid-specific enhancer-binding protein/NKX2.1 is required for the maintenance of ordered architecture and function of the differentiated thyroid. Mol. Endocrinol. 20, 1796–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gudmundsson J., Sulem P., Gudbjartsson D. F., Jonasson J. G., Sigurdsson A., Bergthorsson J. T., He H., Blondal T., Geller F., Jakobsdottir M., Magnusdottir D. N., Matthiasdottir S., Stacey S. N., Skarphedinsson O. B., Helgadottir H., Li W., Nagy R., Aguillo E., Faure E., Prats E., Saez B., Martinez M., Eyjolfsson G. I., Bjornsdottir U. S., Holm H., Kristjansson K., Frigge M. L., Kristvinsson H., Gulcher J. R., Jonsson T., Rafnar T., Hjartarsson H., Mayordomo J. I., de la Chapelle A., Hrafnkelsson J., Thorsteinsdottir U., Kong A., Stefansson K. (2009) Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat. Genet. 41, 460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kang Y., Hebron H., Ozbun L., Mariano J., Minoo P., Jakowlew S. B. (2004) Nkx2.1 transcription factor in lung cells and a transforming growth factor-β1 heterozygous mouse model of lung carcinogenesis. Mol. Carcinog. 40, 212–231 [DOI] [PubMed] [Google Scholar]
- 63. Hoshi S., Hoshi N., Okamoto M., Paiz J., Kusakabe T., Ward J. M., Kimura S. (2009) Role of NKX2-1 in N-bis(2-hydroxypropyl)-nitrosamine-induced thyroid adenoma in mice. Carcinogenesis 30, 1614–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Snyder E. L., Watanabe H., Magendantz M., Hoersch S., Chen T. A., Wang D. G., Crowley D., Whittaker C. A., Meyerson M., Kimura S., Jacks T. (2013) Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol. Cell 50, 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. De Vita G., Bauer L., da Costa V. M., De Felice M., Baratta M. G., De Menna M., Di Lauro R. (2005) Dose-dependent inhibition of thyroid differentiation by RAS oncogenes. Mol. Endocrinol. 19, 76–89 [DOI] [PubMed] [Google Scholar]
- 66. Missero C., Pirro M. T., Di Lauro R. (2000) Multiple Ras downstream pathways mediate functional repression of the homeobox gene product TTF-1. Mol. Cell. Biol. 20, 2783–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hosono Y., Yamaguchi T., Mizutani E., Yanagisawa K., Arima C., Tomida S., Shimada Y., Hiraoka M., Kato S., Yokoi K., Suzuki M., Takahashi T. (2012) MYBPH, a transcriptional target of TTF-1, inhibits ROCK1, and reduces cell motility and metastasis. EMBO J. 31, 481–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Saito R. A., Watabe T., Horiguchi K., Kohyama T., Saitoh M., Nagase T., Miyazono K. (2009) Thyroid transcription factor-1 inhibits transforming growth factor-β-mediated epithelial-to-mesenchymal transition in lung adenocarcinoma cells. Cancer Res. 69, 2783–2791 [DOI] [PubMed] [Google Scholar]
- 69. Li C., Zhu N. L., Tan R. C., Ballard P. L., Derynck R., Minoo P. (2002) Transforming growth factor-β inhibits pulmonary surfactant protein B gene transcription through SMAD3 interactions with NKX2.1 and HNF-3 transcription factors. J. Biol. Chem. 277, 38399–38408 [DOI] [PubMed] [Google Scholar]
- 70. Ocaña O. H., Córcoles R., Fabra A., Moreno-Bueno G., Acloque H., Vega S., Barrallo-Gimeno A., Cano A., Nieto M. A. (2012) Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22, 709–724 [DOI] [PubMed] [Google Scholar]
- 71. Tsai J. H., Donaher J. L., Murphy D. A., Chau S., Yang J. (2012) Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Runkle E. A., Rice S. J., Qi J., Masser D., Antonetti D. A., Winslow M. M., Mu D. (2012) Occludin is a direct target of thyroid transcription factor-1 (TTF-1/NKX2-1). J. Biol. Chem. 287, 28790–28801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Niimi T., Nagashima K., Ward J. M., Minoo P., Zimonjic D. B., Popescu N. C., Kimura S. (2001) claudin-18, a novel downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor, encodes lung- and stomach-specific isoforms through alternative splicing. Mol. Cell. Biol. 21, 7380–7390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Runkle E. A., Mu D. (2013) Tight junction proteins: from barrier to tumorigenesis. Cancer Lett. 337, 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bhaskaran M., Wang Y., Zhang H., Weng T., Baviskar P., Guo Y., Gou D., Liu L. (2009) MicroRNA-127 modulates fetal lung development. Physiol. Genomics 37, 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lizé M., Herr C., Klimke A., Bals R., Dobbelstein M. (2010) MicroRNA-449a levels increase by several orders of magnitude during mucociliary differentiation of airway epithelia. Cell Cycle 9, 4579–4583 [DOI] [PubMed] [Google Scholar]
- 77. Qi J., Mu D. (2012) MicroRNAs and lung cancers: from pathogenesis to clinical implications. Front Med. 6, 134–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sozzi G., Pastorino U., Croce C. M. (2011) MicroRNAs and lung cancer: from markers to targets. Cell Cycle 10, 2045–2046 [DOI] [PubMed] [Google Scholar]
- 79. Yu S. L., Chen H. Y., Chang G. C., Chen C. Y., Chen H. W., Singh S., Cheng C. L., Yu C. J., Lee Y. C., Chen H. S., Su T. J., Chiang C. C., Li H. N., Hong Q. S., Su H. Y., Chen C. C., Chen W. J., Liu C. C., Chan W. K., Chen W. J., Li K. C., Chen J. J., Yang P. C. (2008) MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 13, 48–57 [DOI] [PubMed] [Google Scholar]
- 80. Dong J., Jiang G., Asmann Y. W., Tomaszek S., Jen J., Kislinger T., Wigle D. A. (2010) MicroRNA networks in mouse lung organogenesis. PLoS ONE 5, e10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Qi J., Rice S. J., Salzberg A. C., Runkle E. A., Liao J., Zander D. S., Mu D. (2012) MiR-365 regulates lung cancer and developmental gene thyroid transcription factor 1. Cell Cycle 11, 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kang S. M., Lee H. J., Cho J. Y. (2013) MicroRNA-365 regulates NKX2-1, a key mediator of lung cancer. Cancer Lett. 335, 487–494 [DOI] [PubMed] [Google Scholar]
- 84. Rice S. J., Lai S. C., Wood L. W., Helsley K. R., Runkle E. A., Winslow M. M., Mu D. (2013) MicroRNA-33a mediates the regulation of high mobility group AT-hook 2 gene (HMGA2) by thyroid transcription factor 1 (TTF-1/NKX2-1). J. Biol. Chem. 288, 16348–16360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Brown M. S., Goldstein J. L. (1997) The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 [DOI] [PubMed] [Google Scholar]
- 86. Horton J. D., Goldstein J. L., Brown M. S. (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gerin I., Clerbaux L. A., Haumont O., Lanthier N., Das A. K., Burant C. F., Leclercq I. A., MacDougald O. A., Bommer G. T. (2010) Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J. Biol. Chem. 285, 33652–33661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Marquart T. J., Allen R. M., Ory D. S., Baldán A. (2010) miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. U.S.A. 107, 12228–12232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Najafi-Shoushtari S. H., Kristo F., Li Y., Shioda T., Cohen D. E., Gerszten R. E., Näär A. M. (2010) MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rayner K. J., Suárez Y., Dávalos A., Parathath S., Fitzgerald M. L., Tamehiro N., Fisher E. A., Moore K. J., Fernández-Hernando C. (2010) MiR-33 contributes to the regulation of cholesterol homeostasis. Science 328, 1570–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Smith R., Owen L. A., Trem D. J., Wong J. S., Whangbo J. S., Golub T. R., Lessnick S. L. (2006) Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell 9, 405–416 [DOI] [PubMed] [Google Scholar]
- 92. Muraguchi T., Tanaka S., Yamada D., Tamase A., Nakada M., Nakamura H., Hoshii T., Ooshio T., Tadokoro Y., Naka K., Ino Y., Todo T., Kuratsu J., Saya H., Hamada J., Hirao A. (2011) NKX2.2 suppresses self-renewal of glioma-initiating cells. Cancer Res. 71, 1135–1145 [DOI] [PubMed] [Google Scholar]
- 93. Barlési F., Pinot D., Legoffic A., Doddoli C., Chetaille B., Torre J. P., Astoul P. (2005) Positive thyroid transcription factor 1 staining strongly correlates with survival of patients with adenocarcinoma of the lung. Br. J. Cancer 93, 450–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Berghmans T., Paesmans M., Mascaux C., Martin B., Meert A. P., Haller A., Lafitte J. J., Sculier J. P. (2006) Thyroid transcription factor 1–a new prognostic factor in lung cancer: a meta-analysis. Ann. Oncol. 17, 1673–1676 [DOI] [PubMed] [Google Scholar]
- 95. Pruitt K. D., Tatusova T., Brown G. R., Maglott D. R. (2012) NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Morishita A., Zaidi M. R., Mitoro A., Sankarasharma D., Szabolcs M., Okada Y., D'Armiento J., Chada K. (2013) HMGA2 is a driver of tumor metastasis. Cancer Res. 10.1158/0008-5472.CAN-12-3848 [DOI] [PMC free article] [PubMed] [Google Scholar]


